Phosphoregulation accommodates Type III secretion and assembly of a tether of ER-Chlamydia inclusion membrane contact sites
Figures
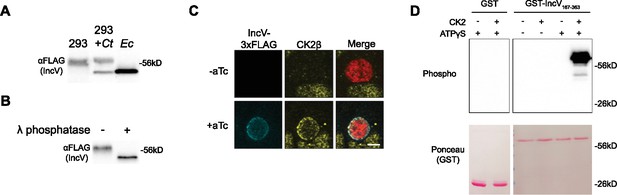
CK2 localizes to the inclusion and phosphorylates IncV.
(A) Western blot of IncV-3xFLAG from lysates of HEK293 cells expressing IncV-3xFLAG (293), HEK293 cells infected with C. trachomatis expressing IncV-3xFLAG (293+Ct), or E. coli expressing IncV-3xFLAG (Ec). (B) Western blot of IncV-3xFLAG purified from lysates of HEK293 cells infected with C. trachomatis expressing IncV-3xFLAG and treated with lambda (λ) phosphatase (+) or phosphatase buffer alone (−). (C) Three-dimensional reconstruction of confocal images of HeLa cells infected with C. trachomatis expressing mCherry constitutively (red) and IncV-3xFLAG (blue) under the control of an anhydrotetracycline (aTc)-inducible promoter in the absence (−aTc) or presence (+aTc) of aTc and stained to detect endogenous CK2β (yellow). The merge is shown on the right. Scale bar is 5 μm. (D) In vitro kinase assay using GST or GST-IncV167-363 purified from E. coli as a substrate in the presence (+) or absence (−) of recombinant CK2 and in the presence (+) or absence (−) of ATPγS. The top panel shows phosphorylated proteins detected with anti-thiophosphate antibodies and the bottom panel is the same membrane stained with Ponceau S to detect total proteins.
-
Figure 1—source data 1
Uncropped, labeled blots for Figure 1A.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig1-data1-v1.psd
-
Figure 1—source data 2
Uncropped, labeled blots for Figure 1B.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig1-data2-v1.psd
-
Figure 1—source data 3
Uncropped, labelled blots for Figure 1D.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig1-data3-v1.psd
-
Figure 1—source data 4
Raw data for FLAG blot in Figure 1A.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig1-data4-v1.tif
-
Figure 1—source data 5
Raw data for FLAG blot in Figure 1B.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig1-data5-v1.tif
-
Figure 1—source data 6
Raw data for thiophosphate blot 1 in Figure 1D.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig1-data6-v1.tif
-
Figure 1—source data 7
Raw data for thiophosphate blot 2 in Figure 1D.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig1-data7-v1.tif
-
Figure 1—source data 8
Raw data for Ponceau S blot 1 in Figure 1D.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig1-data8-v1.tif
-
Figure 1—source data 9
Raw data for Ponceau S blot 2 in Figure 1D.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig1-data9-v1.tif
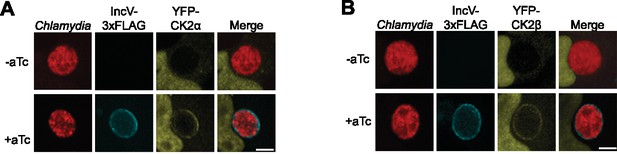
IncV recruits CK2 to the inclusion membrane.
Three-dimensional reconstruction of confocal images of HeLa cells overexpressing YFP-CK2α (A) or YFP-CK2β (B) (yellow) and infected with C. trachomatis expressing mCherry constitutively (red) and IncV-3xFLAG (blue) under the control of an anhydrotetracycline (aTc)-inducible promoter in the absence (−aTc) or presence (+aTc) of aTc. The merge is shown on the right. Scale bar is 5 μm.
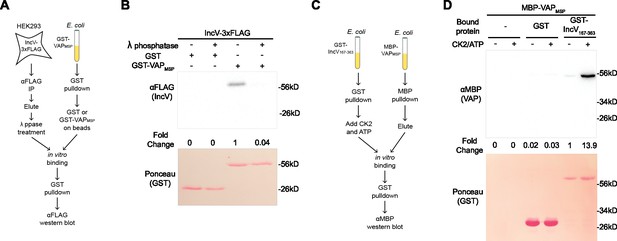
Phosphorylation of IncV is necessary and sufficient to promote the IncV–VAP interaction in vitro.
(A) Schematic depicting the experimental setup for results in B. (B) In vitro binding assay using IncV-3xFLAG purified from HEK293 lysates and treated with lambda (λ) phosphatase (+) or phosphatase buffer alone (−) combined with GST or GST-VAPMSP purified from E. coli and immobilized on glutathione beads. The top panel shows proteins detected with anti-FLAG antibodies and the bottom panel is the same membrane stained with Ponceau S to detect total protein. (C) Schematic depicting the experimental setup for results in D. (D) In vitro binding assay using GST or GST-IncV167–363 purified from E. coli, and immobilized on glutathione beads, as a substrate for CK2 in the presence (+) or absence (−) of CK2 and ATP, combined with MBP-VAPMSP purified from E. coli. The top panel was probed with anti-MBP and the bottom panel was the same membrane stained with Ponceau S to detect the GST construct.
-
Figure 2—source data 1
Quantification of blot densities for Figure 2.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Uncropped, labeled blots for Figure 2B.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig2-data2-v1.psd
-
Figure 2—source data 3
Uncropped, labelled blots for Figure 2D.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig2-data3-v1.psd
-
Figure 2—source data 4
Raw data for FLAG blot in Figure 2B.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig2-data4-v1.tif
-
Figure 2—source data 5
Raw data for Ponceau S blot in Figure 2B.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig2-data5-v1.tif
-
Figure 2—source data 6
Raw data for MBP blot in Figure 2D.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig2-data6-v1.tif
-
Figure 2—source data 7
Raw data for Ponceau S blot in Figure 2D.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig2-data7-v1.tif
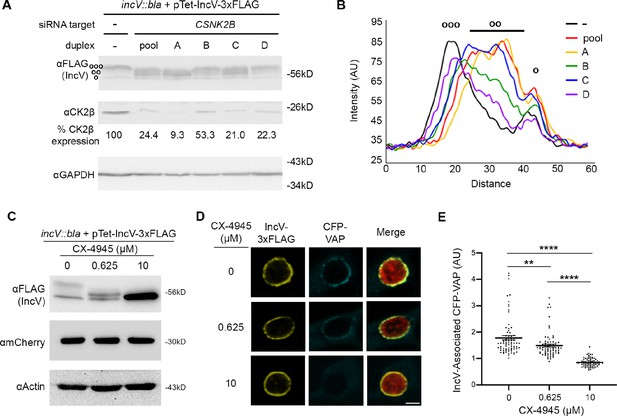
CK2 plays a role in the IncV–VAP interaction during infection.
(A) Western blot of lysates of HeLa cells treated with siRNA buffer alone (−) or with siRNA duplexes targeting CSNK2B (pool of four duplexes or individual duplexes A, B, C, or D) and infected with a C. trachomatis incV mutant expressing IncV-3xFLAG. The top panel was probed with anti-FLAG. The middle panel was probed with anti-CK2β. The bottom panel was probed with anti-GAPDH. Relative expression levels of CK2β normalized to GAPDH loading controls are shown as a percentage of no siRNA control expression. (ooo) hyperphosphorylated IncV, (oo) intermediate hypophosphorylated IncV, and (o) unphosphorylated IncV. (B) Line scan analysis of FLAG signal detected in A. The peak on the left (ooo) corresponds to the hyperphosphorylated species of IncV, and the peak on the right (o) corresponds to the unphosphorylated species of IncV. Intermediate hypophosphorylated species are indicated by any peak between the left and right peaks (oo). Each line represents a different condition: Control, black; siRNA pool of duplexes A–D, red; siRNA duplex A, yellow; siRNA duplex B, green; siRNA duplex C, blue; siRNA duplex D, purple. (C–E) HeLa cells, expressing CFP-VAP (D and E only), were infected with C. trachomatis incV mutant expressing IncV-3xFLAG under the control of the anhydrotetracycline (aTc)-inducible promoter and treated with increasing concentrations of the CK2 inhibitor CX-4945 (0, 0.625, 10 μM) for 2 hr at 18 h pi and prior to the induction of IncV-3xFLAG expression at 20 h pi. The samples were processed 24 h pi for western blot (C) or confocal microscopy (D, E). (C) Cell lysates were probed with anti-FLAG (top blot), anti-mCherry (middle blot), or anti-actin (bottom blot) antibodies. (D) Single plane confocal micrographs of HeLa cells expressing CFP-VAP (blue), infected with incV mutant expressing IncV-3xFLAG (yellow) and mCherry (red). The merge is shown on the right. Scale bar is 5 μm. (E) Quantification of the mean intensity of the CFP-VAP signal within an object generated from the IncV-3xFLAG signal and normalized to the mean intensity of CFP-VAP in the ER. Each dot represents one inclusion. Data show the mean and SEM of a combination of three independent experiments. One-way ANOVA and Tukey’s post hoc test were performed. **p < 0.01, ****p < 0.0001.
-
Figure 3—source data 1
Quantification of blot densities, line scan analysis, and IncV-associated CFP-VAP for Figure 3.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Uncropped, labeled blots for Figure 3A.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-data2-v1.psd
-
Figure 3—source data 3
Uncropped, labelled blots for Figure 3C.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-data3-v1.psd
-
Figure 3—source data 4
Raw data for FLAG blot in Figure 3A.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-data4-v1.tif
-
Figure 3—source data 5
Raw data for CK2 blot in Figure 3A.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-data5-v1.tif
-
Figure 3—source data 6
Raw data for GAPDH blot in Figure 3A.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-data6-v1.tif
-
Figure 3—source data 7
Raw data for FLAG blot in Figure 3C.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-data7-v1.tif
-
Figure 3—source data 8
Raw data for mCherry blot in Figure 3C.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-data8-v1.tif
-
Figure 3—source data 9
Raw data for Actin blot in Figure 3C.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-data9-v1.tif
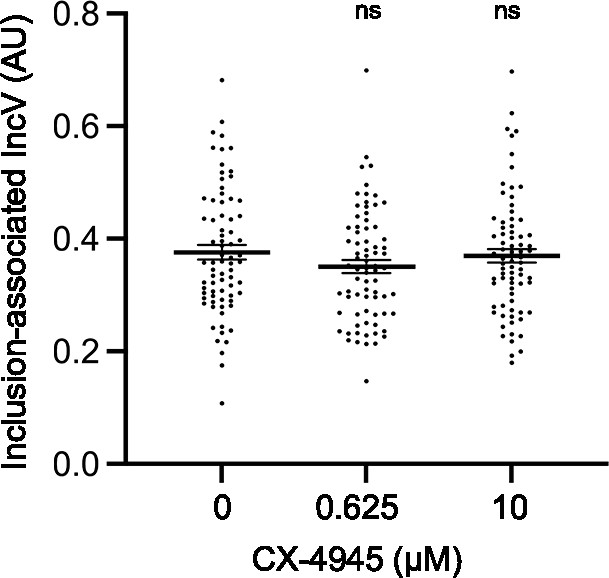
IncV inclusion localization is not affected upon CX-4945 treatment.
Quantification of the volume of the indicated IncV-3xFLAG signal associated with the inclusion normalized to the volume of an object generated from the mCherry signal of the bacteria. Each dot represents one inclusion. The mean and SEM are shown. One-way ANOVA and Tukey’s post hoc test were performed comparing drug treated cells to control cells. ns: nonsignificant.
-
Figure 3—figure supplement 1—source data 1
Quantification of inclusion-associated IncV for Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-figsupp1-data1-v1.xlsx
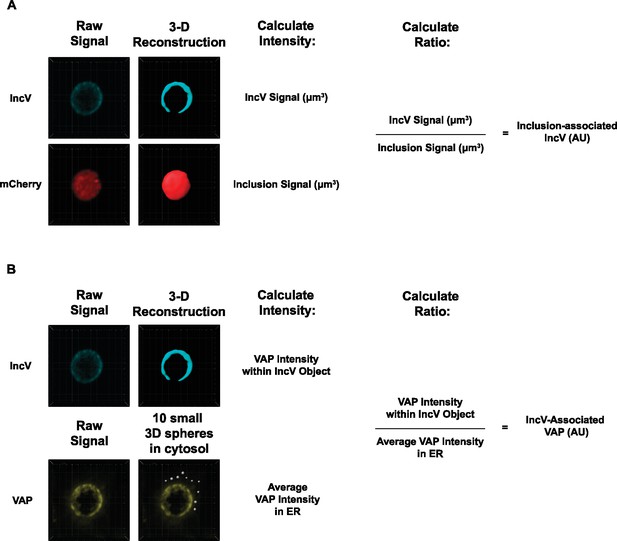
Method to quantify the inclusion association of a given marker.
(A) Three-dimensional (3D) objects were reconstructed from the raw signal corresponding to the IncV (blue) and the mCherry-positive inclusion (red) signals. The ratio of the IncV signal over the inclusion signal was calculated to determine IncV association with the inclusion in arbitrary units [AU]. (B) The signal intensity of a given marker (VAP in yellow is shown here) within the reconstituted 3D IncV object (blue), and within the cytosol (determined by the average of 10 small 3D spheres throughout the ER), was determined. The IncV association of the given marker was determined by normalizing the signal intensity of the marker of interest within the IncV 3D object to the average signal intensity of the marker of interest within the ER (VAP) or cytosol (CK2).
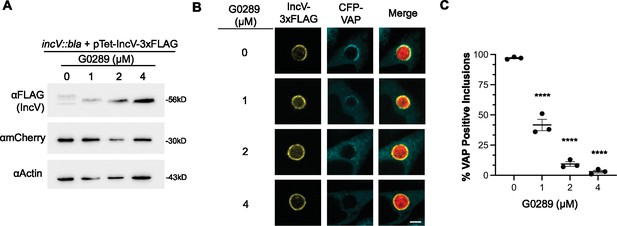
The CK2 kinase inhibitor GO289 inhibits IncV phosphorylation and VAP recruitment to the inclusion.
(A) Cell lysates of HeLa cells infected for 24 hr with a C. trachomatis incV mutant expressing IncV-3xFLAG under the control of the anhydrotetracycline (aTc)-inducible promoter and treated with increasing concentrations of the CK2 inhibitor G0289 (0, 1, 2, and 4 μM) for 2 hr at 18 h pi and prior to the aTc induction of IncV-3xFLAG expression at 20 h pi, probed with anti-FLAG (top blot), anti-mCherry (middle blot), or anti-actin (bottom blot) antibodies. (B) Single plane confocal micrographs of HeLa cells expressing CFP-VAP (blue), infected with incV mutant expressing IncV-3xFLAG (yellow) and mCherry (red) and treated with GO289 as described in A. The merge is shown on the right. Scale bar is 5 μm. (C) Quantification of the percentage on inclusions exhibiting VAP recruitment to the inclusion. Data show the mean and SEM of three independent experiments. One-way ANOVA and Tukey’s post hoc test were performed. ****p < 0.0001.
-
Figure 3—figure supplement 3—source data 1
Uncropped, labelled blots for Figure 3—figure supplement 3A.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-figsupp3-data1-v1.psd
-
Figure 3—figure supplement 3—source data 2
Raw data for FLAG blot in Figure 3—figure supplement 3A.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-figsupp3-data2-v1.tif
-
Figure 3—figure supplement 3—source data 3
Raw data for mCherry blot in Figure 3—figure supplement 3A.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-figsupp3-data3-v1.tif
-
Figure 3—figure supplement 3—source data 4
Raw data for Actin blot in Figure 3—figure supplement 3A.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-figsupp3-data4-v1.tif
-
Figure 3—figure supplement 3—source data 5
Quantification of percentage of CFP-VAP-positive inclusions in Figure 3—figure supplement 3C.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig3-figsupp3-data5-v1.xlsx
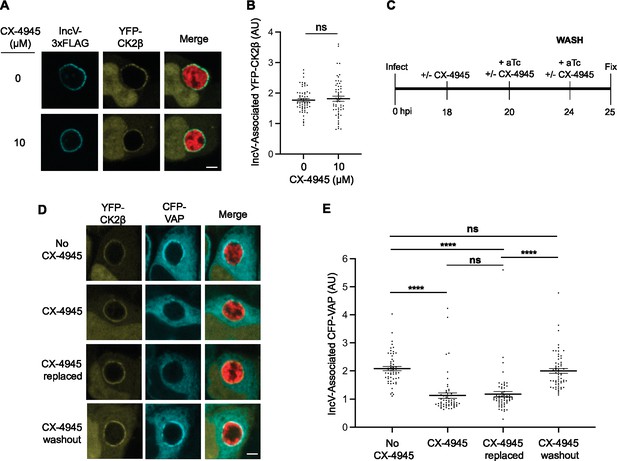
CK2 kinase activity is dispensable for CK2 localization to the inclusion and reversing CK2 inhibition allows for VAP recruitment.
(A) Single plane confocal micrographs of HeLa cells expressing YFP-CK2β (yellow), infected with an incV mutant expressing IncV-3xFLAG (blue) and mCherry (red), and treated with 10 μM CX-4945 (bottom panels), or not (top panels), for 2 hr at 18 h pi and prior to the induction of IncV-3xFLAG expression at 20 h pi. The merge is shown on the right. Scale bar is 5 μm. (B) Quantification of the mean intensity of the YFP-CK2β signal within an object generated from the IncV-3xFLAG signal and normalized to the mean intensity of YFP-CK2β in the cytosol. Each dot represents one inclusion. Data show the mean and SEM of a combination of three independent experiments. ns: nonsignificant (Student’s t-test). (C) Schematic depicting the experimental setup for the results in D and E. (D) Single plane confocal micrographs of HeLa cells expressing CFP-VAP (blue) and YFP-CK2β (yellow), infected with an incV mutant expressing IncV-3xFLAG under the control of the anhydrotetracycline (aTc)-inducible promoter and mCherry (red). Infections were performed in the absence of CX-4945 (No CX-4945) or presence of 10 μM CX-4945 for the duration of the experiment (CX-4945). Alternatively, CX-4945 was present until 24 h pi, when the cells were washed and incubated with media containing either aTc and CX-4945 (CX-4945 replaced) or aTc only (CX-4945 washout) for an additional hour. The merge is shown on the right. Scale bar is 5 μm. (E) Quantification of the mean intensity of the CFP-VAP signal within an object generated from the YFP-CK2β signal and normalized to the mean intensity of CFP-VAP in the ER. Each dot represents one inclusion. Data show the mean and SEM of a combination of three independent experiments. One-way ANOVA and Tukey’s post hoc test were performed. ****p < 0.0001.
-
Figure 4—source data 1
Quantification of IncV-associated YFP-CK2β and IncV-associated CFP-VAP for Figure 4.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig4-data1-v1.xlsx
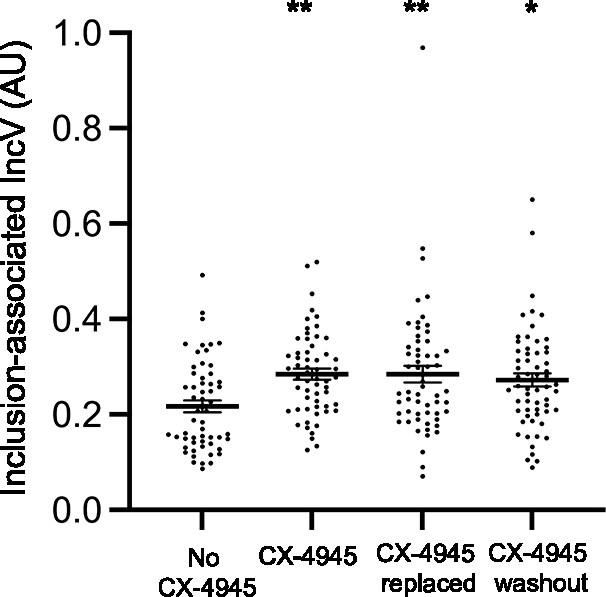
IncV inclusion localization is not affected upon CX-4945 replacement/washout treatment.
Quantification of the volume of the indicated IncV-3xFLAG signal associated with the inclusion normalized to the volume of an object generated from the mCherry signal of the bacteria. Each dot represents one inclusion. The mean and SEM are shown. One-way ANOVA and Tukey’s post hoc test were performed comparing drug treated cells to control cells. *p < 0.05, **p < 0.01.
-
Figure 4—figure supplement 1—source data 1
Quantification of inclusion-associated IncV for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig4-figsupp1-data1-v1.xlsx
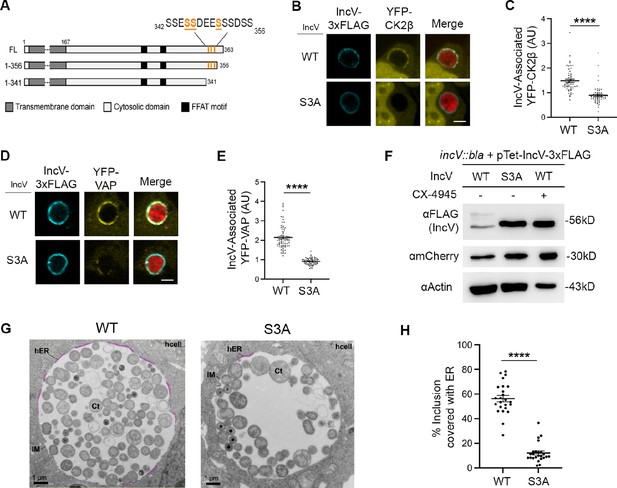
Three serine residues in a C-terminal domain of IncV mediate CK2 and VAP recruitment to the inclusion, IncV phosphorylation, and ER-Inclusion membrane contact site (MCS) formation.
(A) Schematic depicting truncated IncV constructs. The numbers indicate the amino acid position within the IncV protein sequence. CK2 phosphorylation sites that do not require priming are indicated in orange. Single plane confocal images of HeLa cells expressing YFP-CK2β (B) or YFP-VAP (D) (yellow), infected with a C. trachomatis incV mutant expressing mCherry (red) and IncVWT- (WT) or IncVS345A/S346A/S350A-3xFLAG (S3A) (blue). The merge is shown on the right. Scale bar is 5 μm. Quantification of the mean intensity of YFP-CK2β (C) and YFP-VAP (E) within the IncV object normalized to the mean intensity of YFP-CK2β in the cytosol and YFP-VAP in the ER, respectively. Data show the mean and SEM of a combination of three independent experiments. ****p < 0.0001 (Student’s t-test). (F) Western blot of lysates of HeLa cells infected with a C. trachomatis incV mutant expressing IncVWT-3xFLAG (WT), IncVS3A-3xFLAG (S3A), or expressing IncVWT-3xFLAG and treated with 10 μM CX-4945 as described in Figure 3C (WT; CX-4945 +) and probed with anti-FLAG (top blot), anti-mCherry (middle blot), and anti-actin (bottom blot) antibodies. (G) Transmission electron micrographs of sections of HeLa cells infected with a C. trachomatis incV mutant expressing IncVWT- (WT) or IncVS3A-3xFLAG (S3A). Representative sections across a whole inclusion are shown. ER-Inclusion MCS are highlighted in pink. Corresponding images without the highlighted MCS are shown in Figure 5—figure supplement 3C, D. Ct: Chlamydia trachomatis; IM: inclusion membrane; hER: host endoplasmic reticulum; hcell: host cell cytosol. Scale bars are 1 µm. (H) Quantification of the percentage of the inclusion membrane covered with host ER. Each dot represents one section across a whole inclusion. Data show the mean and SEM for 24 representative electron micrographs per condition. ****p < 0.0001 (Student’s t-test).
-
Figure 5—source data 1
Quantification of IncV-associated YFP-CK2β and IncV-associated CFP-VAP for Figure 5.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig5-data1-v1.xlsx
-
Figure 5—source data 2
Uncropped, labeled blots for Figure 5F.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig5-data2-v1.psd
-
Figure 5—source data 3
Raw data for FLAG and mCherry blots in Figure 5F.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig5-data3-v1.tif
-
Figure 5—source data 4
Raw data for Actin blot in Figure 5F.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig5-data4-v1.tif
-
Figure 5—source data 5
Quantification of percentage of inclusion membrane associated with host ER for Figure 5H.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig5-data5-v1.xlsx
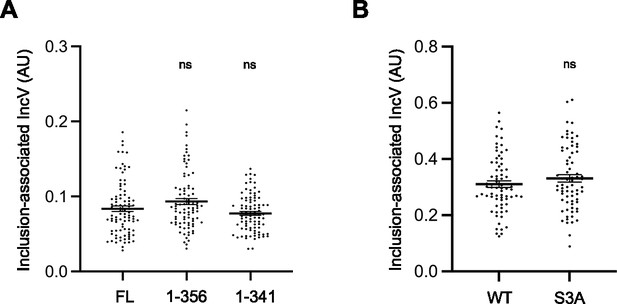
Truncation and alanine substitution of S245, S346, and S350 does not affect IncV localization to the inclusion membrane.
Quantification of the volume of the indicated IncV-3xFLAG signal associated with the inclusion normalized to the volume of an object generated from the mCherry signal of the bacteria. Each dot represents one inclusion. The mean and SEM are shown. One-way ANOVA and Tukey’s post hoc test (A) or Student’s t-test (B) were performed comparing truncations to full length (A), and alanine substitution mutant to wild-type (B). ns: nonsignificant.
-
Figure 5—figure supplement 1—source data 1
Quantification of inclusion-associated IncV for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig5-figsupp1-data1-v1.xlsx
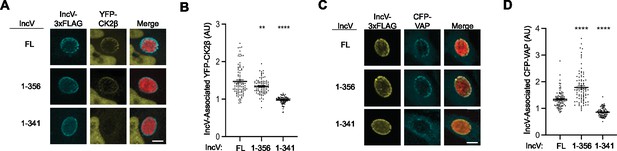
A C-terminal domain of IncV mediates VAP recruitment to the inclusion.
(A) Three-dimensional reconstruction of confocal images of HeLa cells expressing YFP-CK2β (yellow) and infected with a C. trachomatis incV mutant expressing mCherry constitutively (red) and IncV-3xFLAG full length (FL) or truncated (1–356, or 1–341) (blue). The merge is shown on the right. Scale bar is 5 μm. (B) Quantification of the mean intensity of YFP-CK2β within an object generated from the IncV-3xFLAG signal and normalized to the mean intensity of YFP-CK2β in the cytosol. Each dot represents one inclusion. Data show the mean and SEM of a combination of three independent experiments. One-way ANOVA and Tukey’s post hoc test were performed comparing truncations to full length. **p < 0.01, ****p < 0.0001. (C) Three-dimensional reconstruction of confocal images of HeLa cells expressing CFP-VAP (blue) and infected with a C. trachomatis incV mutant expressing mCherry constitutively (red) and IncV-3xFLAG full length (FL) or truncated (1–356 or 1–341) (yellow). The merge is shown on the right. Scale bar is 5 μm. (D) Quantification of the mean intensity of CFP-VAP within an object generated from the IncV-3xFLAG signal and normalized to the mean intensity of CFP-VAP in the cytosol. Each dot represents one inclusion. Data show the mean and SEM of a combination of three independent experiments. One-way ANOVA and Tukey’s post hoc test were performed comparing truncations to full length. ****p < 0.0001.
-
Figure 5—figure supplement 2—source data 1
Quantification of IncV-associated YFP-CK2β and IncV-associated CFP-VAP for Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig5-figsupp2-data1-v1.zip
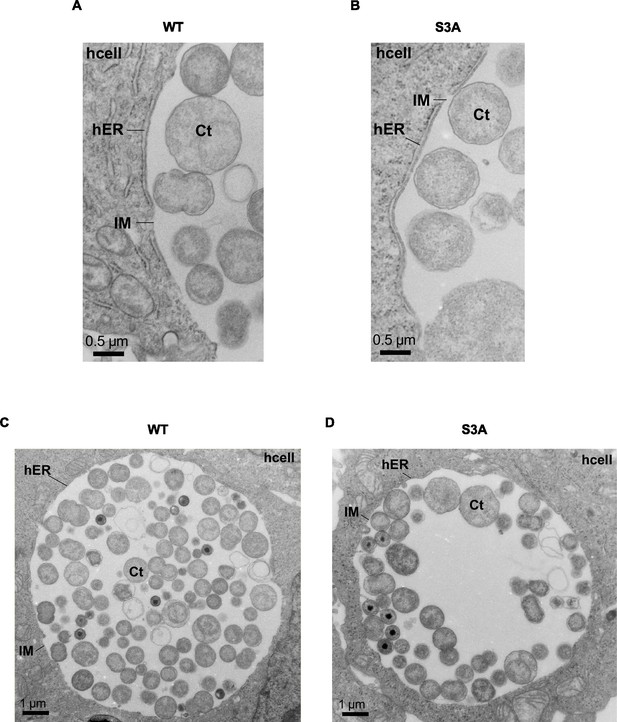
Ultrastructural analysis of ER-Inclusion membrane contact site (MCS).
(A, B) High magnification transmission electron micrographs of sections of MCS between the ER and the inclusion of C. trachomatis incV mutant expressing IncVWT- (WT) or IncV-S3A3xFLAG (S3A). Scale bar, 0.5 µm. (C, D) Transmission electron micrographs of sections of HeLa cells infected with a C. trachomatis incV mutant expressing IncVWT- (WT) or IncVS3A-3xFLAG (S3A) as shown in Figure 5G but without the mask highlighting the ER-Inclusion MCS. Section across a whole inclusion is shown. Scale bar is 1 µm. Ct: Chlamydia trachomatis; IM: inclusion membrane; hER: host endoplasmic reticulum; hcell: host cell cytosol.
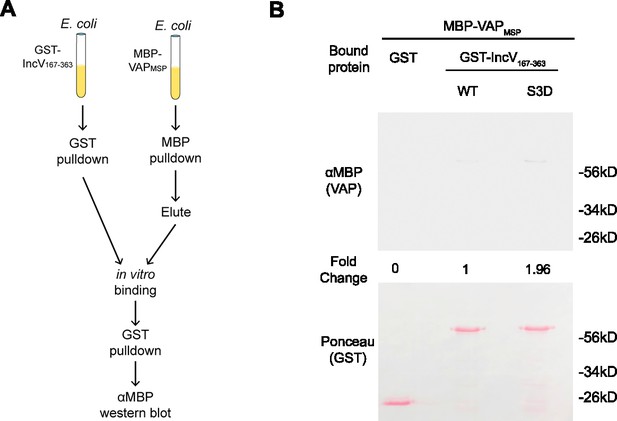
Phosphomimetic mutation of three serine residues in the C-terminal domain of IncV is not sufficient to promote the IncV–VAP interaction in vitro.
(A) Schematic depicting the experimental setup for results in B. (B) In vitro binding assay using GST, GST-IncVWT, or GST-IncVS3D purified from E. coli, immobilized on glutathione beads, and combined with MBP-VAP purified from E. coli. The top panel was probed with anti-MBP and the bottom panel was the same membrane stained with Ponceau S to detect the GST construct. Note that the IncV and VAP constructs, only include the cytosolic domain of IncV (aa 167–363) and the MSP domain of VAP, respectively.
-
Figure 5—figure supplement 4—source data 1
Quantification of blot densities for Figure 5—figure supplement 4B.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig5-figsupp4-data1-v1.xlsx
-
Figure 5—figure supplement 4—source data 2
Uncropped, labeled blots for Figure 5—figure supplement 4B.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig5-figsupp4-data2-v1.psd
-
Figure 5—figure supplement 4—source data 3
Raw data for MBP blot in Figure 5—figure supplement 4B.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig5-figsupp4-data3-v1.tif
-
Figure 5—figure supplement 4—source data 4
Raw data for Ponceau S blot in Figure 5—figure supplement 4B.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig5-figsupp4-data4-v1.tif
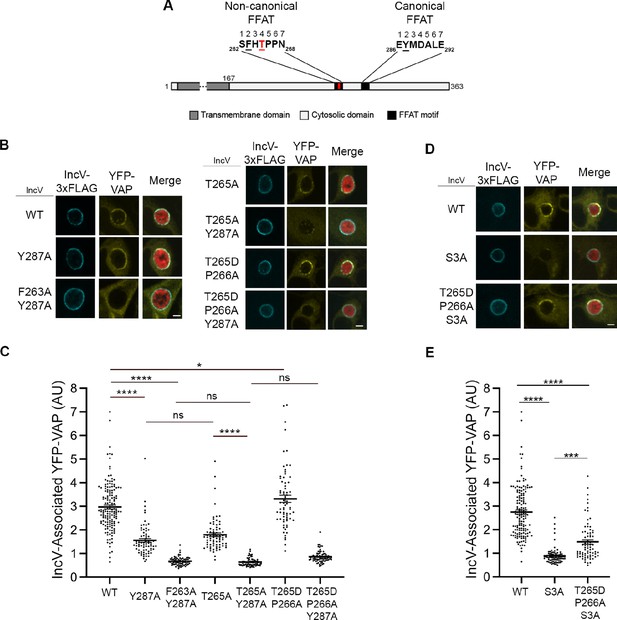
Phosphorylation of T265 in the noncanonical FFAT motif of IncV contributes to the IncV–VAP interaction.
(A) Schematic depicting the IncV protein. The transmembrane domain, the cytosolic domain, and the noncanonical and the canonical FFAT motif cores are indicated in dark gray, light gray, and black, respectively. The amino acid sequence of the FFAT motif cores is shown. Numbers 1–7 indicate the amino acid position within the FFAT motif cores, other numbers indicate the amino acid position within the IncV protein sequence. Residues at position 2 of the FFAT motif cores are in black and underlined. Threonine 265 at position 4 of the noncanonical FFAT is in red and underlined. (B) Single plane confocal images of HeLa cells expressing YFP-VAP (yellow), infected with a C. trachomatis incV mutant expressing mCherry constitutively (red) and IncVWT- (WT), IncVY287A- (Y287A), IncVF263A/Y287A (F263A/Y287A), IncVT265A- (T265A), IncVT265A/Y287A- (T265A/Y287A), IncVT265D/P266A- (T265D/P266A), or IncVT265D/P266A/Y287A-3xFLAG (T265D/P266A/Y287A) (blue). The merge is shown on the right. Scale bar is 5 μm. (C, E) Quantification of the mean intensity of YFP-VAP within an object generated from the IncV-3xFLAG signal corresponding to the indicated IncV constructs, and normalized to the mean intensity of YFP-VAP in the ER. Each dot represents one inclusion. Data show the mean and SEM of three to six independent experiments. One-way ANOVA and Tukey’s post hoc test were performed. ns: nonsignificant, *p < 0.05, ***p < 0.001, ****p < 0.0001. (D) Single plane confocal images of HeLa cells expressing YFP-VAP (yellow), infected with a C. trachomatis incV mutant expressing mCherry constitutively (red) and IncVWT- (WT), IncVS3A- (S3A), or IncVT265D/P266A/S3A-3xFLAG (T265D/P266A/S3A) (blue). The merge is shown on the right. Scale bar is 5 μm.
-
Figure 6—source data 1
Quantification of IncV-associated YFP-VAP for Figure 6.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig6-data1-v1.xlsx
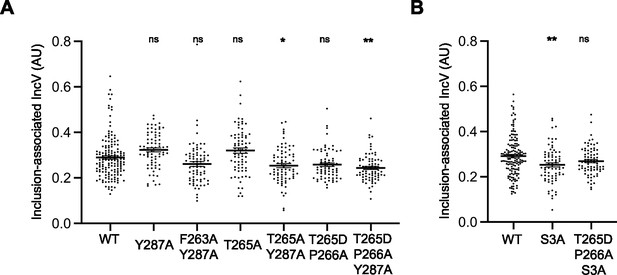
Inclusion localization of IncV variants with amino acid substitutions in the canonical and/or noncanonical FFAT motifs.
(A, B) Quantification of the volume of the indicated IncV-3xFLAG signal associated with the inclusion normalized to the volume of an object generated from the mCherry signal of the bacteria. Each dot represents one inclusion. The mean and SEM are shown. One-way ANOVA and Tukey’s post hoc test were performed comparing alanine or aspartic acid substitution to wild-type. ns: nonsignificant, *p < 0.05, **p < 0.01.
-
Figure 6—figure supplement 1—source data 1
Quantification of inclusion-associated IncV for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig6-figsupp1-data1-v1.xlsx
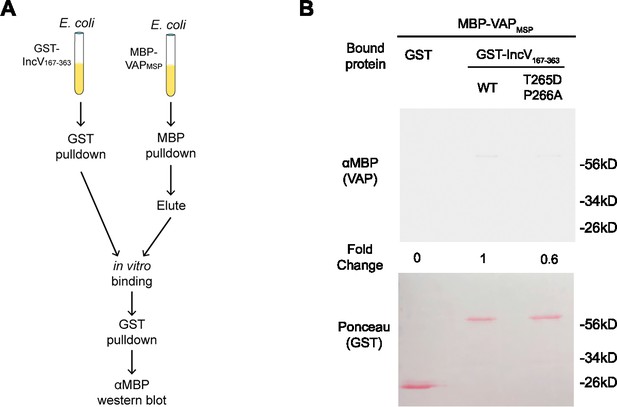
Phosphomimetic mutation of T265 in the IncV noncanonical FFAT is not sufficient to promote the IncV–VAP interaction in vitro.
(A) Schematic depicting the experimental setup for results in B. (B) In vitro binding assay using GST, GST-IncVWT, or GST-IncVT265D/P266A purified from E. coli, immobilized on glutathione beads, and combined with MBP-VAP purified from E. coli. The top panel was probed with anti-MBP and the bottom panel was the same membrane stained with Ponceau S to detect the GST construct. Note that the IncV and VAP constructs, only include the cytosolic domain of IncV (aa 167–363) and the MSP domain of VAP, respectively.
-
Figure 6—figure supplement 2—source data 1
Quantification of blot densities for Figure 6—figure supplement 2B.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig6-figsupp2-data1-v1.xlsx
-
Figure 6—figure supplement 2—source data 2
Uncropped, labeled blots for Figure 6—figure supplement 2B.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig6-figsupp2-data2-v1.psd
-
Figure 6—figure supplement 2—source data 3
Raw data for MBP blot in Figure 6—figure supplement 2B.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig6-figsupp2-data3-v1.tif
-
Figure 6—figure supplement 2—source data 4
Raw data for Ponceau S blot in Figure 6—figure supplement 2B.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig6-figsupp2-data4-v1.tif
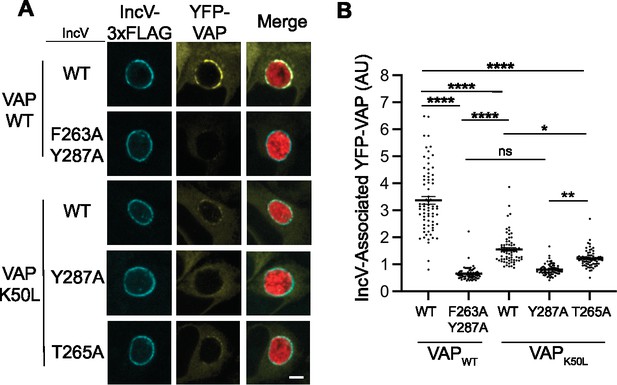
Recruitment of VAPWT and VAPK50L to C. trachomatis inclusions displaying wild-type or mutated IncV.
(A) Single plane confocal images of HeLa cells expressing YFP-VAPWT (VAPWT) or -VAPK50L (VAPK50L) (yellow), infected with a C. trachomatis incV mutant expressing mCherry constitutively (red) and IncVWT- (WT), IncVF263A/Y287A- (F263A/Y287A), IncVY287A- (Y287A), or IncVT265A-3xFLAG (T265A) (blue). The merge is shown on the right. Scale bar is 5 μm. (B) Quantification of the mean intensity of YFP-VAP within an object generated from the IncV-3xFLAG signal corresponding to the indicated IncV constructs and normalized to the mean intensity of YFP-VAP in the ER. Each dot represents one inclusion. Data show the mean and SEM of three independent experiments. One-way ANOVA and Tukey’s post hoc test were performed. ns: nonsignificant, *p < 0.05, **p < 0.01, ****p < 0.0001.
-
Figure 6—figure supplement 3—source data 1
Quantification of IncV-associated YFP-VAP for Figure 6—figure supplement 3.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig6-figsupp3-data1-v1.xlsx
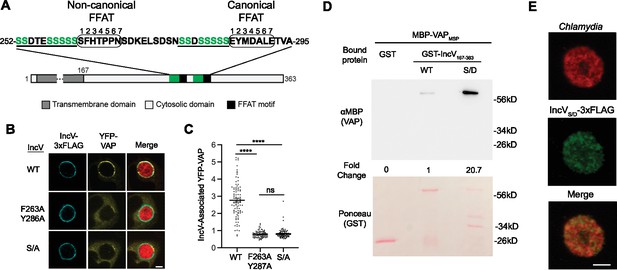
Phosphorylation of the serine tracts upstream of the IncV FFAT motifs facilitates the IncV–VAP interaction.
(A) Schematic depicting the IncV protein. The transmembrane domain, the cytosolic domain, and the noncanonical and canonical FFAT motif cores are indicated in dark gray, light gray, and black, respectively. The amino acid sequence of the FFAT motif cores (circled) and their respective upstream sequence is shown. The serine-rich tracts are underlined. Serine residues are in green. Numbers 1–7 indicate the amino acid position within the FFAT motif cores, other numbers indicate the amino acid position within the IncV protein sequence. (B) Single plane confocal images of HeLa cells expressing YFP-VAP (yellow), infected with a C. trachomatis incV mutant expressing mCherry constitutively (red) and IncVWT-3xFLAG (WT), IncVF263A/Y287A-3xFLAG (F263A/Y287A), or IncVS/A-3xFLAG (S/A) (blue). The merge is shown on the right. Scale bar is 5 μm. (C) Quantification of the mean intensity of YFP-VAP within an object generated from the IncV-3xFLAG signal and normalized to the mean intensity of YFP-VAP in the ER. Each dot represents one inclusion. Data show the mean and SEM of a combination of three independent experiments. One-way ANOVA with Tukey’s post hoc test was performed. ****p < 0.0001. (D) In vitro binding assay using GST, GST-IncVWT, or GST-IncVS/D purified from E. coli, and immobilized on glutathione beads and combined with MBP-VAP purified from E. coli. The top panel was probed with anti-MBP and the bottom panel was the same membrane stained with Ponceau S to detect the GST construct. Note that the IncV and VAP constructs, only include the cytosolic domain of IncV (aa 167–363) and the MSP domain of VAP, respectively. (E) Single plane confocal images of HeLa cells infected with a C. trachomatis incV mutant expressing mCherry consitutively (red) and IncVS/D-3xFLAG (green). The merge is shown on the bottom. Scale bar is 5 μm.
-
Figure 7—source data 1
Quantification of IncV-associated YFP-VAP and blot densities for Figure 7.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig7-data1-v1.xlsx
-
Figure 7—source data 2
Uncropped, labeled blots for Figure 7D.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig7-data2-v1.psd
-
Figure 7—source data 3
Raw data for MBP blot in Figure 7D.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig7-data3-v1.tif
-
Figure 7—source data 4
Raw data for Ponceau S blot in Figure 7D.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig7-data4-v1.tif
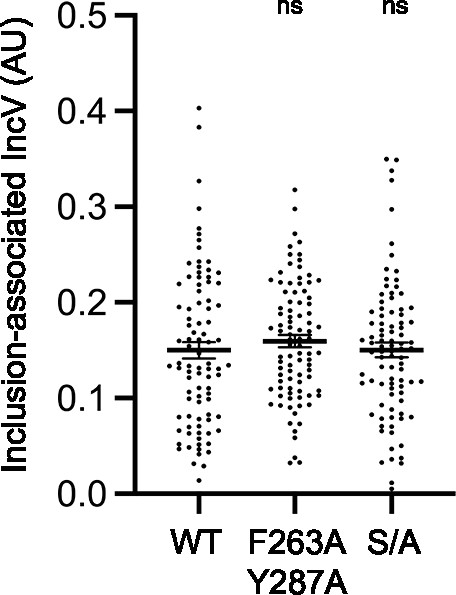
Alanine substitution of IncV serine tracts does not affect IncV inclusion localization.
Quantification of the volume of the indicated IncV-3xFLAG signal associated with the inclusion normalized to the volume of an object generated from the mCherry signal of the bacteria. Each dot represents one inclusion. The mean and SEM are shown. One-way ANOVA and Tukey’s post hoc test were performed comparing alanine substitution to wild-type. ns: nonsignificant.
-
Figure 7—figure supplement 1—source data 1
Quantification of inclusion-associated IncV for Figure 7—figure supplement 1.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig7-figsupp1-data1-v1.xlsx
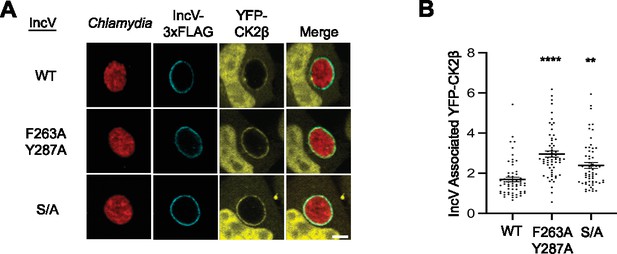
Alanine substitution of residues in position 2 of IncV FFAT motifs or of the serine-rich tracts upstream of IncV FFAT motifs does not affect IncV-dependent CK2 recruitment to the inclusion.
(A) Single plane confocal images of HeLa cells expressing YFP-CK2β (yellow) and infected with a C. trachomatis incV mutant expressing mCherry (red) and IncVWT-3xFLAG (WT), IncVF263A/Y287A-3xFLAG (F263A/Y287A), or IncVS/A-3xFLAG (S/A) (blue). The merge is shown on the right. Scale bar is 5 μm. (B) Quantification of the mean intensity of the YFP-CK2β within the IncV object normalized to the mean intensity of YFP-CK2β in the cytosol. Data show the mean and SEM of a combination of three independent experiments. One-way ANOVA with Tukey multiple comparisons test was performed to compare IncVF263A/Y287A and IncVS/A to IncVWT. **p < 0.01, ****p < 0.0001.
-
Figure 7—figure supplement 2—source data 1
Quantification of IncV-associated YFP-CK2β for Figure 7—figure supplement 2.
- https://cdn.elifesciences.org/articles/74535/elife-74535-fig7-figsupp2-data1-v1.xlsx
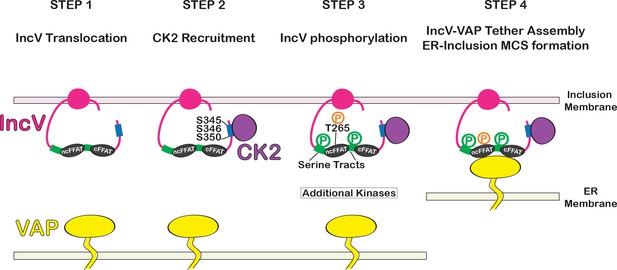
Model of assembly of the IncV–VAP tether at ER-Inclusion membrane contact site (MCS).
Step 1: After Type III-mediated translocation, unphosphorylated IncV (pink) is inserted into the inclusion membrane. Step 2: IncV recruits CK2 (purple) via three serine residues S345, S346, and S350 (blue) that are part of CK2 recognition motifs and located in a C-terminal domain of IncV. Step 3: IncV becomes hyperphosphorylated, including phosphorylation of the noncanonical FFAT on threonine residue T265 (orange) and the serine-rich tract (green) immediately upstream of FFAT core motifs (black). Additional kinases contribute to IncV phosphorylation. Step 4: IncV phosphorylation leads to full mimicry of FFAT motifs and binding to VAP (yellow) resulting in ER-Inclusion MCS formation. P: phosphorylated residues; ncFFAT: noncanonical FFAT; cFFAT: canonical FFAT. Light pink: inclusion membrane. Light yellow: ER membrane.
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/74535/elife-74535-transrepform1-v1.docx
-
Supplementary file 1
Primers and cloning strategies.
- https://cdn.elifesciences.org/articles/74535/elife-74535-supp1-v1.xlsx





