Cnidarian hair cell development illuminates an ancient role for the class IV POU transcription factor in defining mechanoreceptor identity
Figures
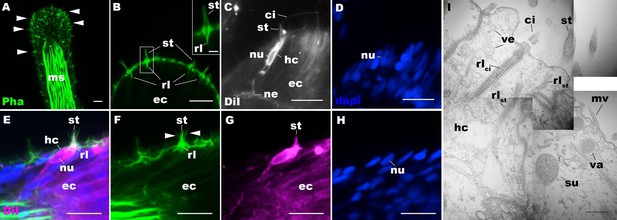
Morphology of sea anemone hair cells.
(A–F) Confocal sections of oral tentacles of Nematostella vectensis at the primary polyp stage. Filamentous actin is labeled with phalloidin (Pha), and nuclei are labeled with DAPI (dapi). DiI is used to label cell membrane of a subset of hair cells. In A, the distal end of the tentacle is to the top, and in B–I, the apical surface of the ectodermal epithelium is to the top. A: sections through the tentacle. Numerous hair bundles (arrowheads) are evident on the tentacle surface. (B) Sections through the hair bundles at the tentacle tip, showing stereovilli (st) and their prominent rootlets (rl) of central hair cells. (C–D) Sections through a DiI-labeled hair cell (hc) at the tentacle tip. Note that the hair cell has an apical cilium (ci) surrounded at its base by stereovilli (st), and basally extended thin neurites (ne). An empty space within the cell body shows the location of a nucleus (nu), as evidenced by DAPI staining (D). (E–H) Sections through a DiI-labeled hair cell (hc) located near the tip of a tentacle. Arrowheads in F point to microvilli of the mechanoreceptor hair bundle contributed by peripheral support cells, which are DiI-negative. (I) Electron microscopic section of an apical region of the tentacular ectodermal epithelium of N. vectensis polyp, showing a hair cell (hc) and a support cell (su). The hair cell has stereovilli that extend dense filaments into the cytoplasm, forming 2–3 µm-long rootlets (rlst), as well as numerous clear vesicles (ve), while the support cell has apical microvilli (mv) and electron-dense vacuoles (va). Abbreviations: ms muscle fibers; rlci ciliary rootlet; ec ectoderm. Scale bar: 10 µm (A–H); 2 µm (inset in B); 500 nm (I).
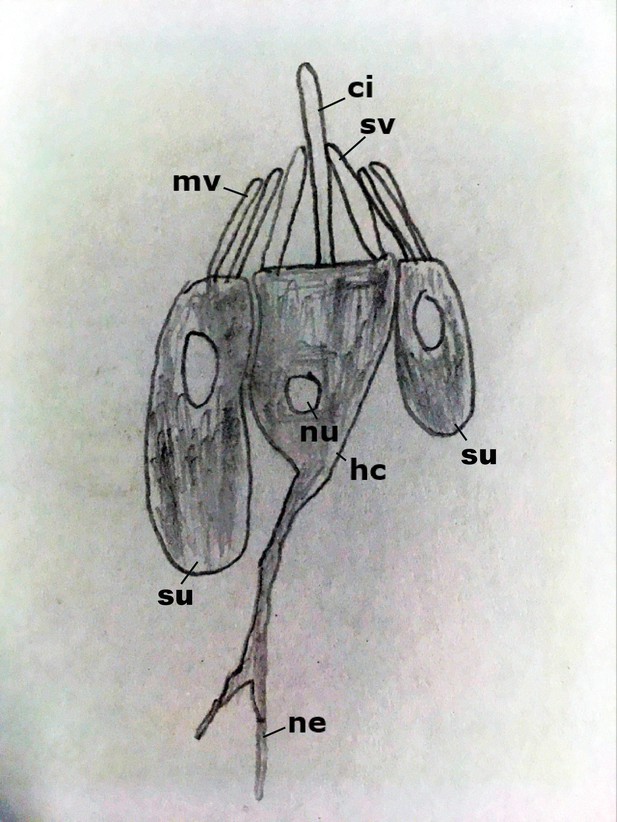
Diagram of the hair bundle mechanoreceptor of sea anemones.
A cilium (ci) extends from a central hair cell (hc). It is encircled by stereovilli (sv), also produced by the hair cell, as well as smaller-diameter microvilli (mv) contributed by surrounding support cells (su), forming a cone-shaped structure. Abbreviations: ne neurites; nu nucleus.
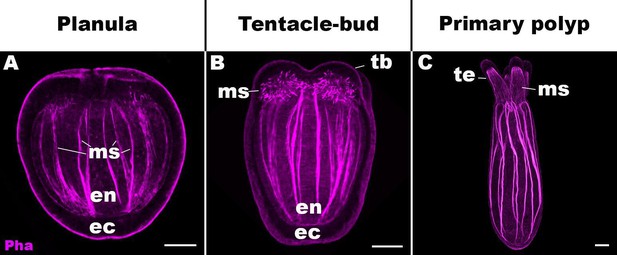
Life cycle transition in the sea anemone cnidarian Nematostella vectensis.
Confocal sections of N. vectensis at metamorphosis from a free-swimming planula (A; 3–5 days post-fertilization [dpf]), through the tentacle-bud stage (B; 5–7 dpf), into a primary polyp (C; 7–10 dpf). Filamentous actin is labeled with phalloidin (Pha). All panels show side views of the animal with the oral pole facing up. (A) The planula consists of ectoderm (ec) and endoderm (en) separated by an extracellular matrix, and develops muscle fibers (ms) in the endoderm. (B) At the tentacle-bud stage, four tentacle primordia known as the tentacle buds (tb) emerge in the circumoral ectoderm. (C) Four primary tentacles (te) and the body column then elongate along the oral-aboral axis, forming a primary polyp. Scale bar: 50 µm.
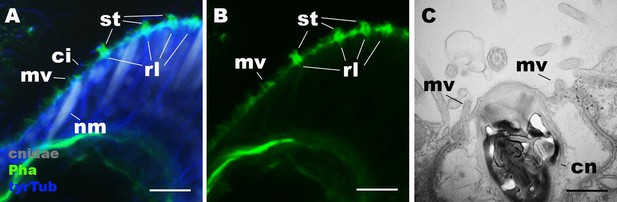
Morphology of nematocytes in the tentacles of Nematostella vectensis polyps.
(A, B) Confocal sections of N. vectensis primary polyp, labeled with an antibody against tyrosinated ∂-tubulin (‘tyrTub’). Filamentous actin is labeled with phalloidin (Pha), and mature cnidocysts (cnidae) are labeled with a high concentration of DAPI in the presence of EDTA (Szczepanek et al., 2002). The section shows the ectoderm at the tentacle tip, with the epithelial surface facing up. Note that nematocytes (nm) have an apical cilium (ci) surrounded by microvilli (mv) without rootlets. Stereovilli (st) and their prominent rootlets (rl) of hair cells are also shown. (C) An electron micrograph of a section of N. vectensis primary polyp, showing an apical structure of a cnida(cn)-containing nematocyte in the tentacular ectoderm. Microvilli (mv) without rootlets occur on the apical cell surface. Scale bar: 10 µm (A, B); 500 nm (C).
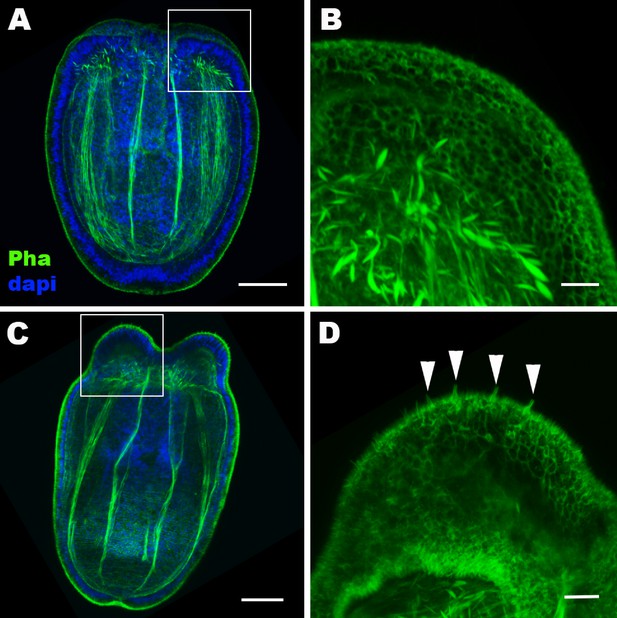
Hair cell development begins in the ectoderm of tentacle primordia at metamorphosis in sea anemones.
Confocal sections of Nematostella vectensis at the late planula (A, B) and tentacle-bud (C, D) stages. Filamentous actin is labeled with phalloidin (Pha), and nuclei are labeled with DAPI (dapi). All panels show side views of animals with the blastopore/mouth facing up. A and C show longitudinal sections through the center, and B and D show surface ectoderm of tentacular primordia boxed in A and C, respectively. Note that 3–5 µm-long stereovilli characteristic of hair cells become evident at the tentacle-bud stage (arrowheads in D), indicative of hair cell differentiation. Scale bar: 50 µm (A, C); 10 µm (B, D).
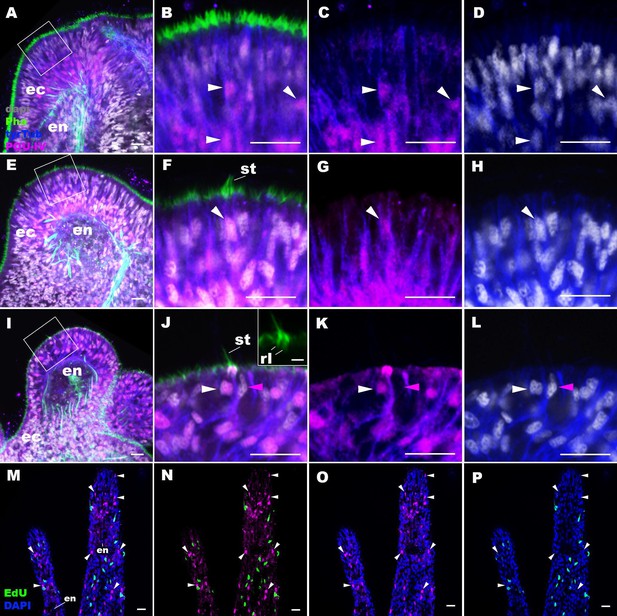
POU-IV is postmitotically expressed in hair cells of tentacular ectoderm at metamorphosis in the sea anemone.
Confocal sections of Nematostella vectensis at metamorphosis, labeled with antibodies against POU-IV, and/or tyrosinated ∂-tubulin (‘tyrTub’). Filamentous actin is labeled with phalloidin (Pha), and nuclei are labeled with DAPI (dapi). Proliferative cells are labeled by the thymidine analogue EdU. A shows a section through the presumptive tentacle primordia with the blastopore/mouth facing up. E, I, M-P show sections through developing oral tentacles with the distal end of the tentacle facing up; M–P are tangential sections of tentacles at the level of the surface ectoderm and parts of the endoderm (en). B–D, F–H, and J–L are magnified views of the boxed regions in A, E, and I, respectively, with the apical epithelial surface facing up. A-D: late planula. E–H: tentacle-bud. I–P: primary polyp. At the late planula stage prior to hair cell differentiation, POU-IV-positive nuclei are primarily localized at the basal and middle layers of the ectoderm of presumptive tentacle primordia (arrowheads in B–D); few POU-IV-positive nuclei are detectable at the superficial stratum. At the tentacle-bud stage, hair cells with pronounced stereovilli (st) and POU-IV-positive nuclei begin to develop in the superficial stratum of the ectodermal epithelium in tentacle primordia (arrowheads in F–H). POU-IV-positive nuclei in the superficial layer specifically occur in hair cells (white arrowheads in J–L) and not in adjacent support cells (purple arrowheads in J–L). The inset in J shows a magnified view of stereovilli (st) of a POU-IV-positive hair cell; note the presence of stereovillar rootlets (rl). In addition to hair cells, cnidocytes express POU-IV in the tentacular ectoderm (Figure 2—figure supplement 2; Tournière et al., 2020). POU-IV-positive cells are EdU-negative (arrowheads in I–L), evidencing their postmitotic cell-cycle status. Abbreviations: ec, ectoderm; en, endoderm. Scale bar: 10 µm (A–P); 2 µm (inset in J).
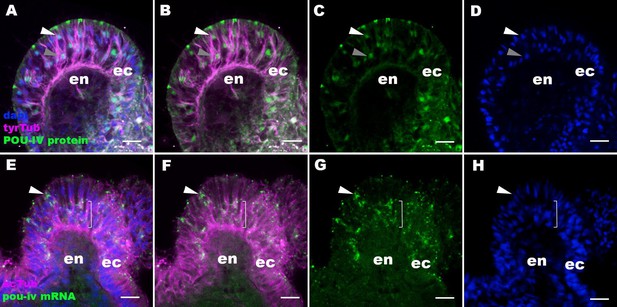
The pattern of immunoreactivity with an anti-POU-IV antibody recapitulates that of pou-iv mRNA expression.
Confocal sections of oral tentacles of Nematostella vectensis at the primary polyp stage, labeled with an anti-POU-IV antibody (‘POU-IV protein’; A−D), or an antisense riboprobe against pou-iv mRNA (‘pou-iv mRNA’; E−H). Microtubules are labeled by antibodies against tyrosinated ∂-tubulin (‘tyrTub’; A, B) or acetylated ∂-tubulin (‘acTub’; E, F). Nuclei are labeled with DAPI (dapi). All panels show sections through oral tentacles with the distal end of the tentacle facing up. Anti-POU-IV immunoreactivity occurs in a subset of apical nuclei (white arrowhead in A−D) and the majority of basal nuclei (gray arrowhead in A−D) within the pseudostratified ectodermal epithelium (ec), suggesting nuclear localization of POU-IV proteins in these cells. Consistent with the pattern of anti-POU-IV immunoreactivity, in situ hybridization signals of pou-iv mRNA occur in a subset of ectodermal epithelial cells whose cell bodies are located apically (white arrowhead in E−H), and in those ectodermal cells whose cell bodies are localized basally (bracketed in E−H). Anti-POU-IV immunoreactivity is absent in nuclei of tentacular endodermal cells (en in A−D), and in situ hybridization signals for pou-iv mRNA are not detectable in these cells (en in E−H), indicating the lack of pou-iv expression in tentacular endoderm. Scale bar: 10 μm.
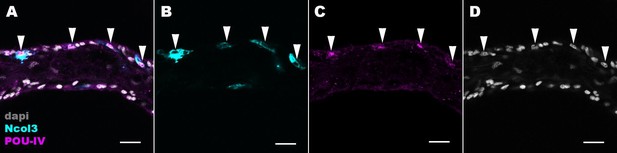
POU-IV localizes to the nuclei of cnidocytes in tentacular ectoderm of the sea anemone.
Confocal sections of an oral tentacle of Nematostella vectensis at the primary polyp stage, labeled with antibodies against POU-IV and/or minicollagen 3 (‘Ncol3’; Zenkert et al., 2011). Nuclei are labeled with DAPI (dapi). All panels show longitudinal sections through the center of the tentacle; the distal end of the tentacle is to the left. Arrowheads show POU-IV-positive nuclei of Ncol3-positive cnidocytes that reside in the tentacular ectoderm. Scale bar: 10 µm.
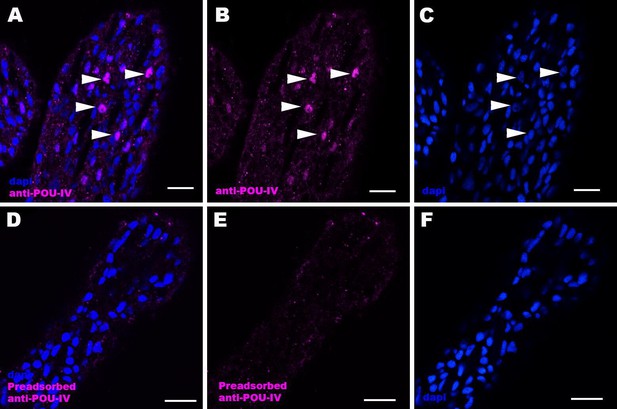
Immunostaining with a preadsorbed anti-POU-IV antibody.
Confocal sections of oral tentacles of Nematostella vectensis at the primary polyp stage, labeled with an anti-POU-IV antibody (‘anti-POU-IV’; A-C) and an anti-POU-IV antibody preadsorbed with the POU-IV antigen (CQPTVSESQFDKPFETPSPINamide) used to generate the antibody (‘preadsorbed anti-POU-IV’; D–F). Nuclei are labeled with DAPI (dapi). In all panels, the distal end of the tentacle is to the top, and sections are at the level of surface ectoderm. Arrowheads indicate nuclear immunoreactivity that is abolished when the preadsorbed antibody is used, indicating that the anti-POU-IV reacts with nuclear POU-IV. Scale bar: 10 μm.
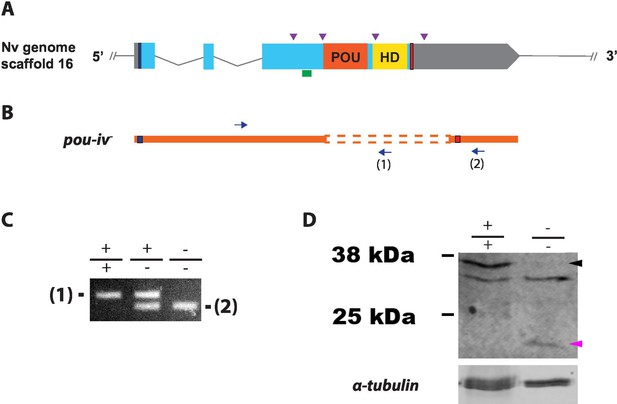
Generation of pou-iv null mutant sea anemones.
(A, B) Diagrams of the pou-iv locus (A) and the disrupted mutant allele (pou-iv-; B). Blue bars show predicted translation start sites; red bars show predicted translation termination sites. In A, filled boxes indicate exons, and the regions that encode the POU- and homeo-domains are highlighted in orange (‘POU’) and yellow (‘HD’), respectively. Purple arrowheads show single guide RNA (sgRNA) target sites. The region that encodes peptides targeted by the antibody generated in this study is indicated by a green line. In B, deletion mutation is boxed in dotted orange lines, and blue arrows mark regions targeted in the PCR analysis shown in C; reverse primers are numbered (1)–(2). (C) Genotyping PCR. Note that the wildtype allele-specific primer (1) generates a 689 bp PCR product from the wildtype allele ('+') but cannot bind to the pou-iv- allele due to deletion mutation. The primer (2) generates a 558 bp PCR product from the pou-iv- allele, and a 1312 bp PCR product from the wildtype allele. (D) Western blotting with an antibody against Nematostella vectensis POU-IV. An antibody against acetylated α-tubulin (‘α-tubulin’; ca. 52 kDa) was used as a loading control. The anti-POU-IV reacts with a protein of expected size for wildtype POU-IV (35.2 kDa) in wildtype (+/+) polyp extracts, but not in pou-iv mutant (-/-) polyp extracts (black arrowhead). Also note that the antibody’s reactivity with a protein of expected size for mutant POU-IV lacking DNA-binding domains (18.7 kDa) is detectable in mutant (-/-) extracts, but not in wildtype (+/+) extracts (purple arrowhead). The band just below the expected size of the wildtype POU-IV occur in both wildtype and mutant protein extracts, and therefore represents non-POU-IV protein(s) that are immunoreactive with the anti-POU-IV antibody.
-
Figure 3—source data 1
An original gel image used to generate Figure 3C and the original image with relevant lanes labeled.
- https://cdn.elifesciences.org/articles/74336/elife-74336-fig3-data1-v2.zip
-
Figure 3—source data 2
An original western blot image used to generate Figure 3D (top; anti-Nematostella vectensis POU-IV) and the original image with relevant lanes labeled.
- https://cdn.elifesciences.org/articles/74336/elife-74336-fig3-data2-v2.zip
-
Figure 3—source data 3
An original western blot image used to generate Figure 3D (bottom; anti-acetylated α-tubulin) and the original image with relevant lanes labeled.
- https://cdn.elifesciences.org/articles/74336/elife-74336-fig3-data3-v2.zip
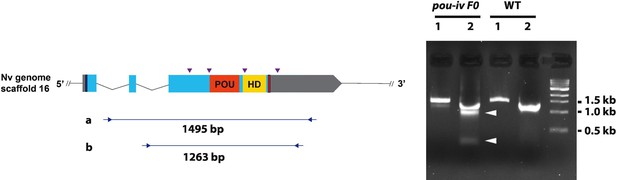
Generation of pou-iv F0 mosaic mutants by CRISPR-Cas9-mediated mutagenesis in Nematostella vectensis.
A schematic view of the pou-iv locus (left), and genomic DNA PCR results of an uninjected wildtype (WT) embryo (‘WT’) and an F0 embryo injected with locus-specific sgRNAs and Cas9 (‘pou-iv F0’) (right). In the schematic depicting the pou-i locus, filled boxes indicate exons. A blue bar shows the predicted translation start site, and a red bar shows the predicted translation termination site. The regions that encode the POU- and homeo-domains are highlighted in orange (‘POU’) and yellow (‘HD’), respectively. Purple arrowheads show single guide RNA (sgRNA) target sites. Blue arrows mark regions targeted in the PCR analysis shown to the right. Note that genomic PCR of the WT embryo shows expected sizes of PCR fragments (1495 bp) for primary PCR (‘a’), and 1263 bp for secondary nested PCR (‘b’), while F0 embryos show additional bands of smaller sizes (arrowheads), indicating that targeted deletions of different sizes have occurred mosaically in each embryo. DNA sequencing of the < 500 bp band (lower arrowhead) indicated that this mutant allele harbored 981 bp deletion encompassing the POU- and homeo-domain-encoding regions.
-
Figure 3—figure supplement 1—source data 1
An original gel image used to generate Figure 3—figure supplement 1 (right) and the original image with relevant lanes labeled.
- https://cdn.elifesciences.org/articles/74336/elife-74336-fig3-figsupp1-data1-v2.zip
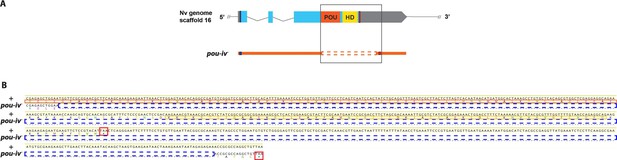
Sequence alignment of wildtype and mutant pou-iv alleles.
(A) Diagrams of the pou-iv locus and the disrupted mutant allele (pou-iv-). Blue bars show predicted translation start sites; red bars show predicted translation termination sites. In the schematic depicting the pou-iv locus, filled boxes indicate exons, and the regions that encode the POU- and homeo-domains are highlighted in orange (‘POU’) and yellow (‘HD’), respectively. In the schematic of the pou-iv- allele, deletion mutation is boxed in orange dotted lines. (B) Alignment of nucleotide and translated amino acid sequences of wildtype (‘+’) and mutant (‘pou-iv-’) alleles boxed in A. POU and homeo-domains are boxed in orange and yellow, respectively. Predicted translation termination sites are boxed in red, and 705 bp deletion mutation is boxed in dotted blue lines. Note that a sequence encoding all but the first four residues of the POU domain and the entire homeodomain is deleted in the pou-iv- allele.
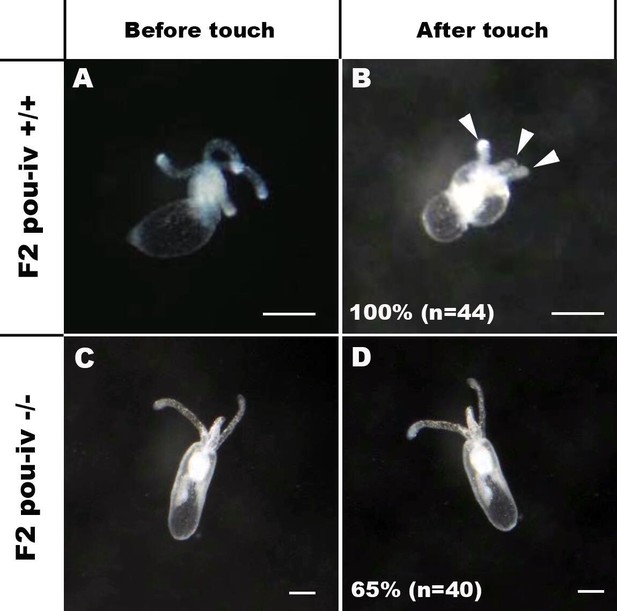
POU-IV is essential for touch-response behavior in the sea anemone.
(A-D) Behavior of wildtype (F2 pou-iv +/+, A, B) and mutant (F2 pou-iv -/-, C, D) Nematostella vectensis polyps in response to tactile stimuli to their oral tentacles. A hair held in a tungsten needle holder was used to touch the distal portion of each tentacle. Animals before (A, C) and after (B, D) tentacle touch are shown. Tactile stimuli to tentacles elicit tentacular retraction in the wildtype individual (100%, n = 44; A, B). In contrast, the majority of pou-iv homozygous mutants were touch-insensitive (65%, n = 40; B, D); only 35% of the animals showed any contractile response to touch stimuli. Arrowheads in B point to retracted tentacles. Scale bar: 1 mm.
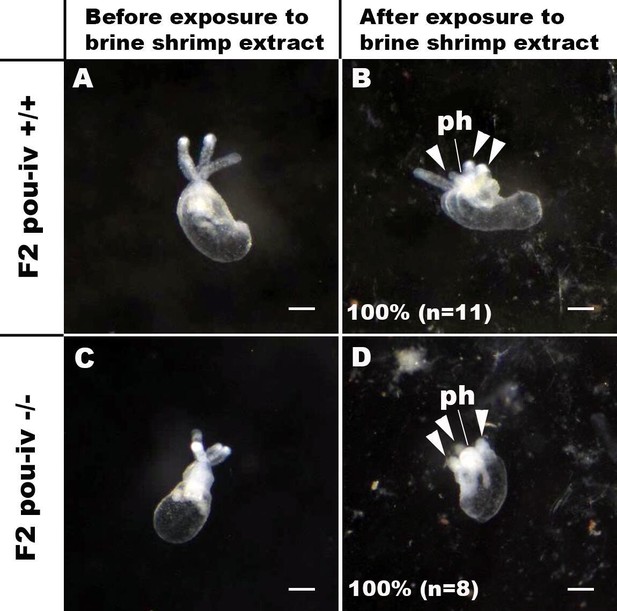
pou-iv mutants respond to brine shrimp extract by tentacular contraction and pharyngeal protrusion.
(A-D) Behavior of wildtype (F2 pou-iv +/+, A, B) and mutant (F2 pou-iv -/-, C, D) Nematostella vectensis polyps in response to exposure to Artemia extract. Both wildtype and mutant animals contracted tentacles (arrowheads in B and D), and protruded the pharynx (ph) within 1 min of exposure to the extract (F2 pou-iv +/+, 100%, n = 3; F2 pou-iv -/-, 100%, n = 9). Scale bar: 1 mm.
Touch-sensitive behavior of a wildtype (F2 pou-iv +/+) polyp.
Touch-insensitive behavior of a pou-iv mutant (F2 pou-iv -/-) polyp.
Behavior of a wildtype (F2 pou-iv +/+) polyp upon exposure to brine shrimp extract.
Behavior of a pou-iv mutant (F2 pou-iv -/-) polyp upon exposure to brine shrimp extract.
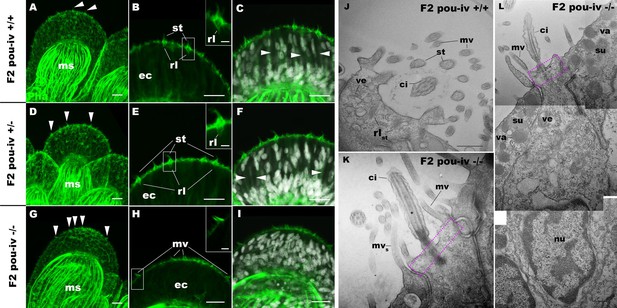
POU-IV is necessary for hair cell differentiation in the sea anemone.
(A-I) Confocal sections of oral tentacles of wildtype (F2 pou-iv +/+, A-C), heterozygous (F2 pou-iv +/-, D-F), and homozygous pou-iv mutant (F2 pou-iv -/-, G-I) Nematostella vectensis polyps. Filamentous actin is labeled with phalloidin (Pha), and nuclei are labeled with DAPI (dapi). In all panels, the distal end of the tentacle is to the top. (A, D, G) Sections through the tentacle. (B, C, E, F, H, I) Sections through hair bundles/ciliary cones at the tip of tentacles. Ciliary cones occur on the epithelial surface of the tentacle regardless of the genotype (arrowheads in A, D, G). However, stereovilli (st) with rootlets (rl) characteristic of mechanosensory hair cells are observed in wildtype (B) and heterozygous (E) siblings, but not in homozygous pou-iv mutants whose ciliary cones contain microvilli without prominent actin rootlets (mv in H). Arrowheads in C and F indicate spaces occupied by cnidocysts in wildtype and heterozygous siblings, respectively, which are absent in pou-iv homozygous mutants (I; Figure 5—figure supplement 1). (J–L) Electron microscopic sections of a hair cell of a F2 pou-iv +/+ polyp (J) and an epithelial cell with hair-cell-like morphologies in an F2 pou-iv -/- polyp (K, L). In all panels, apical cell surfaces face up. K and L are sections of the same cell at different levels. The hair-cell-like epithelial cell of the mutant has a central apical cilium surrounded by a collar of rootlet-less microvilli (mv in K, L), which are encircled by microvilli of the adjacent support cells (mvs in L), forming a ciliary cone. It also has numerous clear vesicles (ve in L) in the cytoplasm, characteristic of hair cells (ve in J; Figure 1G). Support cells of mutants are morphologically indistinguishable from those of wildtype animals, having characteristic large electron-dense vacuoles (va in L) in addition to apical microvilli (mvs in L) that contribute to the ciliary cone/hair bundle. Consistent with light microscopy data (A–C, G–I), stereovillar rootlets (rlst) are absent in the F2 pou-iv -/- polyp, but are present in hair cells of their wildtype siblings (J). In K and L, regions of apical cytoplasm where stereovillar rootlets would normally be observed are boxed with dotted purple lines. Abbreviations: ms muscle fibers; ec ectoderm; st stereovilli; ci cilium; rlst stereovillar rootlets. Scale bar: 10 µm (A-I); 2 µm (insets in B, E, H); 500 nm (J–L).
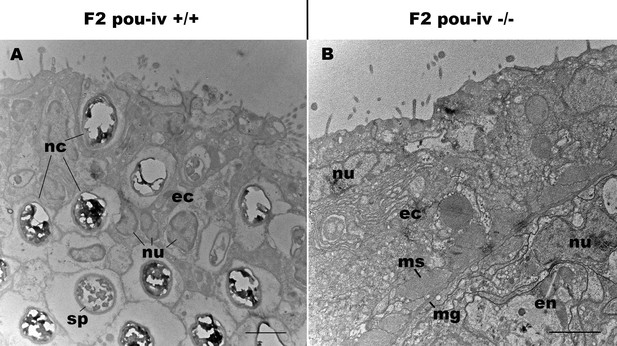
pou-iv mutants lack mature cnidocytes.
Electron microscopic sections of tentacular ectoderm of an F2 pou-iv +/+ polyp (A) and F2 pou-iv -/- polyp (B). Note the occurrence of numerous cnidae – nematocysts (nc) and spirocysts (sp) – in A that are absent in B. Abbreviations: nu, nucleus; ec, ectoderm; en, endoderm; ms, muscle fibers; mg, mesoglea. Scale bar: 2 µm.
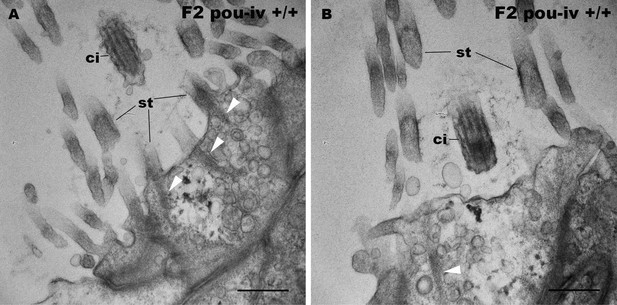
F2 pou-iv wildtype siblings develop hair cells with stereovillar rootlets.
Electron microscopic sections of hair cells of an F2 pou-iv +/+ polyp. In all panels, apical cell surfaces face up. Note the presence of stereovillar rootlets (arrowheads in A and B). Abbreviations: st, stereovilli; ci, cilium. Scale bar: 500 nm.
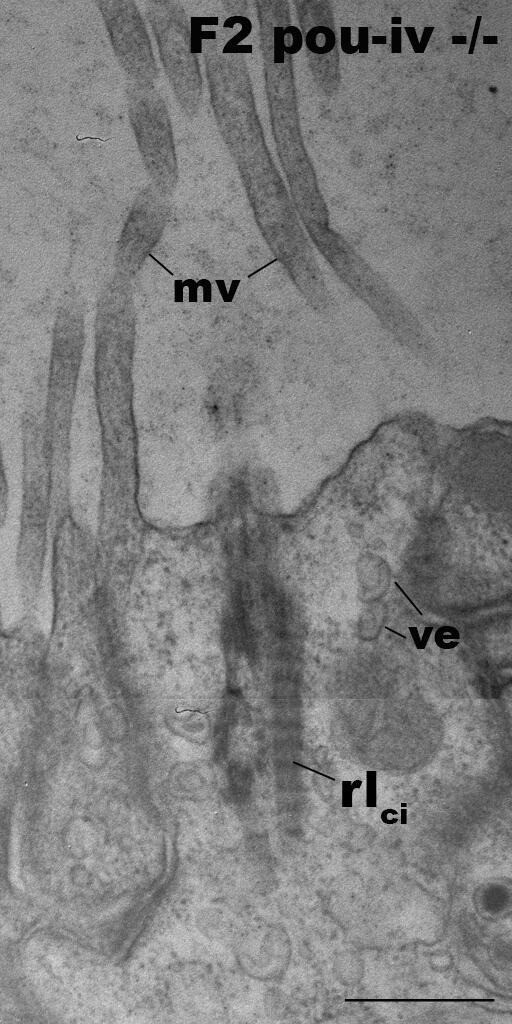
Hair-cell-like cells of pou-iv mutants have ciliary rootlets.
Electron microscopic sections of a hair-cell-like cell of an F2 pou-iv -/- polyp, characterized by ciliary-cone-associated microvilli (mv). The apical cell surface faces up. Note the presence of a ciliary rootlet (rlci). Abbreviations: ve, vesicles. Scale bar: 500 nm.
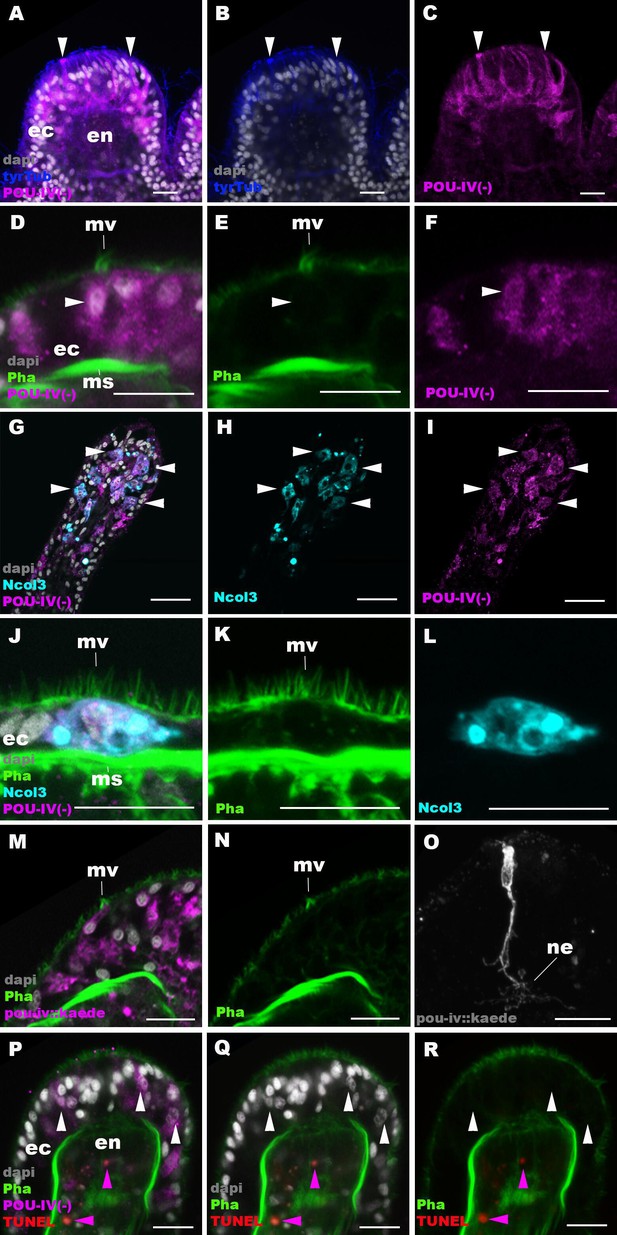
POU-IV is necessary for maturation of hair cells in the sea anemone.
Confocal sections of oral tentacles in F2 pou-iv -/- Nematostella vectensis polyps, labeled with antibodies against tyrosinated ∂-tubulin (‘tyrTub’), minicollagen 3 (‘Ncol3’; Zenkert et al., 2011), mutant POU-IV (‘POU-IV(-)’), and/or Kaede fluorescent protein (‘pou-iv::kaede’). DNA fragmentation is labeled by terminal deoxynucleotidyl transferase dUTP nick end-labeling (TUNEL). Filamentous actin is labeled with phalloidin (Pha), and nuclei are labeled with DAPI (dapi). In all panels, the apical surface of the tentacular ectodermal epithelium is to the top. (A–C) Sections through developing oral tentacles with the distal end of the tentacle facing up. Arrowheads point to a subset of POU-IV(-)-expressing epithelial cells, which are abundant in the tentacular ectoderm (ec). Note the cytoplasmic distribution of the POU-IV(-) likely resulting from the lack of nuclear localization signal. (D–F) Sections showing ciliary cone microvilli (mv)-bearing cells. Ciliary cone-bearing epithelial cells express POU-IV(-) (arrowheads). (G–I) Sections at the level of surface ectoderm of developing oral tentacles with the distal end of the tentacle facing up. A subset of POU-IV(-)-expressing cells are Ncol3-positive (arrowheads), representing immature cnidocytes. (J–L) Sections showing an immature cnidocyte which expresses POU-IV(-) and Ncol-3. Note that the cell bears apical microvilli (mv) that do not form a ciliary cone. (M–O) Sections showing immature hair cells in F2 pou-iv -/- N. vectensis polyps injected with pou-iv::kaede construct. Note the presence of ciliary cone microvilli (mv) and basal neurites (ne). (P–R) Sections through tentacles with the distal end facing up. White arrowheads point to nuclei of POU-IV(-)-expressing ectodermal epithelial cells, which are TUNEL-negative. TUNEL-positive, pyknotic nuclei are observed in the endoderm (purple arrowheads). Abbreviations: ec ectoderm; en endoderm; ms muscle fiber. Scale bar: 10 µm.
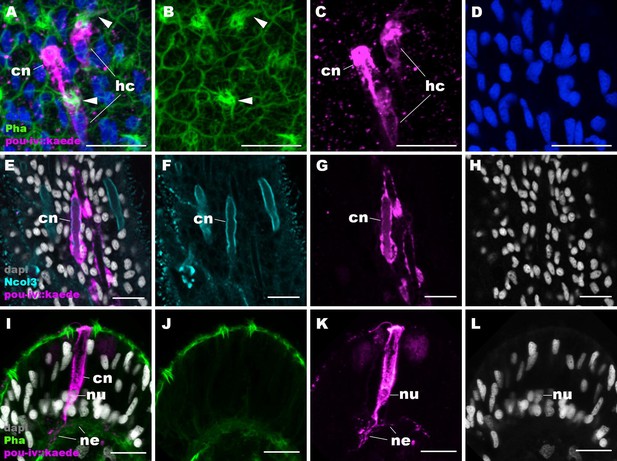
pou-iv::kaede reporter construct drives transgene expression in hair cells and cnidocytes.
Confocal sections of oral tentacles of Nematostella vectensis polyps injected with pou-iv::kaede construct, labeled with antibodies against Kaede (‘pou-iv::kaede’) and minicollagen 3 (‘Ncol3’). Filamentous actin is labeled with phalloidin (Pha), and nuclei are labeled with DAPI (dapi). Panels A–H show tangential sections at the level of the surface ectoderm. Panels I–L show a side view of a cnidocyte (cn), with the apical cell surface facing up. pou-iv::kaede expression occurs in hair cells (hc in A–D) and cnidocytes (cn in A–L). Arrowheads in A–C indicate stereovilli of hair cells. Cnidocytes have basally localized nuclei (nu) and extend basal neurite-like processes (ne). Scale bar: 10 µm.
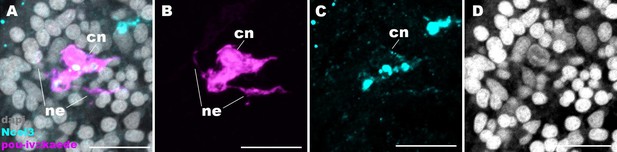
Cnidocytes in pou-iv null mutants develop basal processes.
Confocal sections of a cnidocyte (cn) in the body column of a F2 pou-iv +/+ polyp injected with pou-iv::kaede construct and labeled with antibodies against Kaede (‘pou-iv::kaede’) and minicollagen 3 (‘Ncol3’). Nuclei are labeled with DAPI (dapi). Panels (A-D) show tangential sections at the level of the basal ectoderm. Note that the cnidocyte extends neurite-like processes (ne). Scale bar: 10 µm.
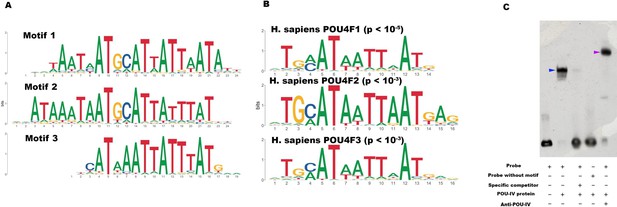
POU-IV-binding motifs are conserved across Cnidaria and Bilateria.
(A) Motifs enriched in Nematostella vectensis POU-IV chromatin immunoprecipitation sequencing (ChIP-seq) peaks. (B) Homo sapiens POU motifs resulting from sequence alignment and comparison against the Jaspar database. The p-value reported corresponds to the highest p-value for any of the three N. vectensis POU4 motifs found. (C) Electrophoretic mobility shift assay (EMSA) using purified N. vectensis POU-IV protein and a 50 bp DNA probe containing the conserved core motif CATTATTAAT. Note that retardation of probe migration occurs in the presence of POU-IV protein (blue arrowhead; lane 2), indicative of formation of the protein-DNA complex. Retardation is inhibited in the presence of an unlabeled competitor probe (‘specific competitor’; lane 3). Removal of the motif sequence in the probe (‘probe without motif’) abolishes retardation of probe migration by POU-IV (lane 4), demonstrating that the motif is necessary for formation of the protein-DNA complex. The mobility of the probe is further decreased in the presence of the anti-POU-IV antibody (purple arrowhead; lane 5), confirming that the protein bound to the probe is POU-IV.
-
Figure 7—source data 1
List of 12,972 genome-wide-binding sites for POU-IV.
- https://cdn.elifesciences.org/articles/74336/elife-74336-fig7-data1-v2.txt
-
Figure 7—source data 2
An original gel image used to generate Figure 7C and the original image with relevant lanes labeled.
- https://cdn.elifesciences.org/articles/74336/elife-74336-fig7-data2-v2.zip
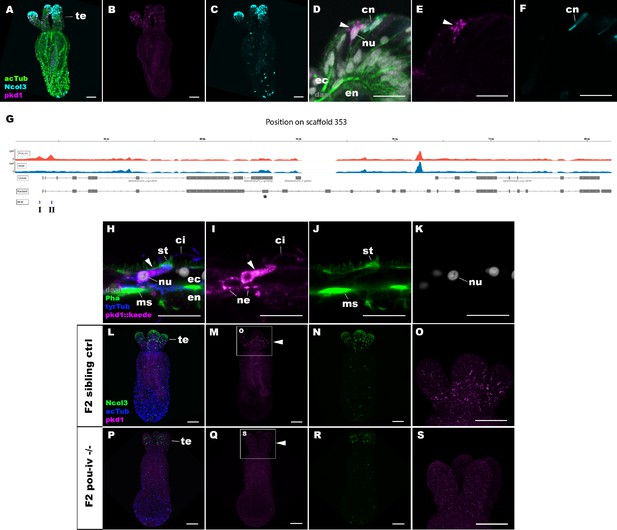
POU-IV activates the expression of polycystin 1 specifically in hair cells.
(A-F) Confocal sections of primary polyps labeled with an antisense riboprobe against polycystin 1 transcript (‘pkd1’) and antibodies against acetylated ∂-tubulin (‘acTub’) and minicollagen 3 (‘Ncol3”; Zenkert et al., 2011). Nuclei are labeled with DAPI (‘dapi’). A–C are side views of the animal with the oral opening facing up. Expression of polycystin 1 occurs exclusively in the ectoderm of the oral tentacles (te). (D–F) Side views of a polycystin 1-expressing epithelial cell (arrowhead) in the tentacular ectoderm (ec) with its apical surface facing up. Note that the cell body is localized apically and lacks minicollagen 3 expression. (G) A schematic of the polycystin 1 locus, showing the distribution of POU-IV ChIP DNA (‘POU-IV’) and input DNA from adult polyps. JGI gene models (‘Genes’) and the revised gene model based on RTPCR (‘Revised’) and the locations of the consensus POU-IV-binding motif – AT(A/T)ATT(A/T)AT – are numbered as I and II. X-axis shows the position along the genomic scaffold, and Y-axis shows the number of reads. * shows an exon whose sequence is missing in the publicly available Nematostella vectensis genome (v1.0; Putnam et al., 2007). (H–K) Confocal sections of an oral tentacle of a primary polyp injected with polycystin 1::kaede construct, labeled with an antibody against Kaede (‘pkd1::kaede’). Filamentous actin is labeled with phalloidin (Pha). The apical surface of the tentacular ectodermal epithelium is to the top. Note that the Kaede-positive cell (arrowhead) has an apical cilium (ci) and stereovilli (st), a central nucleus (nu), and basal neurites (ne), exhibiting morphological hallmarks of a hair cell. No other cell types were found to be Kaede-positive. L–S: Confocal sections of a homozygous pou-iv mutant (‘F2 pou-iv -/-’, P-S) and its sibling control (F2 pou-iv +/+ or pou-iv +/-, ‘F2 sibling ctrl’, L–O) at the primary polyp stage, labeled with an antisense riboprobe against polycystin 1 transcript (‘pkd1’) and antibodies against acetylated ∂-tubulin (‘acTub’) and minicollagen 3 (‘Ncol3’; Zenkert et al., 2011). Panels show side views of the animal with the oral opening facing up. Animals lacking mature cnidocysts based on Ncol3 staining were assumed to be pou-iv -/- mutants; animals with mature cnidocysts were assumed to be pou-iv +/+ or pou-iv +/-. O and S are magnified views of tentacles boxed in M and Q, respectively. Note that cell-type-specific expression of polycystin 1 in tentacular ectoderm (arrowhead in M; O) is absent in the POU-IV null mutant (arrowhead in Q; S), demonstrating that POU-IV is necessary for polycystin 1 expression. Abbreviations: en, endoderm; cn, cnidocyst; nu, nucleus; ms, muscle fiber. Scale bar: 50 µm (A–C, L–S); 10 µm (D–F, H–K).
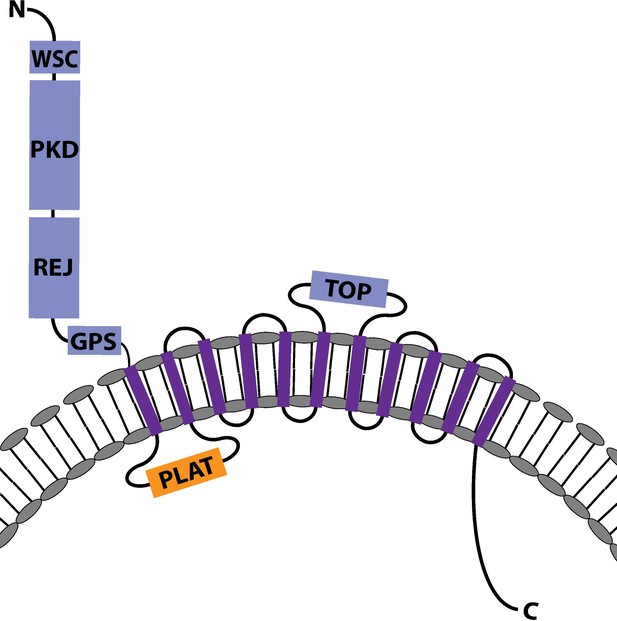
The predicted protein structure of Nematostella vectensis Polycystin 1-like.
The predicted protein structure of N. vectensis Polycystin 1-like. Extracellular domains are shown in blue, the intracellular domains in orange, and the transmembrane domains in purple. Transmembrane-spanning regions were predicted by using Phyre2 (Kelley et al., 2015) and based on alignment with human and Fugu Polycystin 1 sequences. Non-transmembrane-spanning regions were predicted by using NCBI conserved domain search with default Blast search parameters. The PKD domain and the REJ module are specific to Polycystin 1 (PKD1) proteins. Abbreviations: WSC, cell wall integrity and stress response component; REJ, receptor for egg jelly; GPCR, G-protein-coupled receptor; TM, transmembrane domain; PLAT, polycystin-1, lipoxygenase, and alpha toxin; TOP, tetragonal opening for polycystins.
-
Figure 8—figure supplement 1—source data 1
Partial cDNA sequence of Nematostella vectensis polycystin 1-like, in which the start codon and stop codon are highlighted in green and red, respectively.
- https://cdn.elifesciences.org/articles/74336/elife-74336-fig8-figsupp1-data1-v2.docx
-
Figure 8—figure supplement 1—source data 2
Translated amino acid sequence of Nematostella vectensis polycystin 1-like, in which conserved domains are highlighted in light blue for transmembrane-spanning regions and in purple for other domains.
- https://cdn.elifesciences.org/articles/74336/elife-74336-fig8-figsupp1-data2-v2.docx
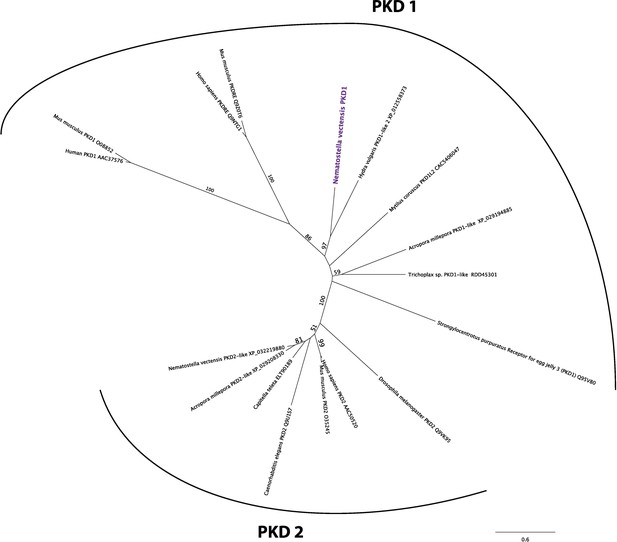
Nematostella vectensis polycystin 1-like belongs to the Polycystin 1/PKD1 group.
Unrooted maximum likelihood phylogeny of Polycystin protein families based on the alignment of Polycystin 1 (PKD1) and Polycystin 2 (PKD2) protein sequences that span the conserved TOP and transmembrane domains over 323 amino acid sites. NCBI accession numbers are shown with the name of each sequence. Bootstrap support values are shown at each node except when lower than 50%. The unit of the branch length is the number of substitutions per site. Note the strong support for the placement of N. vectensis polycystin 1-like (N. vectensis PKD1, highlighted in purple) within the PKD1 group, and for that of N. vectensis PKD2-like within the PKD2 group. The JGI ID for N. vectensis PKD2 (polycystin 2)-like is 160849.
-
Figure 8—figure supplement 2—source data 1
Alignment of polycystin 1 and polycystin 2 amino acid sequences used to construct phylogeny shown in Figure 8—figure supplement 2.
- https://cdn.elifesciences.org/articles/74336/elife-74336-fig8-figsupp2-data1-v2.docx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Nematostella vectensis) | F1 pou-iv +/- | This paper | Nv F1 pou-iv +/-, this paper | Maintained in N. Nakanishi Lab, University of Arkansas |
| Antibody | Anti- Nematostella vectensis POU-IV/Brn-3; rabbit; polyclonal | This paper | RRID:AB_2895562 | IF (1:200); stored in N. Nakanishi Lab, University of Arkansas |
Additional files
-
Supplementary file 1
List of 4188 candidate POU-IV downstream target genes.
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp1-v2.xlsx
-
Supplementary file 2
List of 577 genes significantly downregulated in NvPOU4 mutants relative to their siblings.
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp2-v2.xlsx
-
Supplementary file 3
List of 657 genes significantly upregulated in NvPOU4 mutants relative to their siblings.
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp3-v2.xlsx
-
Supplementary file 4
List of Gene Ontology terms overrepresented in genes downregulated in NvPOU4 mutants relative to their siblings.
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp4-v2.xlsx
-
Supplementary file 5
List of Gene Ontology terms overrepresented in genes upregulated in NvPOU4 mutants relative to their siblings.
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp5-v2.xlsx
-
Supplementary file 6
List of 293 POU-IV-activated genes.
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp6-v2.xlsx
-
Supplementary file 7
List of 178 POU-IV-repressed genes.
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp7-v2.xlsx
-
Supplementary file 8
List of Gene Ontology terms overrepresented in POU-IV-activated genes.
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp8-v2.xlsx
-
Supplementary file 9
List of POU-IV downstream target genes represented in the adult metacell c79 (hair cell).
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp9-v2.xlsx
-
Supplementary file 10
List of Gene Ontology terms overrepresented in POU-IV-activated genes represented in the adult metacell c79 (hair cell).
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp10-v2.xlsx
-
Supplementary file 11
List of POU-IV downstream target genes represented in the cnidocyte metacell c8.
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp11-v2.xlsx
-
Supplementary file 12
List of Gene Ontology terms overrepresented in POU-IV-repressed genes represented in the cnidocyte metacell c8.
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp12-v2.xlsx
-
Supplementary file 13
Lists of POU-IV downstream target genes represented in POU-IV-positive adult metacells c63, c64, c65, c66, c75, c76, c100, c101, and c102.
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp13-v2.xlsx
-
Supplementary file 14
List of GPCR-encoding genes in the adult metacell c79 (hair cell).
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp14-v2.xlsx
-
Supplementary file 15
List of gene-specific primer sequences used to amplify polycystin 1 cDNA.
- https://cdn.elifesciences.org/articles/74336/elife-74336-supp15-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/74336/elife-74336-transrepform1-v2.docx





