Early anteroposterior regionalisation of human neural crest is shaped by a pro-mesodermal factor
Figures
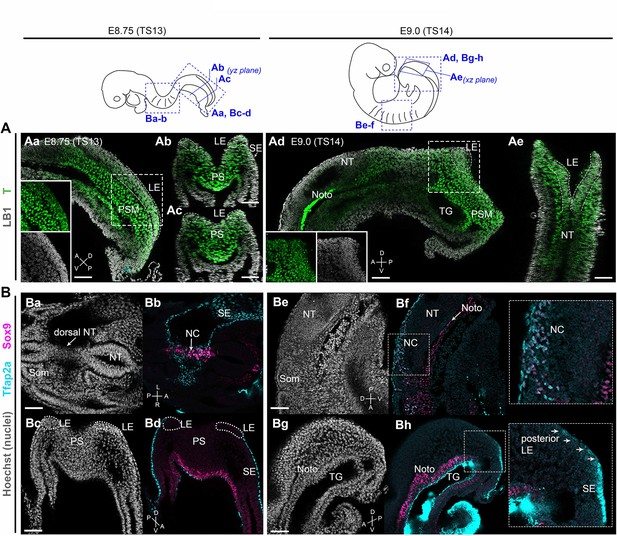
Brachyury expression marks neural crest (NC)-fated axial progenitors.
Top: schematics showing location and orientation of immunostaining data in in embryonic day (E) 8.75 (Theiler stage [TS]13) and E9.0 (TS14) embryos. (A) Confocal sections of wholemount immunostaining showing T (Brachyury) expression (green) in the lateral-most caudal epiblast (LE, indicated by dashed lines). Anti-Lamin B1 (LB1) expression (grey) denotes nuclei. (B) Confocal sections of wholemount immunostaining for Sox9 (magenta) and Tfap2a (cyan) showing NC derivatives (Ba–Bb, Be–Bf) and their progenitors (Bc-Bd, Bg-Bh) in the trunk and tail bud, respectively. At E9.0, Tfap2a was detected in posterior LE progenitors and at a lower level compared to the surface ectoderm (SE; inset in Bg). Noto: notochord; NT: neural tube; PS: primitive streak; PSM: presomitic mesoderm; SE: surface ectoderm; som: somite; TG: tail gut. A: anterior; P: posterior; D: dorsal; V: ventral; L: left; R: right. Scale bars = 50 µm.
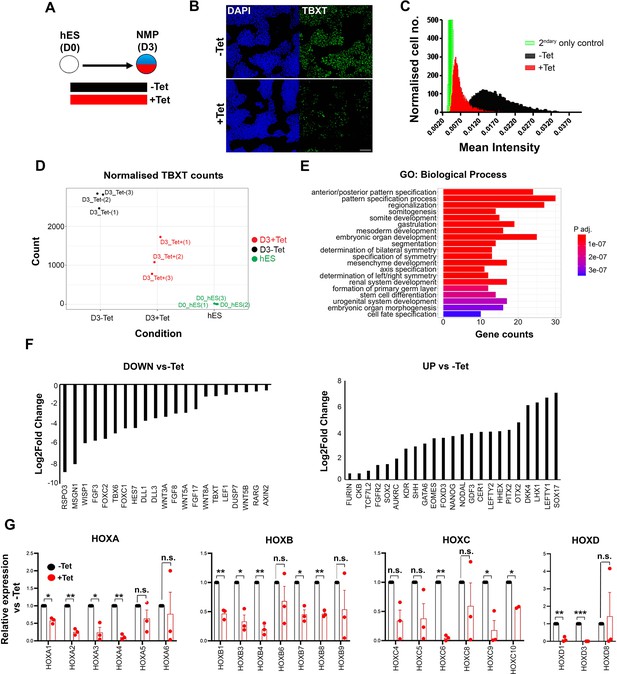
Effect of TBXT reduction on human pembryonic stem cell (hES)-derived neuromesodermal progenitors (NMPs).
(A) Differentiation/tetracycline (Tet) treatment scheme. (B) Immunofluorescence analysis of the expression of TBXT in shRNA hEScell-derived NMPs in the presence and absence of Tet. Scale bar = 100 μm. (C) Mean fluorescence intensity of TBXT protein in Tet-treated and control NMP cultures. (D) Normalised expression values of TBXT transcripts in control, Tet-treated NMPs and undifferentiated hESC samples following RNA sequencing (RNA-seq) analysis. (E) Gene ontology (GO) term analysis of differentially expressed genes in hESC-derived NMPs following TBXT knockdown. (F) Representative significantly downregulated and upregulated transcripts following TBXT depletion. (G) Quantitative PCR (qPCR) expression analysis of indicated HOX genes in control vs Tet-treated NMPs. Error bars represent SD (n=3). *p<0.05, **p<0.005, ***p<0.0005 (paired t-test). n.s. not significant.
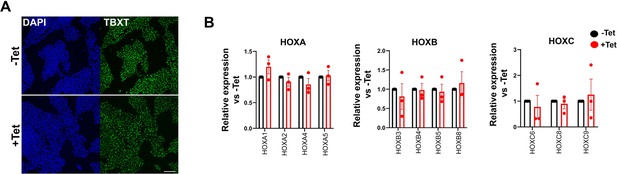
Effect of Tet treatment on control B2M shRNA hESC-NMPs.
(A) Immunofluorescence analysis of the expression of TBXT in control B2M shRNA hESC-derived NMPs in the presence and absence of Tet. Scale bar = 100 μm. (B) qPCR expression analysis of indicated HOX genes in control vs Tet-treated NMPs generated from B2M shRNA hESCs. Error bars represent SD (n=3).
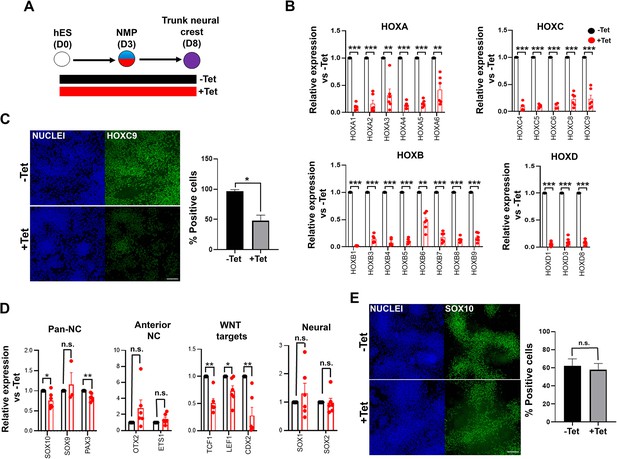
TBXT depletion impairs posterior axial identity acquisition by neural crestNC.
(A) Differentiation/tetracycline (Tet) treatment scheme. (B) qPCR expression analysis of indicated HOX genes in control vs Tet-treated NMP-derived trunk NC cells. Error bars represent SD (n=4–6). *p<0.05, **p<0.005, ***p<0.0005 (paired t-test). (C) Immunofluorescence analysis of the expression of HOXC9 in control vs Tet-treated NMP-derived trunk NC cells. Quantification of HOXC9+ cells in the presence and absence of Tet is also shown. Error bars represent SD (n=3). *p<0.05 (paired t-test). Scale bar = 100 μm. (D) qPCR expression analysis of indicated markers in control vs Tet-treated NMP-derived trunk NC cells. Error bars represent SD (n=6). *p<0.05, **p<0.01, n.s. not significant (paired t-test). (E) Immunofluorescence analysis of the expression of SOX10 in control vs Tet-treated NMP-derived trunk NC cells. Quantification of SOX10+ cells in the presence and absence of Tet is also shown. Error bars represent SD (n=3). Scale bar = 100 μm. n.s. not significant (paired t-test).
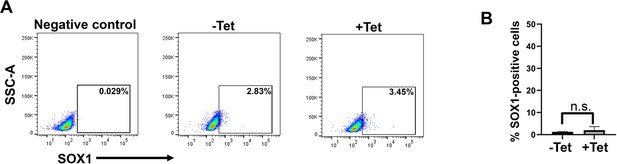
TBXT depletion does not influence the number of neurectodermal cells in trunk NC cultures.
(A) Representative flow cytometry (FACS) plots showing the presence of SOX1-positive cells in trunk NC cultures generated from NMPs in the presence and absence of tetracycline (Tet) following antibody staining. A secondary only negative control is also shown. (B) Quantification of SOX1-positive cells in control vs Tet-treated trunk NC cultures following immunofluorescence and image analysis. Error bars represent SD (n=3). n.s. not significant.

TBXT-driven programming of a posterior neural crest (NC) axial identity is an early event.
(A) Differentiation/tetracycline (Tet) treatment scheme. The temporal Tet treatment regimens employed were: no Tet control (− −); Tet treatment between days 3 and 7 (− +); Tet treatment between days 0 and 7 (+ +); Tet treatment between days 0 and 3 (+−). (B) qPCR expression analysis of representative lineage identity/posterior markers in early spinal cord/NC cells generated following culture in ‘neutral’ basal media in the presence of Tet during the time windows shown in A. Error bars represent SD (n=3). Only statistically significant changes are indicated. *p<0.05, **p<0.005 (paired t-test). (C) Immunofluorescence analysis of the expression of HOXC9, SOX1, and SOX10 in early spinal cord/NC cells generated following culture in ‘neutral’ basal media in the presence of Tet during the time windows shown in A. Scale bar = 100 µm. Quantification of the cell populations expressing these proteins in relation to the different Tet treatment regimens is shown in (D). Error bars represent SD (n=3). H: HOXC9; S1: SOX1; S10: SOX10. (E) qPCR expression analysis of indicated HOX genes in early spinal cord/NC cells generated following culture in ‘neutral’ basal media in the presence of Tet during the time windows shown in A. Error bars represent SD (n=3). Only statistically significant changes are indicated. *p<0.05, **p<0.005 (paired t-test).

Early programming of a posterior axial identity in neuromesodermal progenitor (NMP)-derived neural crest(NC) cells is primarily WNT-dependent.
(A) Scheme of treatments during the differentiation of human pluripotent stem cells (hPSCs) toward NMPs. (B) qPCR expression analysis of representative WNT-FGF targets in NMP cultures treated with the indicated combinations of WNT-FGF agonists/antagonists. Error bars represent SD (n=3–4). In all cases, changes (agonist-antagonist treatments vs corresponding WNT-FGF controls) were significantly different (ratio paired t-test) unless otherwise stated (n.s. not significant). (C–D) qPCR expression analysis of key NMP markers (C) and HOX genes (D) in NMP cultures treated with the indicated combinations of WNT-FGF agonists/antagonists. Error bars represent SD (n=3–4). In all cases, changes (agonist-antagonist treatments vs corresponding WNT-FGF controls) were significantly different (ratio paired t-test). (E) Scheme of treatments during the differentiation of hPSC-derived NMPs toward trunk NC cells. (F–G) qPCR expression analysis of representative lineage-specific, axial identity (F) and HOX genes (G) in NMP-derived trunk NC cultures treated with the indicated combinations of WNT-BMP agonists/antagonists. Error bars represent SD (n=3). Statistically significant changes are indicated. *p<0.05, **p<0.005.
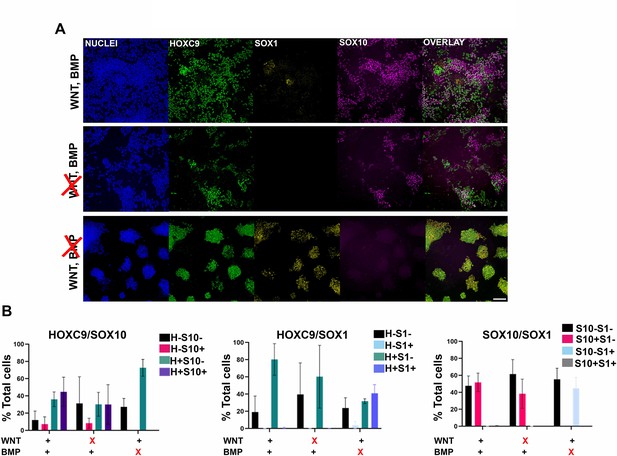
Influence of WNT and BMP signalling on trunk NC specification.
(A) Immunofluorescence analysis of the expression of HOXC9, SOX1, and SOX10 in trunk NC cultures treated with the indicated combinations of WNT-BMP agonists/antagonists. Scale bar = 100 µm. (B) Quantification of cell populations expressing HOXC9, SOX1, and SOX10 protein following treatment with the indicated combinations of WNT-BMP agonists/antagonists. Error bars represent SD (n=3). H: HOXC9; S1: SOX1; S10: SOX10.

WNT signalling dynamics during posterior NC emergence.
(A) Fluorescence analysis of R26-WntVis mouse embryos at embryonic day (E) 8.75 (Theiler stage [TS]13) and E9.0 (TS14) demonstrating EGFP reporter responsiveness to WNT signalling. (B) Sagittal sections of immunostained R26-WntVis tail buds showing anti-GFP (a-GFP) signal in the T+ lateral-most caudal epiblast (LE; dashed lines) at E8.75 (Ba-c) and E9.0 (Bd-f). Nuclei were defined by anti-Lamin B1 staining (LB1). Arrowheads indicate T-negative surface ectoderm cells. (Bg-j) 3D reconstruction of imaging data showing the T+ area (green), nuclei (LB1, grey), and a-GFPhigh volumes, coloured by the intensity of the a-GFP signal. Since the reporter is active at a low level in many cells, a cutoff was made to show only higher expressing a-GFP+ volumes. The Wnthigh T+ cells are found in the LE: posterior view at E8.75 (Bg-h) and lateral view at E9.0 (Bi-j). Noto: notochord; NT: neural tube; PS: primitive streak; TG: tail gut. A,:anterior; P: posterior; D: dorsal; V: ventral; L: left; R: right. Scale bars = 50 µm.

Posterior axial identity acquisition by neuromesodermal progenitor (NMP)-derived, WNT-FGF-induced pre-neural spinal cord cells is TBXT-independent.
(A, D) Differentiation/treatment schemes associated with different time windows of TBXT knockdown during spinal cord differentiation from NMPs. (B–C, E–F) qPCR expression analysis of indicated HOX genes (B, E) and representative NMP/early spinal cord markers (C, F) in control vs tetracycline (Tet)-treated NMP-derived early spinal cord progenitors corresponding to the Tet treatment regimens shown in A and D, respectively. Error bars represent SD (n=3). Statistically significant changes are indicated. *p<0.05, n.s. not significant (paired t-test).
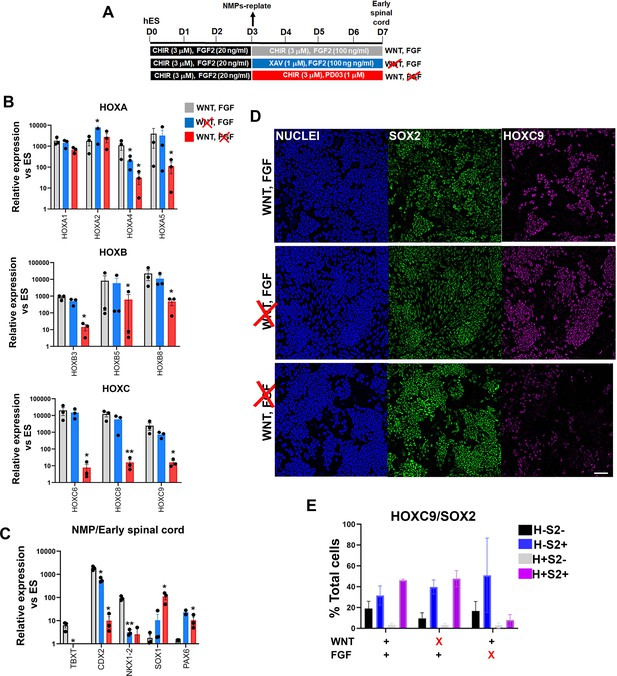
Posterior axial identity acquisition by NMP-derived pre-neural spinal cord progenitors is FGF-dependent.
(A) Scheme of treatments during the differentiation of hPSC-derived NMPs toward early spinal cord progenitors. (B–C) qPCR expression analysis of indicated HOX genes (B) and representative NMP/early spinal cord/neural markers (C) in NMP-derived early spinal cord progenitor cultures generated as depicted in A. Error bars represent SD (n=3). Statistically significant changes are indicated. *p<0.05, **p<0.005. (D) Immunofluorescence analysis of SOX2 and HOXC9 protein expression in NMP-derived early spinal cord progenitor cultures treated with the indicated combinations of WNT-FGF agonists/antagonists. Quantification of the cell populations expressing these proteins in relation to the different treatment regimens is shown in (E). Error bars represent SD (n=3). H: HOXC9; S2: SOX2.
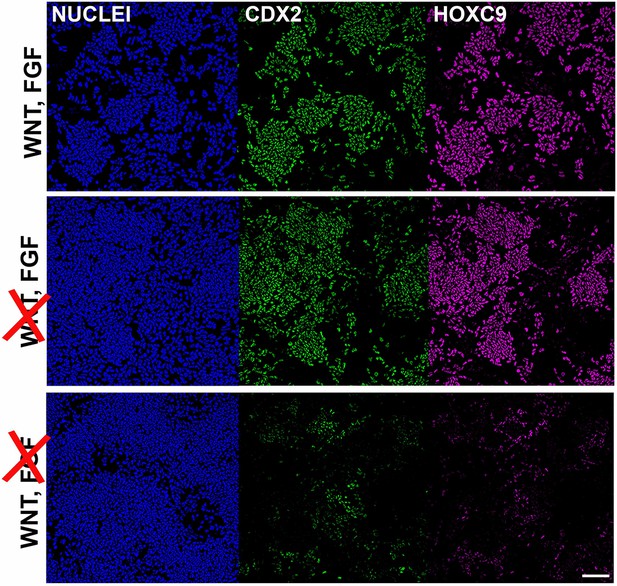
Posterior axial identity acquisition by NMP-derived pre-neural spinal cord progenitors is FGF-dependent.
Immunofluorescence analysis of CDX2 and HOXC9 protein expression in NMP-derived early pre-neural spinal cord progenitor cultures treated with the indicated combinations of WNT-FGF agonists/antagonists.
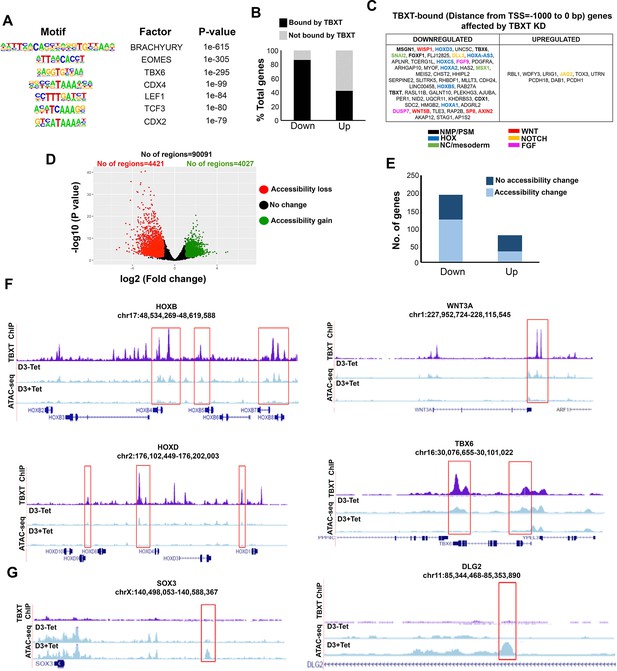
TBXT controls HOX gene expression and WNT signalling in NMPs by influencing chromatin accessibility.
(A) Representative transcription factor-binding motifs enriched in TBXT binding sites. (B) Graph showing the percentages of differentially expressed genes (padj<0.05, log2FC>|1|) following TBXT knockdown during the transition of hESCs toward NMPs that are bound directly by TBXT. (C) Table showing all differentially expressed genes following TBXT knockdown that exhibit TBXT binding within their promoter region. (D) Volcano plot of differentially accessible ATAC-seq peaks between TBXT-depleted and control NMPs. (E) Graph showing the number of significantly upregulated and downregulated direct TBXT targets in relation to changes in chromatin accessibility associated with TBXT knockdown. (F) Correspondence between TBXT binding (ChIP) and chromatin accessibility changes (ATAC-seq) in the presence and absence of tetracycline (Tet) in indicated HOX clusters, WNT/presomitic mesoderm-linked loci. (G) Correspondence between TBXT binding and chromatin accessibility changes in the presence and absence of Tet in indicated neural differentiation-linked loci. Boxed areas highlight TBXT-bound regions marked by loss or gain of chromatin accessibility in the presence of Tet.

Effect of TBXT binding on chromatin accessibility.
(A) Average density plot of tag distributions across peak regions corresponding to the neuromesodermal progenitor (NMP), human embryonic stem cells (hES), and input samples. (B) Genomic distribution of TBXT-bound sites in hESC-derived NMPs. (C) Gene ontology (GO) biological processes enrichment analysis of target genes associated with TXBT binding sites around their transcriptional start site (–2000 to +500 bp). (D) Correspondence between TBXT binding (ChIP) and chromatin accessibility changes (ATAC-seq) in the presence and absence of tetracycline (Tet) in indicated HOX and WNT-linked loci. Boxed areas highlight TBXT-bound regions marked by loss of chromatin accessibility in the presence of Tet. (E) Venn diagram showing the overlap between transcription factor DNA binding motifs in genomic regions associated with chromatin accessibility gain and loss following TBXT knockdown.
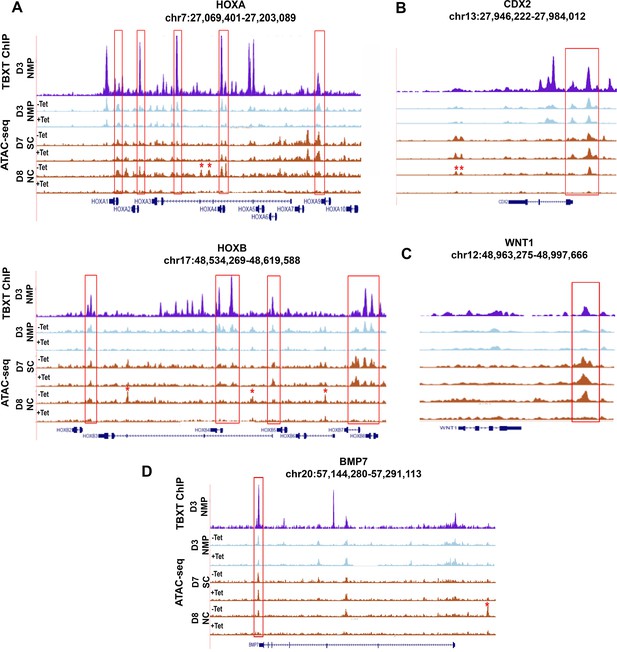
Chromatin accessibility dynamics during NMP differentiation toward trunk NC and pre-neural spinal cord progenitors.
Correspondence between TBXT binding (ChIP) in human pluripotent stem cell (hPSC)-derived NMPs and chromatin accessibility changes (ATAC-seq) in NMPs (D3) and NMP-derived early spinal cord (D7 SC) and trunk neural crest (D8 NC) in the presence and absence of tetracycline (Tet) in the indicated HOX loci (A), CDX2 (B), and WNT/BMP-linked loci (C, D). Boxed areas highlight TBXT-bound regions marked by loss of chromatin accessibility in the presence of Tet. Asterisks mark trunk NC-specific regulatory elements that are not bound by TBXT but exhibit loss of chromatin accessibility upon Tet treatment.
Additional files
-
Supplementary file 1
Significantly up- and downregulated transcripts in Tet-treated, TBXT-depleted hESC-derived NMPs.
- https://cdn.elifesciences.org/articles/74263/elife-74263-supp1-v2.xlsx
-
Supplementary file 2
List of GO terms and corresponding gene lists enriched in Tet-treated, TBXT depleted hESC-derived NMPs.
- https://cdn.elifesciences.org/articles/74263/elife-74263-supp2-v2.xlsx
-
Supplementary file 3
List of all genomic regions (Intervals) with peak p-value below the applied threshold bound by TBXT in hESC-derived NMPs and undifferentiated hESCs.
- https://cdn.elifesciences.org/articles/74263/elife-74263-supp3-v2.xlsx
-
Supplementary file 4
List of GO terms and corresponding gene lists associated with TBXT binding sites in hPSC-derived NMPs.
- https://cdn.elifesciences.org/articles/74263/elife-74263-supp4-v2.xlsx
-
Supplementary file 5
List of known HOMER database motifs enriched in TBXT binding sites in hESC-derived NMPs.
- https://cdn.elifesciences.org/articles/74263/elife-74263-supp5-v2.xlsx
-
Supplementary file 6
List of TBXT target genes which are differentially expressed following Tet treatment (padj<0.05 log2FC>|0.5|) and Gene Ontology Biological Processes enrichment analysis.
Genes are listed in relation to the genomic position (in relation to TSS) of TBXT binding within their proximity. Blue highlight denotes downregulation while red represents upregulation in expression.
- https://cdn.elifesciences.org/articles/74263/elife-74263-supp6-v2.xlsx
-
Supplementary file 7
List of ATAC-seq peaks associated with gain or loss of chromatin accessibility following TBXT depletion in hESC-derived NMPs.
Gene Ontology Biological Processes enrichment analysis, list of HOX genes as well as other genes (padj<0.05 log2FC>|1|) affected by TBXT depletion and are associated with changes in chromatin accessibility are also included.
- https://cdn.elifesciences.org/articles/74263/elife-74263-supp7-v2.xlsx
-
Supplementary file 8
List of transcription factor DNA binding motifs enriched in ATAC-seq sites associated with chromatin accessibility gain, chromatin accessibility loss or both.
- https://cdn.elifesciences.org/articles/74263/elife-74263-supp8-v2.xlsx
-
Supplementary file 9
ATAC-seq analysis of trunk NC cells generated from hPSCs via NMPs in the presence of Tet.
Includes: (i) Summary: Summary of detected peak numbers; (ii) all_peaks: List of all the peaks with P-value <0.05 (gained open, gained close, no change) with the relative annotations; (iii) Accessibility Loss padj <0.05: Significant Gained close peaks (pvalue adjusted (padj) <0.05); (iv) HOX genes: Peaks in HOX gene regions, yellow colour highlights the significant peaks (padj <0.05); (v) Enrichment-Accessibility Loss: Enrichment of Significant Gained close peaks (pvalue adjusted <0.05); (vi) Accessibility gain padj <0.05: Significant Gained close peaks (pvalue adjusted (padj) <0.05).
- https://cdn.elifesciences.org/articles/74263/elife-74263-supp9-v2.xlsx
-
Supplementary file 10
List of primers used.
- https://cdn.elifesciences.org/articles/74263/elife-74263-supp10-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/74263/elife-74263-transrepform1-v2.docx






