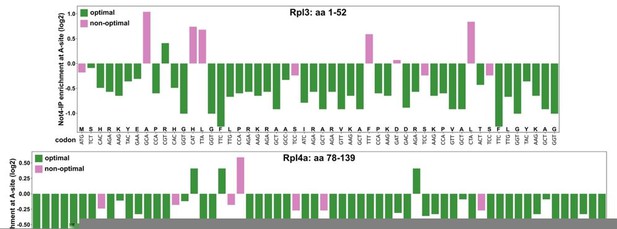
Dedicated chaperones coordinate co-translational regulation of ribosomal protein production with ribosome assembly to preserve proteostasis
Figures
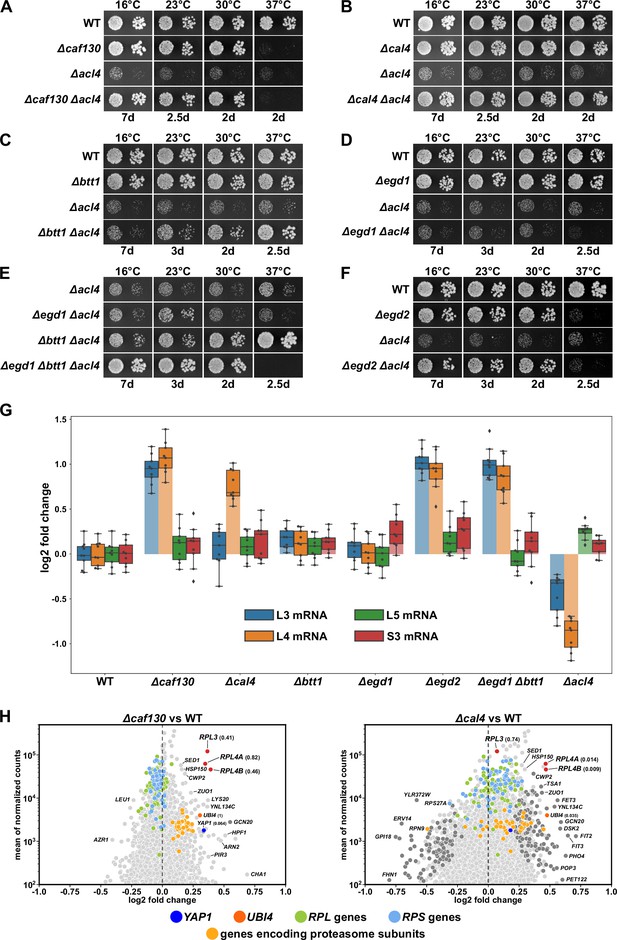
Absence of Caf130, Cal4, or the nascent polypeptide-associated complex (NAC) suppresses the ∆acl4 growth defect by increasing RPL4 mRNA levels.
(A–F) Suppression of the ∆acl4 growth defect. The indicated wild-type (WT), single, double, and triple deletion strains, all derived from tetratype tetrads, were spotted in 10-fold serial dilution steps onto YPD plates, which were incubated for the indicated times at 16, 23, 30, or 37°C. (G) Cells lacking Caf130, Cal4, or the NAC exhibit increased RPL4 mRNA levels. Cells of the indicated genotype were grown in YPD medium at 30°C to an OD600 of around 0.6, and relative changes in mRNA levels were determined by qRT-PCR (see Materials and methods). The data shown were obtained from three independent strains of the same genotype (biological triplicates), in each case consisting of a technical triplicate. The darker-colored boxes highlight the quartiles of each dataset, while the whiskers indicate the minimal and maximal limits of the distribution; outliers are shown as diamonds. The horizontal line in the quartile box represents the median log2 fold change of each dataset. (H) Christmas tree representation of differential gene expression analysis between ∆caf130 (left panel) or ∆cal4 (right panel) and WT cells. The RNA-seq data were generated from the same total RNA samples used for the above qRT-PCRs. Genes exhibiting statistically significant differential mRNA levels are colored in dark gray (adjusted p-value, padj<0.05). The adjusted p-values for the selected mRNAs are indicated in parentheses. Categories of genes or specific genes, regardless of the adjusted p-value, are colored as indicated.
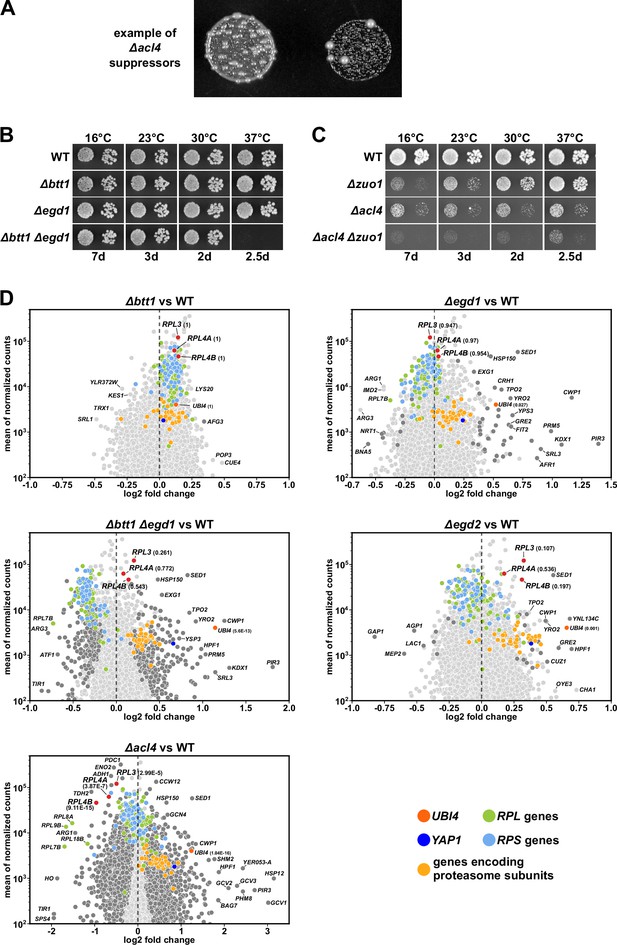
Differential gene expression analysis between nascent polypeptide-associated complex (NAC)-deficient and wild-type (WT) cells.
(A) Example of spontaneous suppression of the ∆acl4 growth phenotype. (B, C) Growth of the indicated WT, single and double deletion strains, all derived from tetratype tetrads, on YPD plates. (D) Christmas tree representation of differential gene expression analysis between the indicated mutant and WT cells. The RNA-seq data were generated from the same total RNA samples used for the qRT-PCRs in Figure 1G. Genes exhibiting statistically significant differential mRNA levels are colored in dark gray (adjusted p-value, padj<0.05). The adjusted p-values for the selected mRNAs are indicated in parentheses. Categories of genes or specific genes, regardless of the adjusted p-value, are colored as indicated.
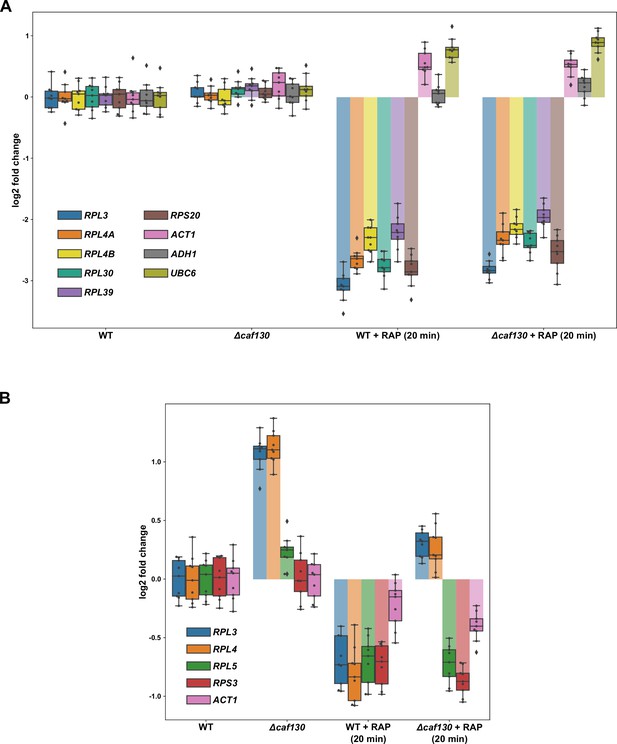
Absence of Caf130 does not lead to increased transcription of the RPL3, RPL4A, and RPL4B genes.
(A) Occupancy of initiating RNA Pol II at the indicated genes was assessed by ChIP-qPCR (see Materials and methods) in wild-type and ∆caf130 cells that were grown at 30°C in YPD medium and either not treated or treated for 20 min with 200 ng/ml rapamycin (RAP). Note that the tested RPGs contain either category I (RPL30 and RPL39), II (RPS20), or III (RPL3, RPL4A, and RPL4B) RPG promoters. The data shown were obtained from three independent wild-type and ∆caf130 strains (biological triplicates) and, for each of these, the qPCRs were performed in technical triplicates. Data are represented as described in the legend to Figure 1G. (B) Effects of TORC1 inhibition on RPL3 and RPL4 mRNA levels. Relative changes in mRNA levels were determined by qRT-PCR in wild-type and ∆caf130 cells that were grown at 30°C in YPD medium and either not treated or treated for 20 min with 200 ng/ml RAP. The data shown were obtained from three independent wild-type and ∆caf130 strains (biological triplicates), in each case consisting of a technical triplicate.
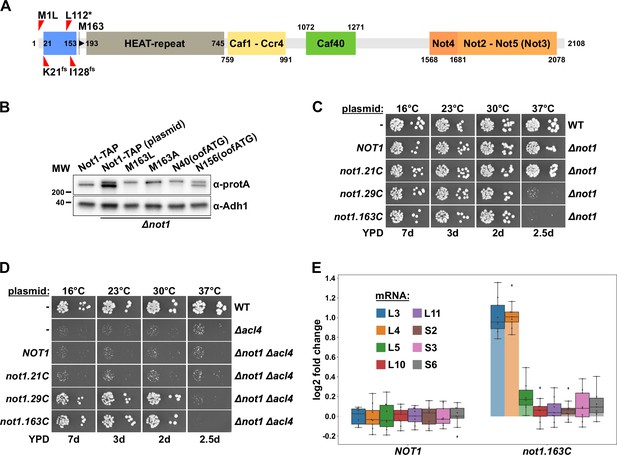
Absence of Not1’s N-terminal domain suppresses the ∆acl4 growth defect and increases RPL3 and RPL4 mRNA levels.
(A) Schematic representation of Not1 highlighting its domain organization and known binding sites of Ccr4-Not core components as revealed by diverse (co-)crystal structures (PDB: 4B8B and 4B8A [Basquin et al., 2012], 4CV5 [Mathys et al., 2014], 5AJD [Bhaskar et al., 2015], and 4BY6 [Bhaskar et al., 2013]). As shown in Figure 3H, the N-terminal Not1 segment encompassing amino acids 21–153 corresponds to the minimal Caf130-interacting domain (CaInD). Note that Ccr4 does not directly bind to Not1, it is recruited via its interaction with Caf1. The position and nature of the ∆acl4 suppressor mutations are indicated: M1L (ATG start codon changed to cTG), K21fs (AAA codon with deletion of one A, resulting in a frameshift), L112* (TTG codon changed to TaG stop codon), and I128fs (ATT codon with A deleted, resulting in a frameshift). M163 denotes the second methionine within Not1, it is encoded by the first occurring ATG trinucleotide after the start codon. (B) The shorter, major isoform of Not1 is generated by utilization of the ATG coding for M163 as the start codon. Total protein extracts, derived from cells expressing Not1-TAP, either from the genomic locus or from plasmid in a ∆not1 strain, and the indicated variants, were analyzed by Western blotting using anti-protA and anti-Adh1 (loading control) antibodies. The N40(oofATG) and N156(oofATG) constructs contain an out-of-frame ATG (oofATG) owing to the silent mutagenesis of the N40 and N156 codons from AAC to AAt, which, together with the first position of the subsequent Asp-encoding codons, forms an ATG trinucleotide. (C, D) Growth phenotype of and suppression of the ∆acl4 growth defect by N-terminal deletion variants of Not1. Plasmids harboring full-length NOT1 or the indicated not1 deletion variants, expressed under the control of the NOT1 promoter, were transformed into a NOT1 shuffle strain (C) or a NOT1/ACL4 double shuffle strain (D). After plasmid shuffling on 5-fluoroorotic acid (5-FOA)-containing plates, cells were restreaked on YPD plates and then, alongside a wild-type (WT) and ∆acl4 control strain, spotted in 10-fold serial dilution steps onto YPD plates. Note that the not1.21C, not1.29C, and not1.163C alleles express N-terminally truncated Not1 variants starting at amino acids 21, 29, and 163, respectively. (E) Absence of Not1’s N-terminal domain increases RPL3 and RPL4 mRNA levels. Relative changes in mRNA levels between ∆not1 cells complemented with either plasmid-borne NOT1 or not1.163C were determined by qRT-PCR. Cells were grown in YPD medium at 30°C. The data shown were obtained with three independent NOT1 shuffle strains (biological triplicates), in each case consisting of a technical triplicate, and they are represented as described in the legend to Figure 1G.
-
Figure 2—source data 1
Original image files of the Western blots shown in Figure 2B, including a PDF file showing the full blots and indicating the cropped areas.
- https://cdn.elifesciences.org/articles/74255/elife-74255-fig2-data1-v1.zip
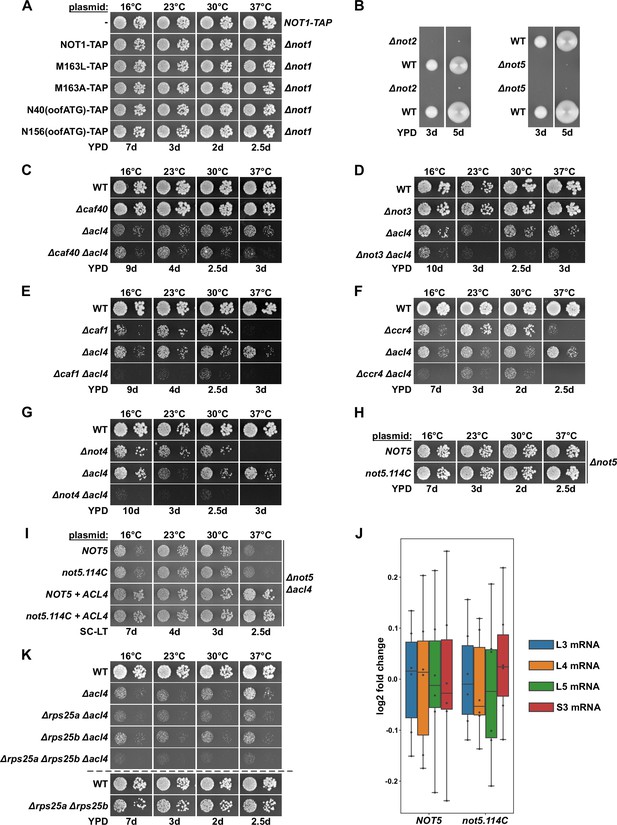
Absence of other Ccr4-Not components does not suppress the ∆acl4 growth defect.
(A) Growth phenotype of cells expressing Not1-TAP, either from the genomic locus or from plasmid in a ∆not1 strain, and the indicated variants. The same strains were used in Figure 2B. (B) Cells lacking Not2 or Not5 display a severe slow-growth phenotype. The growth of spore clones, after dissection of NOT2/∆not2 or NOT5/∆not5 diploids, is shown on YPD plates at 30°C. (C–G) Absence of other Ccr4-Not components does not suppress the ∆acl4 growth defect. The indicated wild-type (WT), single and double deletion strains, all derived from tetratype tetrads, were spotted in 10-fold serial dilution steps onto YPD plates. (H) Plasmids expressing full-length Not5 or the Not5.114C variant were transformed into a NOT5 shuffle strain and, after plasmid shuffling, cells were spotted in 10-fold serial dilution steps onto YPD plates. (I) The same plasmids were co-transformed with either an empty vector or an ACL4-harboring plasmid into a NOT5/ACL4 double shuffle strain. After plasmid shuffling, cells were spotted in 10-fold serial dilution steps onto synthetic complete medium lacking leucine and tryptophan (SC-LT) plates. (J) Absence of Not5’s N-terminal domain does not increase RPL3 and RPL4 mRNA levels. Relative changes in mRNA levels between ∆not5 cells complemented either with plasmid-borne NOT5 or not5.114C were determined by qRT-PCR. Cells were grown in YPD medium at 30°C. The data shown were obtained with two independent ∆not5 strains (biological duplicates), in each case consisting of a technical triplicate, and they are presented as described in the legend to Figure 1G. (K) The indicated single, double, and triple deletion strains, all derived from tetratype tetrads, were spotted, alongside a wild-type (WT) and a ∆rps25a/∆rps25b reference strain, in 10-fold serial dilution steps onto YPD plates.
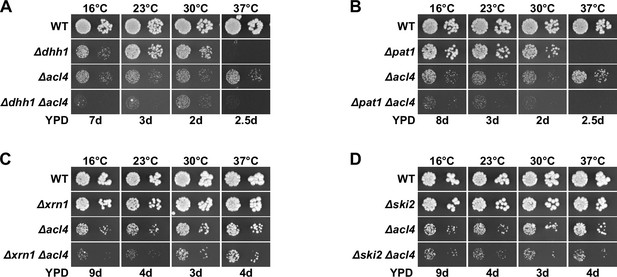
Absence of general mRNA decay factors does not suppress the ∆acl4 growth defect.
(A–D) The indicated wild-type (WT), single and double deletion strains, all derived from tetratype tetrads, were spotted in 10-fold serial dilution steps onto YPD plates, which were incubated for the indicated times at 16, 23, 30, or 37°C.
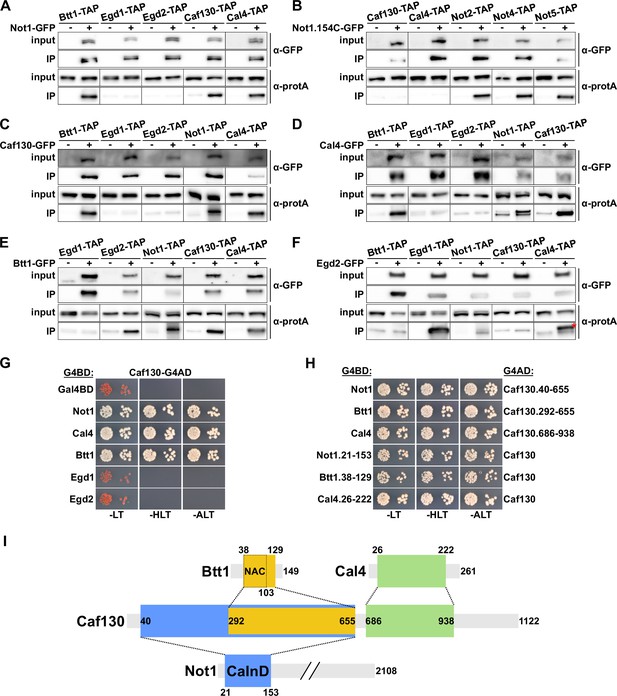
Caf130 connects Cal4 and Btt1 to Ccr4-Not by exclusively interacting with the full-length translational isoform of Not1.
(A–F) Assessment of in vivo interactions by GFP-Trap co-immunoprecipitation. Cells expressing nontagged (-) or C-terminally GFP-tagged (+) versions of Not1 (A), Not1.154C (B), Caf130 (C), Cal4 (D), Btt1 (E), and Egd2 (F) together with the indicated C-terminally TAP-tagged prey proteins were grown in YPD medium at 30°C. All fusion proteins were expressed from their genomic locus, except GFP-tagged Not1 and Not1.154C as well as their nontagged counterparts, which were expressed from plasmid under the control of the NOT1 promoter in ∆not1 cells. Cell lysates (input; 1/1000 of IP input) and GFP-Trap affinity purifications (IP; 1/5 of complete IP) were analyzed by Western blotting using anti-GFP and anti-protA antibodies. Since Not1, Egd1, and Egd2 are expressed at higher levels, the inputs for detection of Not1-TAP were diluted twofold and those of Egd1-TAP and Egd2-TAP 20-fold to keep all Western signals in a similar range. Note that the band marked with a red asterisk corresponds to the Egd2-GFP bait protein, which is, due to its abundance in the IP, nonspecifically recognized by the anti-protA antibody. (G, H) Assessment of protein–protein interactions by yeast 2-hybrid (Y2H). (G) Caf130 interacts with Not1, Cal4, and Btt1. Plasmids expressing full-length Not1, Cal4, Btt1, Egd1, or Egd2, fused to the C-terminal Gal4 DNA-binding domain (G4BD), and full-length Caf130, fused to the C-terminal Gal4 activation domain (G4AD), were co-transformed into the Y2H reporter strain PJ69-4A. Cells were spotted in 10-fold serial dilution steps onto SC-Leu-Trp (-LT), SC-His-Leu-Trp (-HLT), and SC-Ade-Leu-Trp (-ALT) plates, which were incubated for 3 days at 30°C. (H) Minimal interaction surfaces mediating the binary Caf130-Not1, Caf130-Btt1, and Caf130-Cal4 association. Plasmids expressing the indicated C-terminally G4BD-tagged Not1, Cal4, or Btt1 and G4AD-tagged Caf130 full-length proteins or respective minimal interaction fragments thereof were co-transformed into the Y2H reporter strain PJ69-4A. (I) Schematic representation of the binary interactions and the determined minimal interaction surfaces. The respective minimal interaction surfaces, as determined by Y2H mapping, are highlighted by colored rectangles. The borders of the NAC domain, as defined in Liu et al., 2010, are also indicated. The Caf130-interacting domain of Not1 is abbreviated as CaInD.
-
Figure 3—source data 1
Original image files of the Western blots shown in Figure 3A and B, including a PDF file showing the full blots and indicating the cropped areas.
- https://cdn.elifesciences.org/articles/74255/elife-74255-fig3-data1-v1.zip
-
Figure 3—source data 2
Original image files of the Western blots shown in Figure 3C, including a PDF file showing the full blots and indicating the cropped areas.
- https://cdn.elifesciences.org/articles/74255/elife-74255-fig3-data2-v1.zip
-
Figure 3—source data 3
Original image files of the Western blots shown in Figure 3D, including a PDF file showing the full blots and indicating the cropped areas.
- https://cdn.elifesciences.org/articles/74255/elife-74255-fig3-data3-v1.zip
-
Figure 3—source data 4
Original image files of the Western blots shown in Figure 3E, including a PDF file showing the full blots and indicating the cropped areas.
- https://cdn.elifesciences.org/articles/74255/elife-74255-fig3-data4-v1.zip
-
Figure 3—source data 5
Original image files of the Western blots shown in Figure 3F, including a PDF file showing the full blots and indicating the cropped areas.
- https://cdn.elifesciences.org/articles/74255/elife-74255-fig3-data5-v1.zip
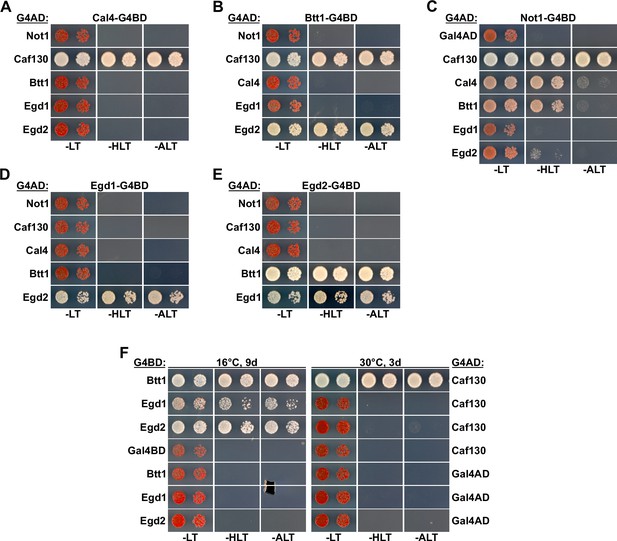
Caf130 interacts with Not1, Cal4, and Btt1.
(A–E) Assessment of protein–protein interactions by Y2H between full-length Not1, Caf130, Cal4, Btt1, Egd1, and Egd2. Plasmids expressing full-length Cal4 (A), Btt1 (B), Not1 (C), Egd1 (D), or Egd2 (E), fused to the C-terminal G4BD, and full-length Not1, Caf130, Cal4, Btt1, Egd1, and Egd2, fused to the C-terminal G4AD, were co-transformed into the Y2H reporter strain PJ69-4A. (F) Comparison of the Y2H interaction between Caf130 and NAC components at 16 and 30°C.
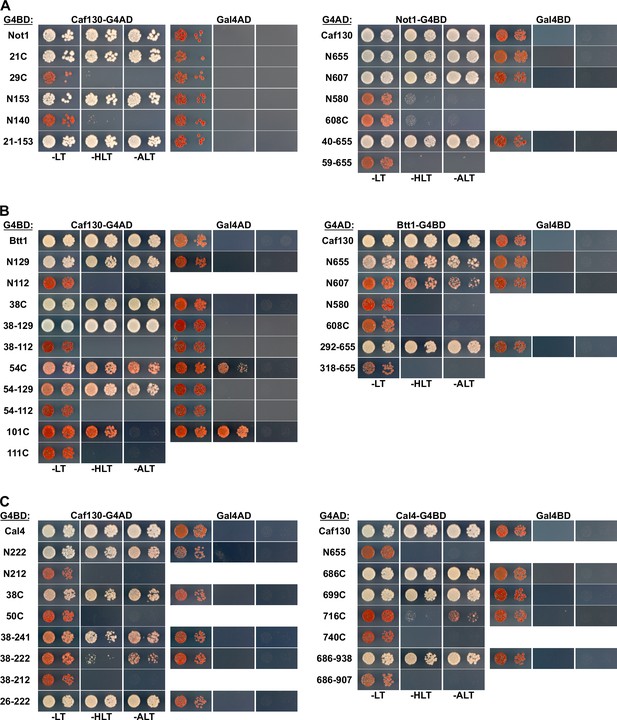
Mapping of the minimal interaction surfaces on Caf130, Not1, Cal4, and Btt1.
Mapping of the respective minimal interaction surfaces mediating the association between Caf130 and its direct interactors (Not1, Btt1, and Cal4). On the left panels, the Y2H reporter strain PJ69-4A was co-transformed with plasmids expressing full-length Caf130, fused to the C-terminal G4AD, and the indicated variants of Not1 (A), Btt1 (B), or Cal4 (C), fused to the C-terminal G4BD. On the right panels, PJ69-4A was co-transformed with plasmids expressing the indicated variants of Caf130, fused to the C-terminal G4AD, and full-length Not1 (A), Btt1 (B), and Cal4 (C), fused to the C-terminal G4BD.
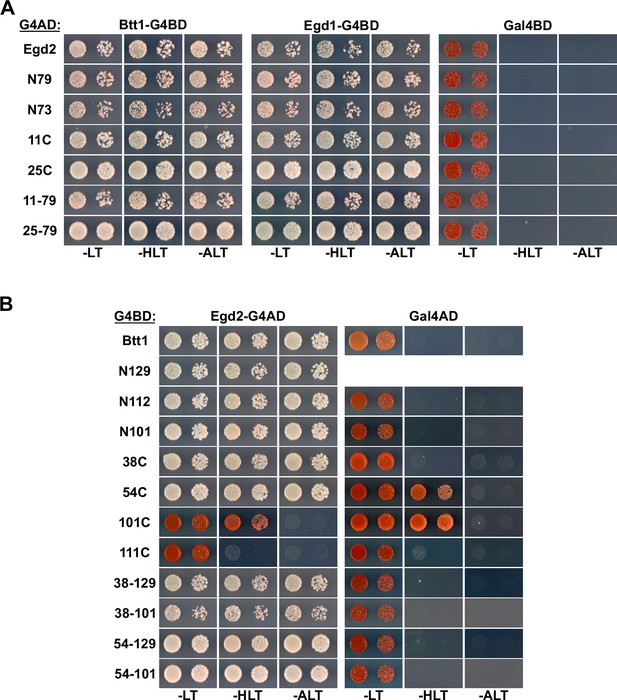
Mapping of the Egd2-binding surface on Btt1.
(A, B) Mapping of the interaction surface mediating the association of Egd2 with full-length Egd1 or Btt1 (A) and of Btt1 with full-length Egd2 (B). The Y2H reporter strain PJ69-4A was co-transformed with plasmids expressing the indicated full-length proteins, deletion variants, and internal fragments, fused to either the C-terminal G4BD or G4AD.
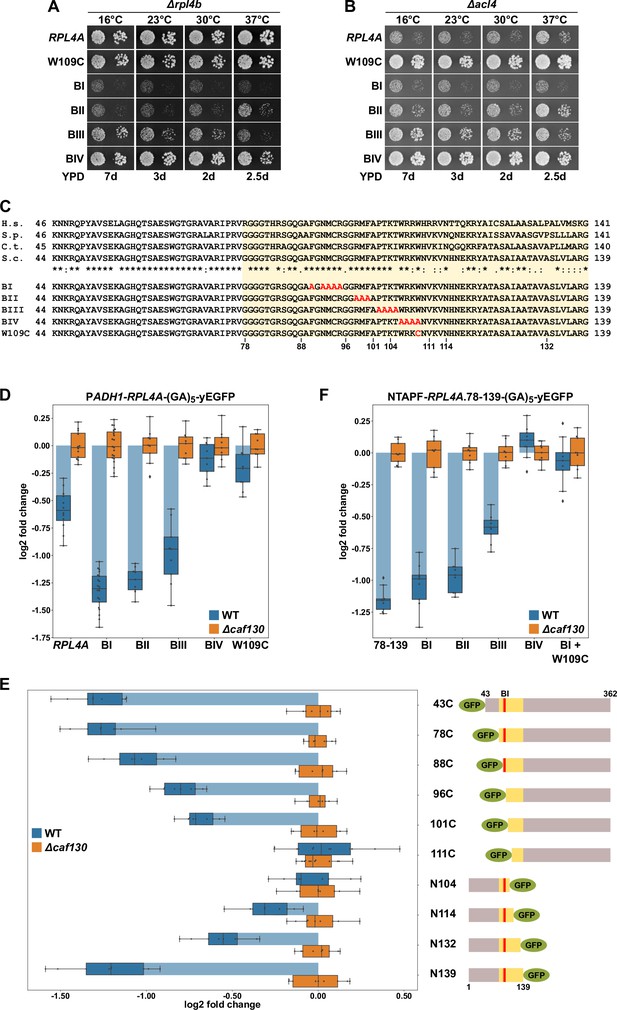
The Rpl4 protein harbors the regulation-conferring signal.
(A, B) Suppression of the ∆acl4 growth defect by the rpl4a.W109C allele. Cells harboring wild-type (WT) RPL4A or the indicated rpl4a alleles, expressed from the genomic locus, in addition to either the deletion of RPL4B (∆rpl4b) (A) or ACL4 (∆acl4) (B) were spotted in 10-fold serial dilution steps onto YPD plates. (C) Amino acid sequences of the long internal loop (amino acids 44–113), extended to the C-terminal border of the minimal segment conferring full RPL4A mRNA regulation (amino acids 78–139; highlighted by a light yellow background color), of Rpl4 from different eukaryotic species (H.s., Homo sapiens; S.p., Schizosaccharomyces pombe; C.t., Chaetomium thermophilum; S.c., Saccharomyces cerevisiae). Conserved (*), strongly similar (:), and weakly similar (.) amino acids are indicated below the alignment. The nonoverlapping, consecutive alanine substitutions within this Rpl4a segment are depicted in the lower part: block-I mutant (BI): F90A/N92A/M93A/C94A/R95A; block-II mutant (BII): R98A/M99A/F100A; block-III mutant (BIII): P102A/T103A/K104A/T105A; and block-IV mutant (BIV): W106A/R107A/K108A/W109A. The W109C exchange is also indicated. (D) Negative regulation of RPL4A mRNA levels is strongly diminished by the rpl4a.W109C mutation. Levels of RPL4A-yEGFP fusion mRNAs were determined in WT (blue bars) or ∆caf130 (orange bars) cells by qRT-PCR with a primer pair specifically amplifying a part of the yEGFP coding sequence fused to the 3′-end of the RPL4A ORF. Cells harboring RPL4A or the indicated rpl4a alleles, expressed from the ADH1 promoter, on plasmid were grown at 30°C in SC-Leu medium. The data shown were obtained from at least three different WT and ∆caf130 strains (biological replicates), in each case consisting of a technical triplicate. Changes in mRNA levels of each assayed RPL4A allele between WT (negative regulation on) and ∆caf130 (negative regulation off) cells have been normalized to their maximal abundance in ∆caf130 cells. The data are represented as described in the legend to Figure 1G. (E) Mapping of the minimal regulation-conferring region on RPL4A. Levels of fusion mRNAs containing different regions of the RPL4A coding sequence were determined in WT (blue bars) or ∆caf130 (orange bars) cells by qRT-PCR. Cells expressing the indicated N-terminal deletion variants, fused to an N-terminal yEGFP tag, or C-terminal deletion variants, fused to a C-terminal yEGFP tag, from plasmid under the transcriptional control of the ADH1 promoter were grown at 30°C in SC-Leu medium. To avoid any effect on mRNA levels of co-translational Acl4 binding to the nascent Rpl4a polypeptides, the BI mutations were introduced into those constructs comprising this region of the RPL4A coding sequence. The yEGFP-fused Rpl4a variants, encoded by the assayed constructs, are schematically represented. The Rpl4a segment encoded by the minimal regulation-conferring RPL4A region is highlighted in yellow and the position of the BI alanine substitutions by a red bar. The data shown were obtained from at least three different WT and ∆caf130 strains (biological replicates), in each case consisting of a technical triplicate. (F) The rpl4a.W109C mutation within the minimal regulation-conferring region strongly diminishes negative regulation of RPL4A mRNA levels. Levels of fusion mRNAs were determined in WT (blue bars) or ∆caf130 (orange bars) by qRT-PCR. Cells expressing the Rpl4a(78-139) fragment harboring the wild-type sequence or the indicated mutations, fused to an N-terminal TAP-Flag (NTAPF) and a C-terminal yEGFP tag, from plasmid under the transcriptional control of the ADH1 promoter were grown at 30°C in SC-Leu medium. The data shown were obtained from three different WT and ∆caf130 strains (biological triplicates), in each case consisting of a technical triplicate.
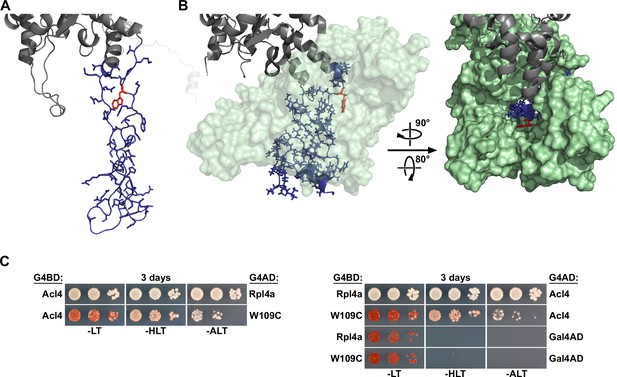
Residue W109 of Rpl4 is facing the inner surface of Acl4 and the W109C exchange reduces the interaction of Rpl4 with Acl4.
(A) Structure of Rpl4’s long internal loop in the ribosome-bound state, extracted from PDB 4V88 (Ben-Shem et al., 2011). The long internal loop of Rpl4 is colored in blue (amino acids 43–113) with side chains in stick representation. The W109 residue is highlighted in red, and the rest of Rpl4 is colored in gray. (B) Co-structure of the C. thermophilum Acl4-Rpl4 complex (PDB 5TQB, Huber and Hoelz, 2017). CtRpl4 is depicted in cartoon representation with the long internal loop colored in blue (amino acids 44–114) and with side chains in stick representation. The W110 residue (corresponding to the S. cerevisiae W109 residue) is highlighted in red, and the rest of ctRpl4 is colored in gray. CtAcl4 is shown in surface representation (light green), either in a semi-transparent manner (left panel) or completely opaque (right panel). (C) Effect of the W109C mutation on the interaction between Rpl4a and Acl4. The Y2H reporter strain PJ69-4A was co-transformed with plasmids expressing full-length Acl4, C-terminally fused to the G4BD (left panel) or the G4AD (right panel), and Rpl4a or Rpl4a.W109C, C-terminally fused to the G4AD (left panel) or the G4BD (right panel).
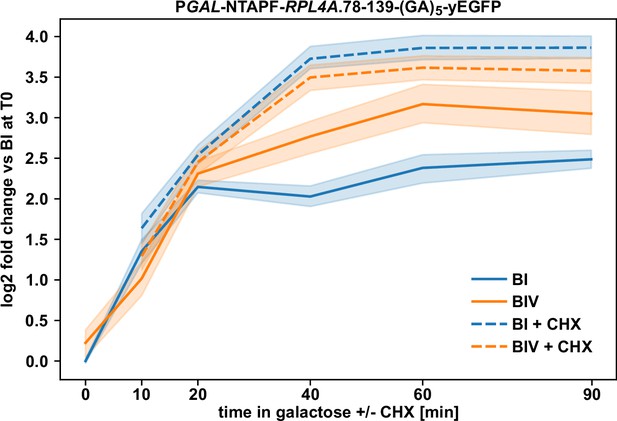
Ongoing translation is required for efficient negative regulation.
Wild-type cells expressing the minimal regulation-conferring Rpl4a segment (residues 78–139) harboring the BI or BIV mutations, fused to an N-terminal NTAPF tag and a C-terminal yEGFP moiety, from plasmid under the transcriptional control of the GAL1-10 promoter were pre-grown in SGal-Leu medium at 30°C to an OD600 of around 0.4, transferred for 1.5 hr to glucose-containing SC-Leu medium (time point 0), and then, after one wash in sugar-free medium, placed again into SGal-Leu medium to enable rapid induction of mRNA production. Cycloheximide (CHX) was added to one half of each culture at a final concentration of 200 µg/ml to inhibit translation elongation. Then, samples were taken after 10, 20, 40, 60, and 90 min of culturing in SGal-Leu medium either lacking or containing CHX. Levels of the two reporter mRNAs were determined by qRT-PCR with a primer pair specifically amplifying a part of the yEGFP coding sequence and normalized to the average abundance of the BI-containing reporter mRNA at time point 0. Data are shown as line plots and were derived from five independent experiments, conducted with four different wild-type strains and in each case performed in technical triplicates. Lines represent the mean values and the shaded areas the 95% bootstrapped confidence interval.
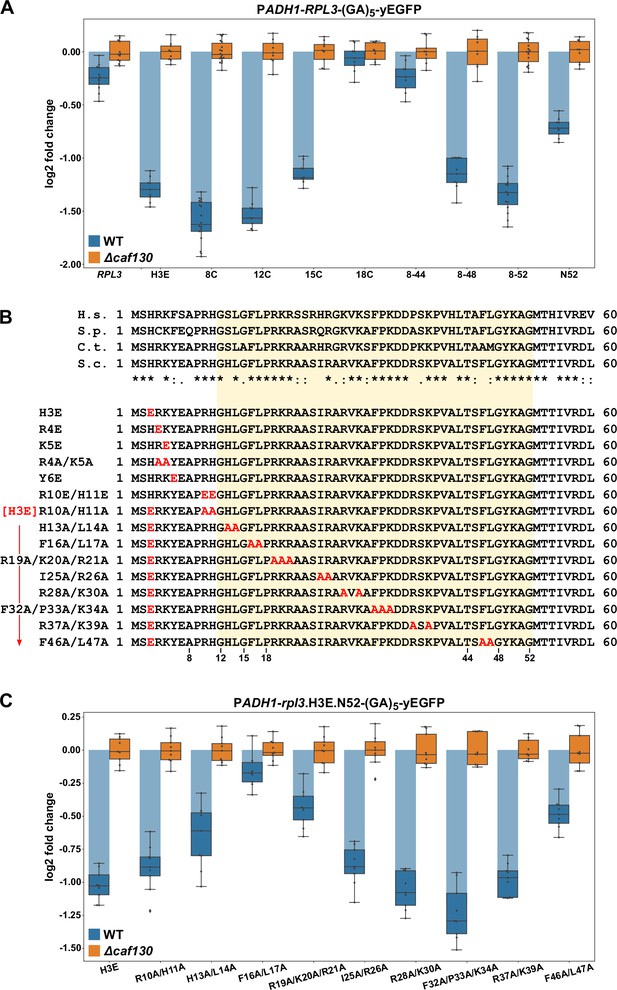
The regulation-conferring Rpl3 segment is adjacent to the Rrb1-binding site.
(A) Mapping of the minimal regulation-conferring region on RPL3. Levels of fusion mRNAs containing different regions of the RPL3 coding sequence were determined in wild-type (WT; blue bars) or ∆caf130 (orange bars) cells by qRT-PCR with a primer pair specifically amplifying a part of the yEGFP coding sequence. Cells expressing full-length Rpl3 or the indicated substitution and deletion variants, fused to a C-terminal yEGFP tag, from plasmid under the control of the ADH1 promoter were grown at 30°C in SC-Leu medium. The data shown were obtained from three different WT and ∆caf130 strains (biological replicates; note that some strains were used more than once), in each case consisting of a technical triplicate. The data are represented as described in the legend to Figure 4D. (B) Amino acid sequences of the N-terminal region of Rpl3, containing the minimal Rrb1-interacting region (amino acids 1–15; Pausch et al., 2015) and extended to the C-terminal border of the minimal segment conferring full RPL3 mRNA regulation (amino acids 12–52; highlighted by a light yellow background color), from different eukaryotic species (H.s., H. sapiens; S.p., S. pombe; C.t., C. thermophilum; S.c., S. cerevisiae). Conserved (*), strongly similar (:), and weakly similar (.) amino acids are indicated below the alignment. The glutamate and alanine substitutions, contained in the Rpl3 variants used in this study, within the N-terminal region of Rpl3 are depicted in the lower part. (C) Residues F16 and L17 are main determinants for efficient negative regulation of RPL3 mRNA levels. Levels of fusion mRNAs were determined in WT (blue bars) or ∆caf130 (orange bars) cells expressing the Rpl3.N52 fragment harboring the indicated mutations, fused to a C-terminal yEGFP tag, from plasmid under the transcriptional control of the ADH1 promoter. To avoid any effect on mRNA levels of co-translational Rrb1 binding to the nascent Rpl3 polypeptides, the H3E mutation was introduced into all assayed constructs. The data shown were obtained from three different WT and ∆caf130 strains (biological triplicates), in each case consisting of a technical triplicate.
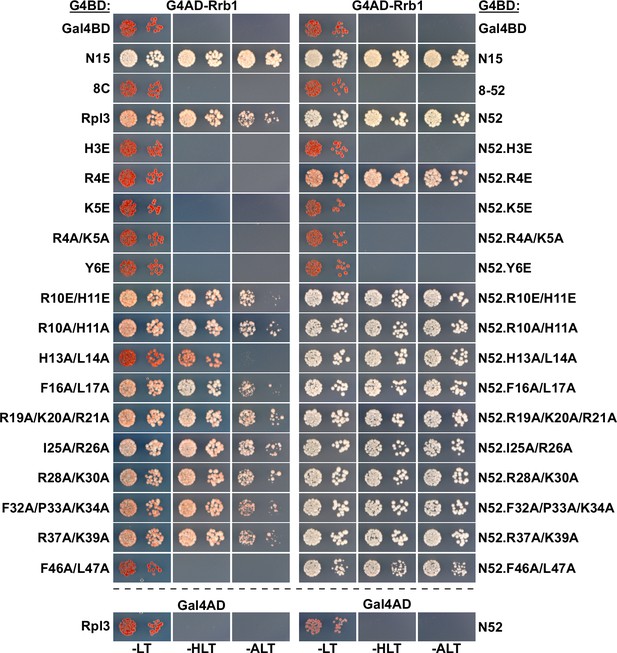
Effect of mutations within Rpl3’s N-terminal region on the Y2H interaction with Rrb1.
The Y2H reporter strain PJ69-4A was co-transformed with plasmids expressing full-length Rrb1, N-terminally fused to the G4AD, and full-length Rpl3 or the indicated substitution and deletion variants, C-terminally fused to the G4BD. Cells were spotted in 10-fold serial dilution steps onto SC-Leu-Trp (-LT), SC-His-Leu-Trp (-HLT), and SC-Ade-Leu-Trp (-ALT) plates. The effects of the introduced mutations were assessed both in the context of full-length Rpl3 (left panel) or the C-terminally truncated Rpl3.N52 variant (right panel).
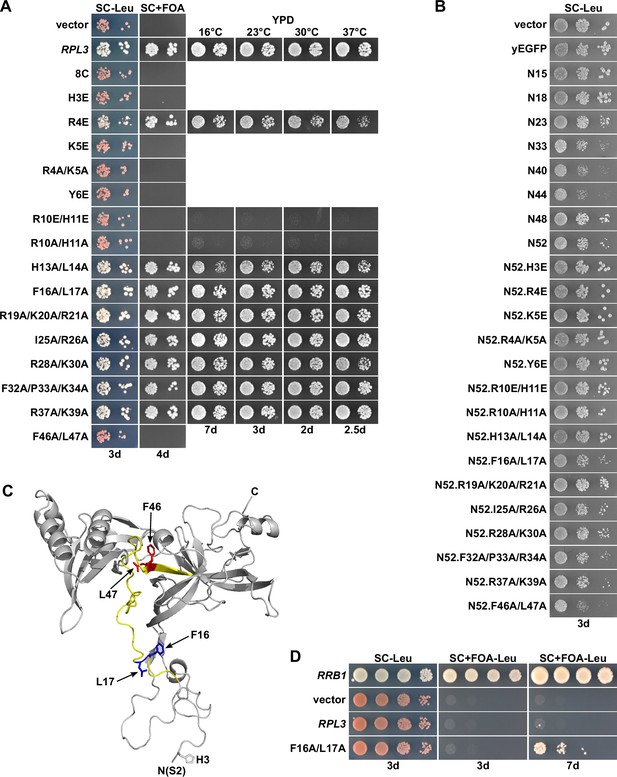
The rpl3.F16A/L17A allele fully complements the absence of endogenous RPL3 and suppresses the lethality of ∆rrb1 cells.
(A) Effect of mutations within Rpl3’s N-terminal region on yeast growth. Empty vector (YCplac111) and plasmid-borne wild-type RPL3 or the indicated rpl3 mutants, expressed under the control of the cognate promoter, were transformed into an RPL3 shuffle strain. Transformants were restreaked on SC-Leu plates, and cells were then spotted in 10-fold serial dilution steps onto SC-Leu and SC + fluoroorotic acid (FOA) plates, which were incubated at 30°C for the indicated number of days. Viable mutants were restreaked from 5-FOA-containing plates on YPD plates and then spotted in 10-fold serial dilution steps onto YPD plates. (B) Effect of the expression of Rpl3’s N-terminal region, when exhibiting different C-terminal borders or including the glutamate and alanine substitutions used in this study, on growth of wild-type cells. The wild-type strain YDK11-5A was transformed with an empty vector (YCplac111) or plasmids expressing nonfused yEGFP or the indicated N-terminal wild-type or mutant Rpl3 fragments, fused to the C-terminal yEGFP tag, under the control of the ADH1 promoter. Transformants were restreaked on SC-Leu plates, and cells were then spotted in 10-fold serial dilution steps onto SC-Leu plates. (C) Cartoon representation of Rpl3’s structure in the ribosome-bound state, extracted from PDB 4V88 (Ben-Shem et al., 2011). The minimal regulation-conferring region of Rpl3 (amino acid 12–52) is colored in light yellow. Residues F16/L17 and F46/L47 are highlighted in blue and red, respectively, with side chains in stick representation. (D) The rpl3.F16A/L17A allele suppresses the lethality of ∆rrb1 null mutant cells. Empty vector (YCplac111) and plasmid-borne RRB1, RPL3, or the rpl3.F16A/L17A mutant, all expressed under the control of their cognate promoters, were transformed into an RRB1 shuffle strain. Transformants were restreaked on SC-Leu plates, and cells were then spotted in 10-fold serial dilution steps onto SC-Leu and SC-Leu + FOA plates, which were incubated at 30°C for the indicated number of days.
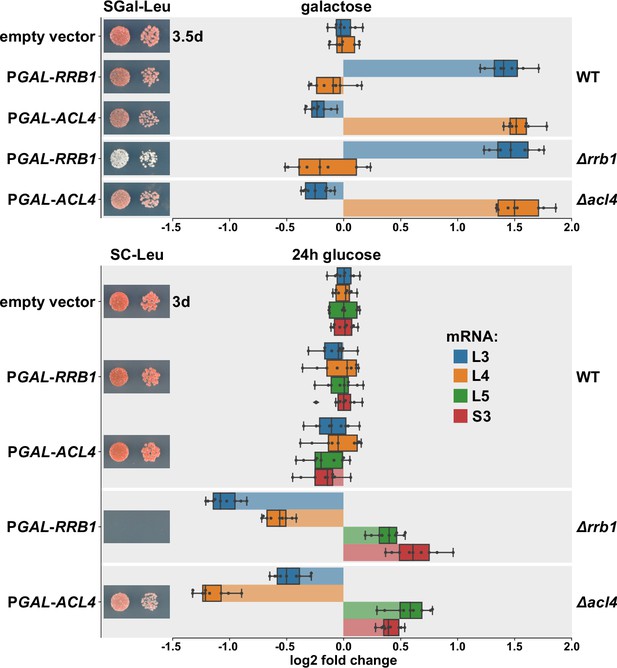
Overexpression of Rrb1 and Acl4 increases RPL3 and RPL4 mRNA levels.
Wild-type (WT), RRB1 shuffle (∆rrb1), and ∆acl4 cells were transformed with an empty vector or plasmids expressing either Rrb1 or Acl4 under the control of the inducible GAL1-10 promoter. Relative levels of the RPL3, RPL4, RPL5, and RPS3 mRNAs were determined by qRT-PCR using total RNA extracted from log-phase cells grown in SGal-Leu medium (galactose; upper panel) or shifted for 24 hr to SC-Leu medium (glucose; lower panel). The relative changes in mRNA levels between the different conditions (Rrb1 and Acl4 overexpression or depletion in WT, ∆rrb1, or ∆acl4 cells) have been normalized to the abundance of each assayed mRNA in WT cells transformed with the empty vector and grown in the same medium. The data shown were obtained from three different WT, RRB1 shuffle, and ∆acl4 strains (biological triplicates), in each case consisting of a technical triplicate, and they are represented as described in the legend to Figure 1G. In addition, the transformed cells were spotted in 10-fold serial dilution steps onto SGal-Leu (galactose) or SC-Leu (glucose) plates, which were incubated at 30°C.
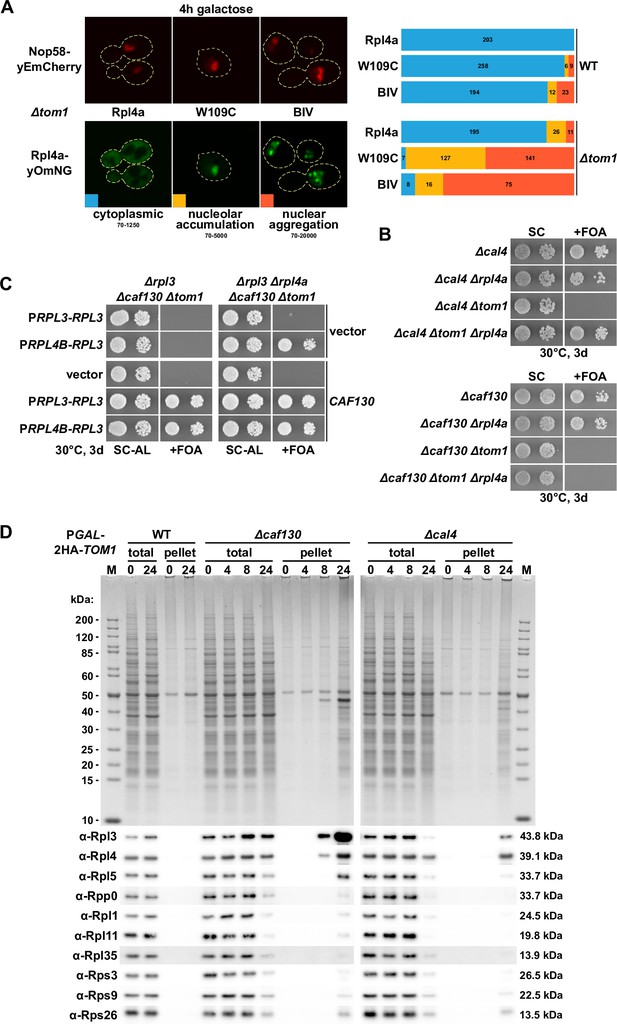
Deregulated expression of Rpl3 and Rpl4 induces their aggregation and abolishes growth in the absence of Tom1.
(A) Overexpressed Rpl4a variants, exhibiting deregulated expression, accumulate in the nucleolus and aggregate in the nucleus in the absence of Tom1. Wild-type (WT) and ∆tom1 strains were co-transformed with plasmids expressing the indicated Rpl4a variants, C-terminally fused to a yeast codon-optimized mNeonGreen (yOmNG), under the control of the inducible GAL1-10 promoter and a plasmid expressing Nop58-yEmCherry to indicate the subcellular position of the nucleolus. Cells were grown at 30°C in SRaf-Leu (raffinose) medium, and expression of the Rpl4a variants was induced for 4 hr with 2% galactose. The left panel shows representative examples of the three types of observed localizations (cytoplasmic, nucleolar accumulation, and nuclear aggregation). The images shown were acquired from ∆tom1 cells expressing wild-type Rpl4a or the two indicated Rpl4a variants and are displayed according to the indicated 16-bit brightness level ranges (min-max); note that the cytoplasmic signal, due to these parameter choices, is not well visible in the examples highlighting the nucleolar accumulation and nuclear aggregation. The right panel shows proportional bar graphs based on the number of counted cells displaying each of the three typical localizations (blue: cytoplasmic; yellow: nucleolar accumulation; red: nuclear aggregation). (B) Reduced expression of Rpl4 suppresses the lethality of ∆cal4/∆tom1 but not of ∆caf130/∆tom1 cells. The indicated single, double, and triple deletion strains, all derived from tetratype tetrads, were spotted in 10-fold serial dilution steps onto SC and SC + fluoroorotic acid (FOA) (+FOA) plates. (C) Reduced expression of both Rpl3 and Rpl4 efficiently suppresses the lethality of ∆caf130/∆tom1 cells. Empty vector (YCplac111) or plasmids harboring RPL3, expressed either from the RPL3 or RPL4B promoter, and empty vector (pASZ11) or a plasmid containing CAF130, expressed from the ADH1 promoter, were co-transformed into RPL3/CAF130 (∆rpl3/∆caf130) double shuffle strains additionally bearing chromosomal deletions of TOM1 (∆tom1; left panel) or both TOM1 and RPL4A (∆tom1/∆rpl4a; right panel). Transformants were restreaked on SC-Ade-Leu plates, and cells were then spotted in 10-fold serial dilution steps onto SC-Ade-Leu (SC-AL) and SC + FOA-Ade-Leu (+FOA) plates. (D) Depletion of Tom1 in Δcaf130 or Δcal4 cells leads to the aggregation of Rpl3 and/or Rpl4, thereby perturbing overall cellular proteostasis. WT, ∆caf130, or ∆cal4 cells, expressing N-terminally 2xHA-tagged Tom1 under the transcriptional control of the GAL1 promoter from the genomic locus (PGAL-2HA-TOM1), were grown at 30°C in YPGal medium and then shifted for up to 24 hr to YPD medium. Cells were harvested after the indicated times of growth in YPD medium (0, 4, 8, or 24 hr). The total extracts (total) and the insoluble pellet fractions (pellet) were analyzed by SDS-PAGE and Coomassie staining (upper panel) and by Western blotting using the indicated antibodies (lower panel).
-
Figure 7—source data 1
Original image files of the Coomassie-stained gel and the Western blots shown in Figure 7D, including a PDF file showing the full gel and blots and indicating the cropped areas.
- https://cdn.elifesciences.org/articles/74255/elife-74255-fig7-data1-v1.zip
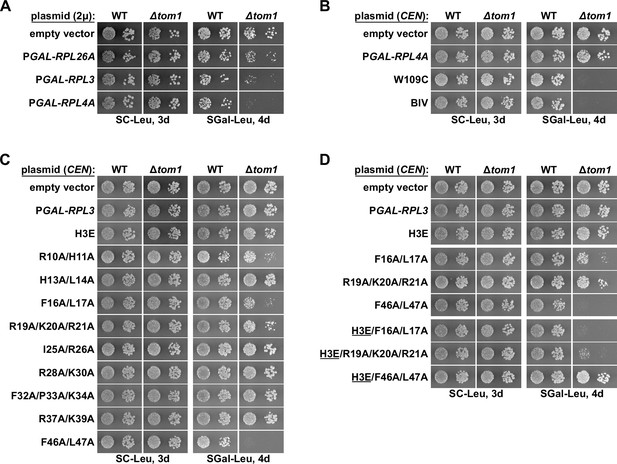
Overexpression of Rpl3 and Rpl4 variants affects the growth of ∆tom1 cells.
(A) Strong overexpression of Rpl3 and Rpl4a negatively affects the growth of ∆tom1 cells. Empty vector or multicopy (2µ) plasmids expressing Rpl26a, Rpl3, or Rpl4a, under the transcriptional control of the inducible GAL1-10 promoter, were transformed into wild-type (WT) and ∆tom1 strains. Transformants were restreaked on SC-Leu plates, and cells were then spotted in 10-fold serial dilution steps onto SC-Leu (glucose) and SGal-Leu (galactose) plates, which were incubated for the indicated times at 30°C. (B–D) Moderate overexpression of Rpl3 and Rpl4a variants, exhibiting deregulated expression, negatively affects the growth of ∆tom1 cells. Empty vector or monocopy (CEN) plasmids expressing wild-type Rpl3 or Rpl4a and the indicated variants thereof, under the transcriptional control of the inducible GAL1-10 promoter, were transformed into WT and ∆tom1 strains. Transformants were restreaked on SC-Leu plates, and cells were then spotted in 10-fold serial dilution steps onto SC-Leu (glucose) and SGal-Leu (galactose) plates, which were incubated for the indicated times at 30°C.
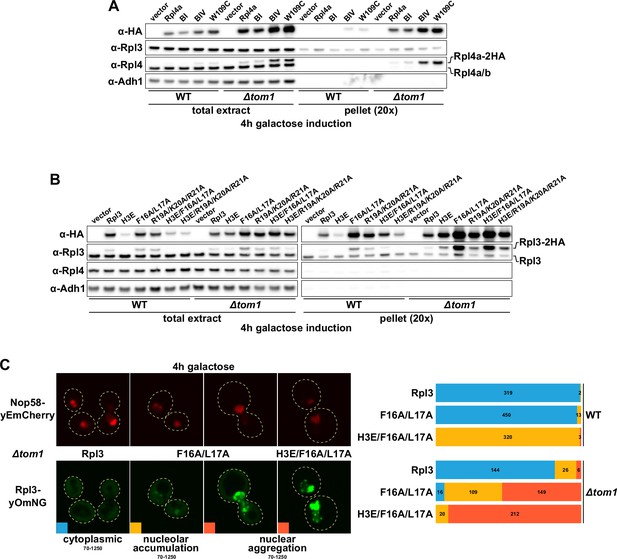
Overexpressed Rpl3 and Rpl4 variants aggregate in ∆tom1 cells.
(A, B) Overexpression of Rpl4a (A) and Rpl3 (B) variants, exhibiting deregulated expression, is prone to aggregation in the absence of Tom1. Empty vector or monocopy (CEN) plasmids expressing C-terminally 2xHA-tagged wild-type Rpl3 or Rpl4a and the indicated variants thereof, under the transcriptional control of the inducible GAL1-10 promoter, were transformed into wild-type (WT) and ∆tom1 strains. Cells were grown at 30°C in SRaf-Leu medium, and expression of the Rpl3 and Rpl4a variants was induced for 4 hr with 2% galactose. The total extracts and the insoluble pellet fractions (pellet) were analyzed by Western blotting using anti-HA, anti-Rpl3, anti-Rpl4, and anti-Adh1 (loading control) antibodies. (C) Overexpressed Rpl3.F16A/L17A, exhibiting deregulated expression, accumulates in the nucleolus and aggregates in the nucleus in the absence of Tom1. WT and ∆tom1 strains were co-transformed with plasmids expressing the indicated Rpl3-yOmNG variants under the control of the inducible GAL1-10 promoter and a plasmid expressing Nop58-yEmCherry. Cells were grown at 30°C in SRaf-Leu medium, and expression of the Rpl3 variants was induced for 4 hr with 2% galactose. The left panel shows representative examples of the three types of observed localizations (cytoplasmic, nucleolar accumulation, and nuclear aggregation). The images shown were acquired from ∆tom1 cells expressing wild-type Rpl3 or the two indicated Rpl3 variants and are displayed according to the indicated 16-bit brightness level ranges (min-max). The right panel shows proportional bar graphs based on the number of counted cells displaying each of the three typical localizations (blue: cytoplasmic; yellow: nucleolar accumulation; red: nuclear aggregation).
-
Figure 7—figure supplement 2—source data 1
Original image files of the Western blots shown in Figure 7—figure supplement 2A, including a PDF file showing the full blots and indicating the cropped areas.
- https://cdn.elifesciences.org/articles/74255/elife-74255-fig7-figsupp2-data1-v1.zip
-
Figure 7—figure supplement 2—source data 2
Original image files of the Western blots shown in Figure 7—figure supplement 2B, including a PDF file showing the full blots and indicating the cropped areas.
- https://cdn.elifesciences.org/articles/74255/elife-74255-fig7-figsupp2-data2-v1.zip
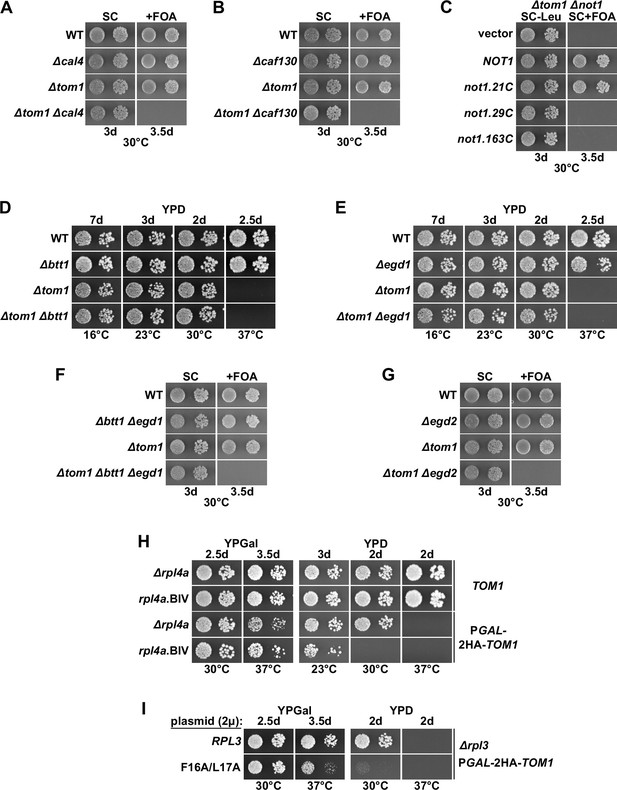
Absence of individual components of the regulatory machinery confers lethality to cells lacking Tom1.
(A, B, F, G) The indicated wild-type (WT), single, double, or triple deletion strains, in each case derived from tetratype tetrads, were spotted in 10-fold serial dilution steps onto SC and SC + fluoroorotic acid (FOA) (+FOA) plates. (C) Empty vector or plasmids harboring full-length NOT1 or the indicated not1 deletion variants, expressed under the control of the NOT1 promoter, were transformed into a NOT1 shuffle strain (∆not1) lacking TOM1 (∆tom1). Transformants were restreaked on SC-Leu plates, and cells were then spotted in 10-fold serial dilution steps onto SC-Leu and SC+ FOA (+FOA) plates. (D, E) The indicated WT, single and double deletion strains, all derived from tetratype tetrads, were spotted in 10-fold serial dilution steps onto YPD plates. (H) WT cells (TOM1) or cells expressing N-terminally 2xHA-tagged Tom1 under the transcriptional control of the GAL1 promoter from the genomic locus (PGAL-2HA-TOM1) and additionally either lacking RPL4A (∆rpl4a) or harboring the genomically integrated rpl4a.BIV allele were spotted in 10-fold serial dilution steps onto YPGal and YPD plates. (I) Multicopy (2µ) plasmids expressing Rpl3 or the Rpl3.F16A/L17A variant were transformed into a RPL3 shuffle strain (∆rpl3) expressing N-terminally 2xHA-tagged Tom1 under the transcriptional control of the GAL1 promoter from the genomic locus (PGAL-2HA-TOM1). After plasmid shuffling on 5-FOA-containing plates, cells were spotted in 10-fold serial dilution steps onto YPGal and YPD plates.
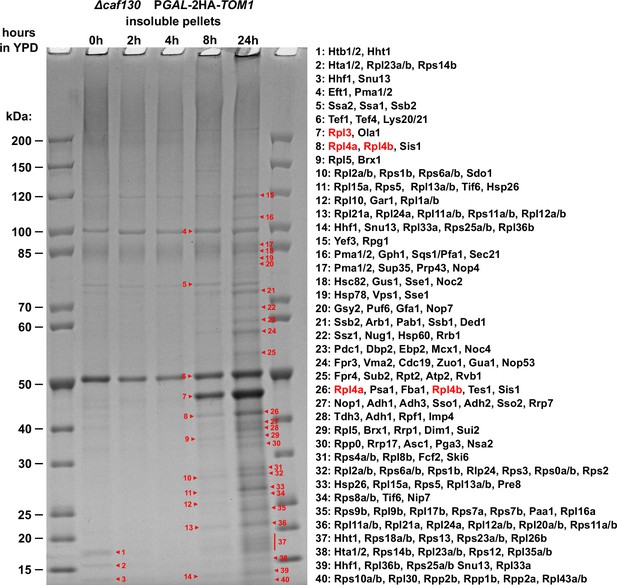
Identification of aggregated proteins in ∆caf130 cells upon genetic depletion of Tom1.
Cells lacking Caf130 (∆caf130) and expressing N-terminally 2xHA-tagged Tom1 under the transcriptional control of the GAL1 promoter from the genomic locus (PGAL-2HA-TOM1) were grown at 30°C in YPGal medium and then shifted for up to 24 hr to YPD medium. Cells were harvested after the indicated times of growth in YPD medium (0, 2, 4, 8, or 24 hr). The insoluble pellet fractions were separated on a NuPAGE gradient gel, which was subsequently stained with Coomassie. The indicated bands (numbered from 1 to 40) were cut out from the gel. Then, the contained proteins were digested in-gel with trypsin and the generated peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Only the most abundant proteins (according to the iBAQ intensity values calculated by MaxQuant), specifically peaking in this gel band and being within the expected molecular mass range, are listed (for the complete list of identified proteins, see Supplementary file 8).
-
Figure 7—figure supplement 4—source data 1
Original image file of the Coomassie-stained gel shown in Figure 7—figure supplement 4, including a PDF file showing the full gel and indicating the cropped area.
- https://cdn.elifesciences.org/articles/74255/elife-74255-fig7-figsupp4-data1-v1.zip
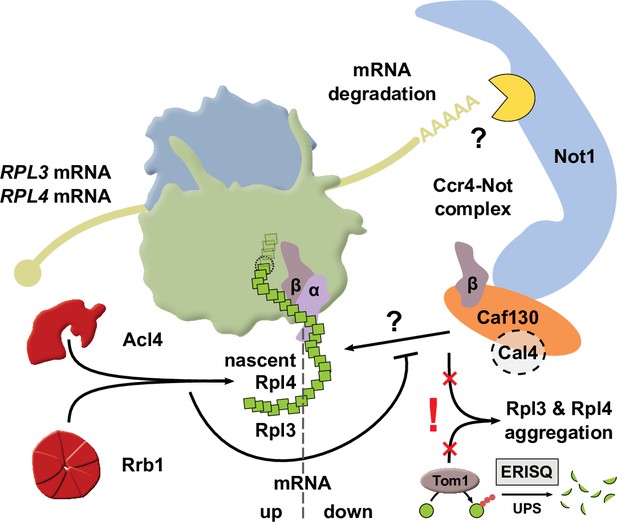
Simplified model showing how availability of the dedicated chaperone Rrb1 or Acl4 and the here uncovered regulatory network cooperate to balance Rpl3 and Rpl4 expression by co-translationally regulating RPL3 and RPL4 mRNA levels.
The question marks indicate that it remains to be determined how nascent Rpl3 or Rpl4 are recognized by the regulatory machinery and how this leads to the degradation, presumably involving a component of the Ccr4-Not complex, of the RPL3 or RPL4 mRNAs. Also included in the model and highlighted by an exclamation mark is the finding that surplus production of Rpl3 and/or Rpl4, for example, elicited by inactivation of the regulatory machinery, may lead to their aggregation when cells lack the E3 ubiquitin ligase Tom1, which is required for mediating the degradation of excess r-proteins by the ubiquitin proteasome system (UPS) via the so-called ERISQ (excess ribosomal protein quality control) pathway. The α and β subunit of the nascent polypeptide-associated complex (NAC) are denoted as α and β, respectively. For more details, see Discussion.
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/74255/elife-74255-transrepform1-v1.docx
-
Supplementary file 1
Yeast strains used in this study.
- https://cdn.elifesciences.org/articles/74255/elife-74255-supp1-v1.xlsx
-
Supplementary file 2
Plasmids used in this study.
- https://cdn.elifesciences.org/articles/74255/elife-74255-supp2-v1.xlsx
-
Supplementary file 3
Identified ∆acl4 and ∆acl4/∆rpl4a suppressor mutations.
- https://cdn.elifesciences.org/articles/74255/elife-74255-supp3-v1.xlsx
-
Supplementary file 4
Results of RNA-Seq.
- https://cdn.elifesciences.org/articles/74255/elife-74255-supp4-v1.xlsx
-
Supplementary file 5
Raw read counts of RNA-Seq.
- https://cdn.elifesciences.org/articles/74255/elife-74255-supp5-v1.txt
-
Supplementary file 6
Metadata of RNA-Seq.
- https://cdn.elifesciences.org/articles/74255/elife-74255-supp6-v1.txt
-
Supplementary file 7
R script for analysis of RNA-Seq raw read counts.
- https://cdn.elifesciences.org/articles/74255/elife-74255-supp7-v1.zip
-
Supplementary file 8
Identified proteins in aggregates of ∆caf130/PGAL-2xHA-TOM1 cells.
- https://cdn.elifesciences.org/articles/74255/elife-74255-supp8-v1.xlsx