Cytotoxic CD4+ T cells driven by T-cell intrinsic IL-18R/MyD88 signaling predominantly infiltrate Trypanosoma cruzi-infected hearts
Figures
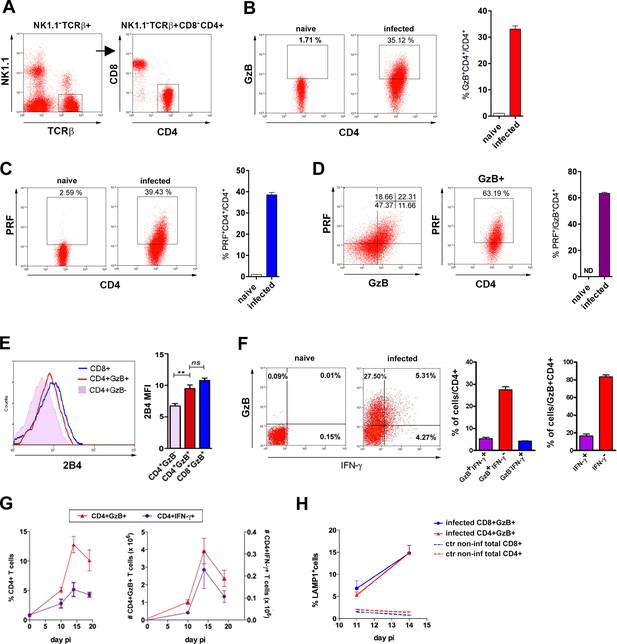
CD4+GzB+PRF+ T cells are present at high frequency in the spleen of mice infected with T. cruzi.
(A) CD4+ T cell gating strategy. (B and C) Representative flow-cytometry dot plots of granzyme B (GzB) (B) or perforin (PRF) (C) staining, gated on CD4+ T cells from non-infected (naïve) or infected B6 mice (left) and mean frequency of CD4+GzB+ (or CD4+PRF+) T cells in the spleen (right). (D) Representative dot plots of PRF and GzB expression in CD4+ T cells gated as in (A) and mean frequencies of PFR+ among GzB+ CD4+ T cells (isotype controls on Figure 1—figure supplement 1A). (E) Representative histogram of 2B4 (CD244) expression on CD8+ (blue), CD4+GzB+ (red) or CD4+GzB- T cells (pink), and correspondent mean fluorescence intensity (mean MFI) values; (F) Representative dot plots of GzB and IFN-γ staining in gated CD4+ T splenocytes (left), mean frequencies of GzB+IFN-γ- (red), GzB+IFN-γ+ (purple) and GzB-IFN-γ+ (blue) subsets among total CD4+ (center) or gated on CD4+GzB+ T cells (right). (G) Kinetics of the mean frequency of CD4+IFN-γ+ (purple) and CD4+GzB+ (red; left), and corresponding absolute numbers (right). (H) Frequency of LAMP-1+(CD107a+) cells among CD4+GzB+ (red) or CD8+GzB+ (blue) T cells in the infected spleen. Data obtained from individually analyzed mice (n=4), at indicated or 14 day post-infection (pi). Error bars = SEM. ND = non-detected. ns = non-significant; **p≤0.01 (Student’s t-test). Data are representative of three or more independent experiments.
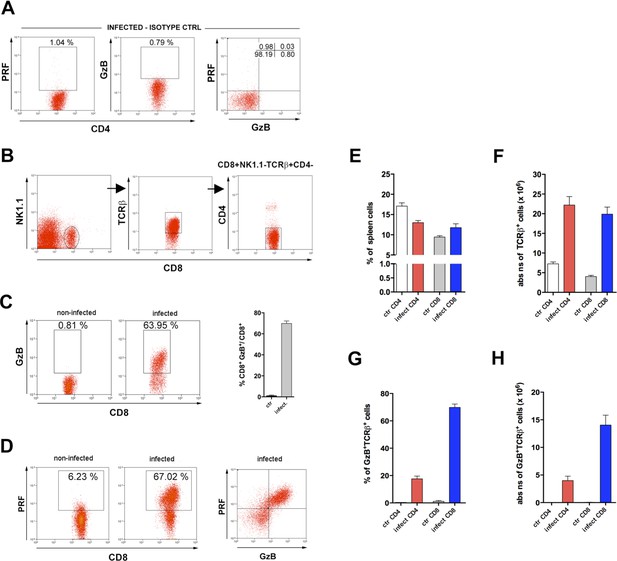
Comparative percentages and absolute numbers of granzyme B-positive (GzB+) and perforin-positive (PRF+) T cell subsets.
(A) Isotype control staining of CD4+ T cells, gated as in Figure 1A, from infected mice. (B) Gating strategy for (C and D) plots. (C) Representative dot plots of GzB staining (on the left) and mean frequency of GzB+CD8+T cells (on the right). (D) Representative dot plots of PRF and GzB staining in CD8+ T cells. (E) Mean frequency and (F) absolute numbers of CD4+ and CD8+ T cells in the spleen of non-infected (ctr) and infected mice. (G) Mean frequency and (H) absolute numbers of GzB+CD4+ and GzB+CD8+ T cells. Mice individually analyzed at day 14 post-infection (pi) (n=4); error bars = SEM. Data are representative of 6 independent experiments.
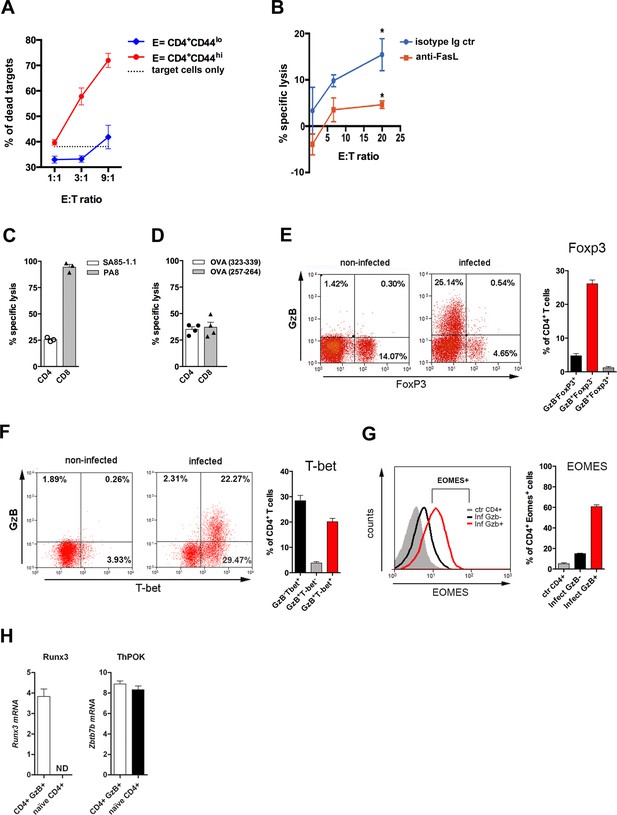
CD4 T cell-mediated Ag-specific cytotoxicity and transcription factor (TF) expression by CD4+GzB+ T cells.
(A) Mean frequencies of dead target cells (LPS-induced B blasts) in redirected cytotoxicity assay employing sorted naïve (blue) or in vivo activated (red) CD4+ T cells as effectors, at different E:T ratios, in triplicates. (B) Specific lysis of IC-21 macrophages (T) loaded with amastigote antigens, co-incubated with highly purified effector CD4+ T cells, at the indicated E:T ratios, in the presence of anti-FasL mAb (red) or isotype control mAb (blue). Mean values of triplicates ± SEM are shown in (A) and (B); *p≤0.05 (two-tailed Student’s t-test). Data are representative of three independent experiments. Gating strategy on Figure 2—figure supplement 1B. (C and D) Percentages of Ag-specific cytotoxicity in vivo; bars represent mean values + SEM (error bars) of individually analyzed mice (n=3) infected with T. cruzi Y in (C) or ovalbumin (OVA)-expressing Y strain trypomastigotes (Y-OVA) strain in (D); class I-restricted (PA8 or OVA257-264) and class II-restricted (SA851.1c+d or OVA323-339) peptides were employed. Data are representative of two independent experiments. Cell survival was measured by flow cytometry (Figure 2—figure supplement 1C) and specific lysis calculated as described in Methods. (E and F) Representative dot plots of CD4+ T cells (gated as in Figure 1A) and corresponding mean frequencies of gated subpopulations: (E) granzyme B (GzB) and FoxP3 staining and (F) GzB and T-bet staining. (G) Representative histogram of Eomes expression in CD4+ T splenocytes of non-infected mice (ctr, gray) and in GzB- (black) and GzB+ (red) CD4+ T cells from infected mice, gated as in Figure 1A and B (left) and mean frequencies of Eomes+ cells among these different subpopulations (right). All experiments were done at day 14 post-infection (pi). Bars represent mean values of non-infected ctr group n=3 or infected group, n=3–5, individually analyzed mice. Error bars = SEM. (H) Runx3d and ThPOK (Zbtb7b) mRNA expression estimated by qRT-PCR in CD4+EYFP+ T cells sorted from infected and tamoxifen-treated GzmbCreERT2/ROSA26EYFP mice, as described in Methods and shown in Figure 2—figure supplement 3A; ND = not detected. Mean values of triplicates + SEM are shown. Data are representative of three independent experiments.
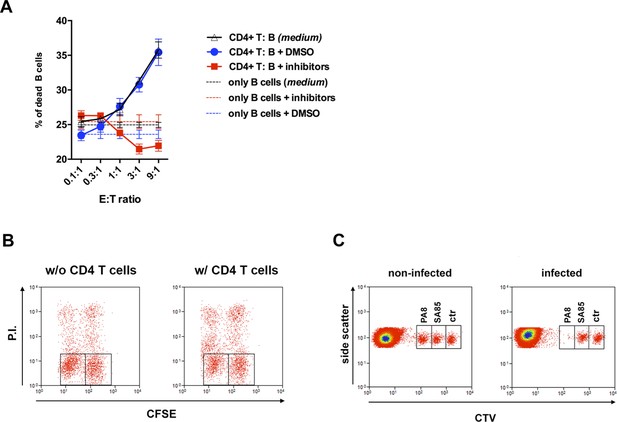
Cytotoxicity assays.
(A) Cytotoxic assays were performed as in Figure 2A (black line and symbols) or in the presence of granzyme B (GzB) (Z-AAD-CMK, 10 μM) and perforin (PRF) (Concamycin A, 0.5 μM) inhibitors (red line and symbols) or DMSO (blue line and symbols). Mean values of triplicates ± SEM are shown. (B–C) Gating strategy of cytotoxic assays: (B) gating strategies and PI staining of IC-21 macrophages loaded with total amastigote protein extract (CFSElo) or not (CFSEhi) and co-cultured with 4 × 105 purified CD4+ T cells (right), or not (left); specific cytotoxicity values are shown in Figure 2B; (C) gating strategies for CTV-stained target splenocytes, loaded with the indicated antigenic peptides or not (ctr), 20 hr after iv injection in non-infected or Y strain-infected mice. The T. cruzi-derived PA8 (class I-restricted) and SA85 (class II-restricted) peptides were employed. Specific cytotoxicity values for the in vivo assay are shown in Figure 2C.
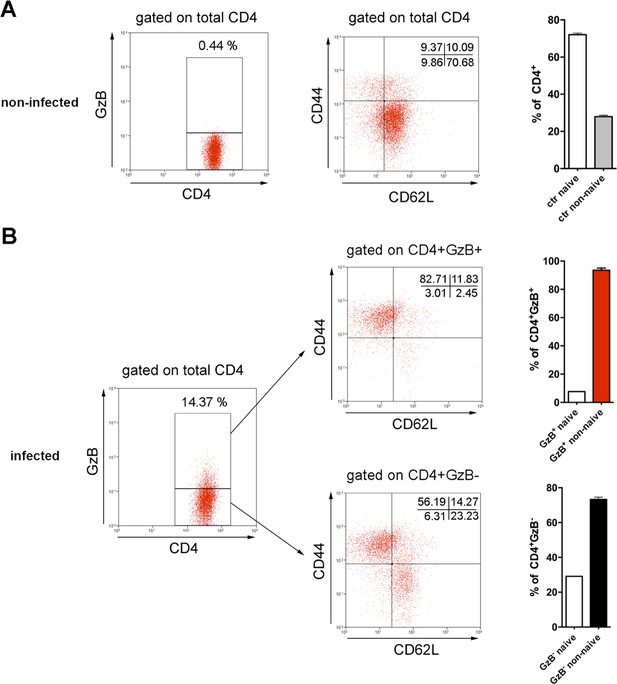
The majority of CD4+ T lymphocytes (GzB+ or GzB-) in the spleen of infected mice are non-naïve (activated effectors and memory) cells.
Representative dot plots and gating strategy of staining for granzyme B (GzB), CD44 and CD62L (on the left) and mean frequency of naive (CD44lo) and non-naive (CD44hi) CD4+ T cells (on the right) in: (A) non-infected (ctr) mice and gated on total CD4+TCRβ+ and (B) infected mice and gated on GzB+ (top) or GzB- (bottom) CD4+TCRβ+ cells, at day 14 post-infection (pi). Bars represent mean values of cell frequency of individually analyzed mice; n=4; Error bars = SEM. Data are representative of three independent experiments.
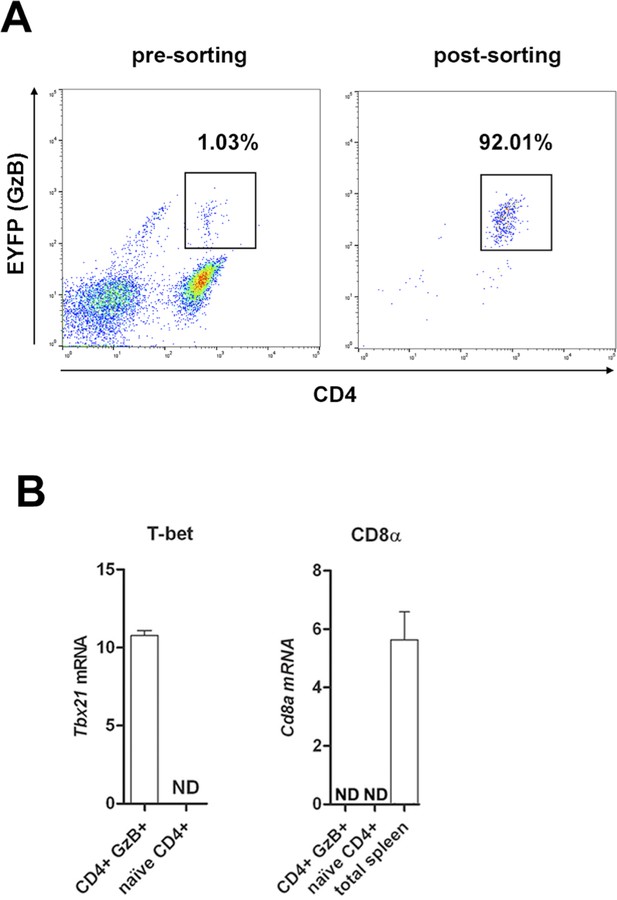
Expression of Tbx21 and Cd8a in sorted GzB+CD4+ T cells.
(A) Sorting gate and frequency of CD4+GzB+ (EYFP)+ T cells obtained from infected and tamoxifen-treated GzmbCreERT2/ROSA26EYFP mice. CD4+ cells were enriched by negative selection with magnetic beads, as described in Methods, and then submitted to FACS-sorting. Pre-sorting CD4+ T cell frequency is shown on the left panel and post-sorting on the right. (B) Sorted cells had RNA extracted and Tbx21, Runx3d, Zbtb7b and Cd8a expression tested by qRT-PCR, as described in Methods. Results for Runx3d and Zbtb7b expression are shown in Figure 2H. ND = not detected. Mean values of triplicates + SEM are shown.
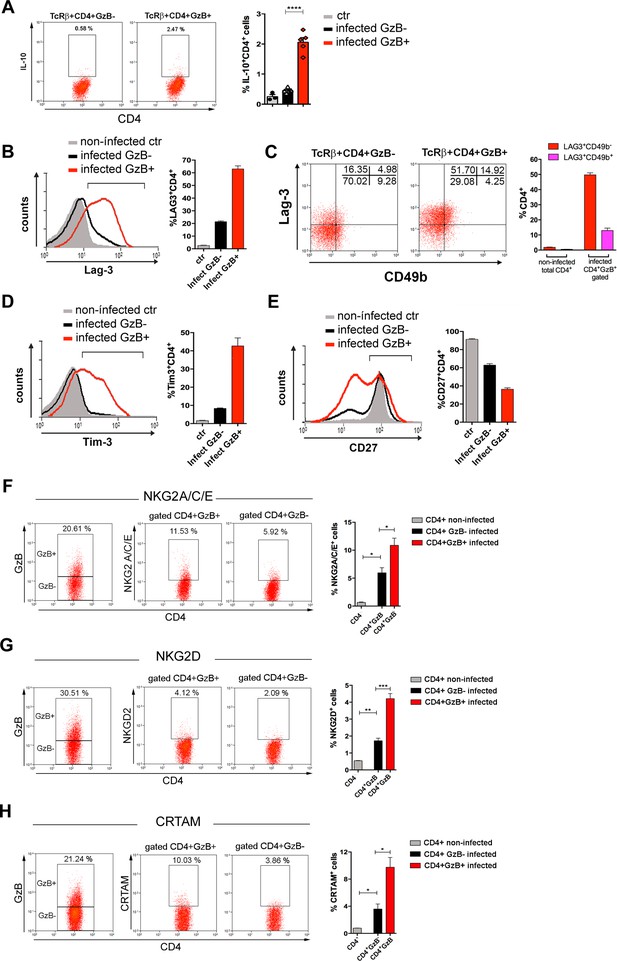
Expression of IL-10, immunoregulatory molecules and cytotoxic markers by CD4+GzB+ T cells.
(A) Representative dot plots of IL-10 staining and mean frequencies of IL-10+ cells among non-infected (ctr, gray) or granzyme B-negative (GzB-, black) and GzB+ (red) CD4+ T cells from infected mice. (B) Representative histogram of Lag-3 staining and mean frequency of Lag-3+ cells. (C) Representative dot plots of Lag-3 and CD49b staining and mean frequency of different cells subsets. (D) Representative histogram of Tim-3 staining and mean frequency of Tim-3+ cells among different cells subsets. (E) Representative histogram of CD27 staining and mean frequency of CD27+ cells. (A–E) CD4+GzB+ T cells gated as in Figure 1A, B; staining controls in Figure 3—figure supplement 1. (F–H) Representative dot plots and mean frequencies of (F) NKG2A/C/E, (G) NKG2D, and (H) CRTAM expression among gated CD4+ T splenocytes. All experiments were done at day 14 post-infection (pi). Non-infected ctr group, n=3; infected group, n=3–5 mice, individually analyzed. Error bars = SEM; *p≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001 (two-tailed Student’s t-test). Data are representative of three independent experiments. Staining controls are shown on Figure 3—figure supplement 2.
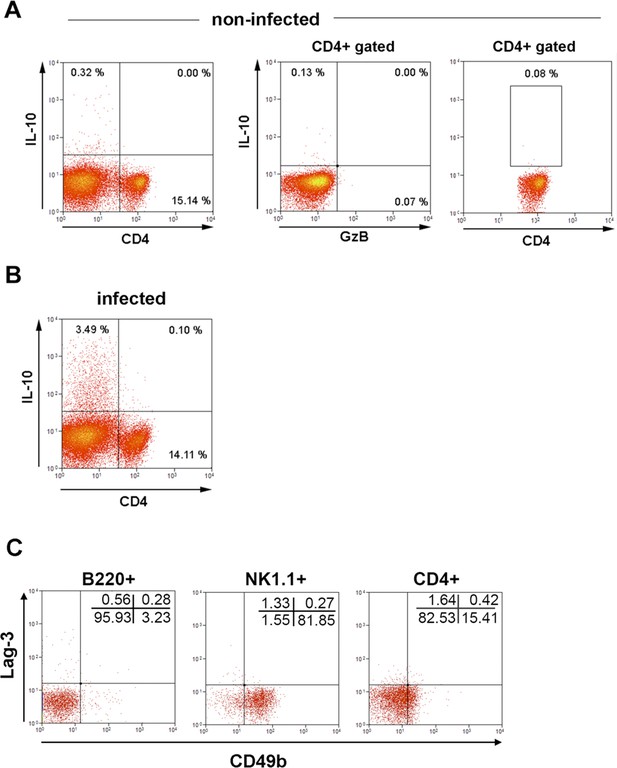
IL-10, Lag-3 and CD49b expression.
(A) Representative dot plots and gating strategy for IL-10 expression. Total spleen cells (left) and gated CD4+ T cells (middle and right panels) of non-infected control mice. (B) Representative dot plot of IL-10 staining in total splenocytes of infected mice. Dot plot and mean frequencies of IL-10+ cells among CD4+GzB- and CD4+GzB+ T cells in infected mice are shown in Figure 3A. (C) Representative dot plot of Lag-3 and CD49b expression on gated B220+, NK1.1+ or total CD4+ T cells from the spleen of non-infected mice (left, middle and right panels, respectively). Mean frequencies of Lag-3+CD49b+ and Lag-3+CD49b- cells among CD4+GzB- and CD4+GzB+ T splenocytes of infected mice are shown in Figure 3C. Data are representative of two independent experiments.
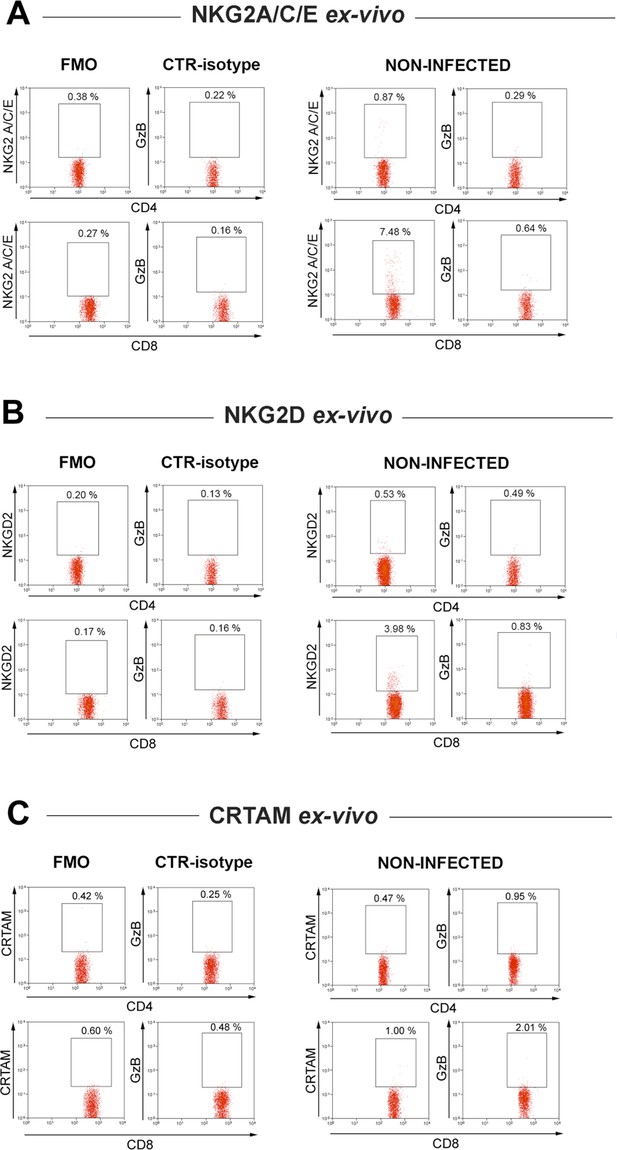
Representative dot plots and gating strategies for NKG2A/C/E, NKG2D and CRTAM staining on CD4+ and CD8+ GzB+ T cells.
Staining of spleen cells from non-infected controls (right panels) and Fluorescence Minus One (FMO) and isotype controls of spleen cells from B6 infected mice (left panels) are shown. Representative dot plots and mean frequencies of CD4+GzB+ and CD8+GzB+ T cells expressing these cytotoxic markers are shown in Figure 3F–H and Figure 3—figure supplement 4, respectively.
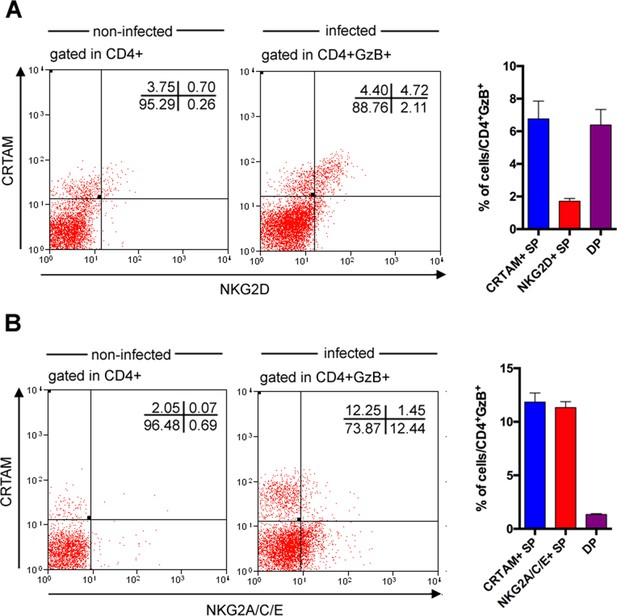
Most CD4+GzB+NKG2D+ T cells express the CRTAM cytotoxic marker while only a minority of CD4+GzB+ T cells co-expresses CRTAM and NKG2A/C/E molecules.
(A and B) Representative dot plots of CRTAM and NDG2D (A) or CRTAM and NKG2A/C/E (B) staining of spleen cells from non-infected and infected mice, gated on total CD4+ or on CD4+GzB+ T cells, respectively, as indicated. Bars graphs on the right represent mean values of individually analyzed infected mice; n=6; error bars = SEM; Data are representative of three independent experiments.
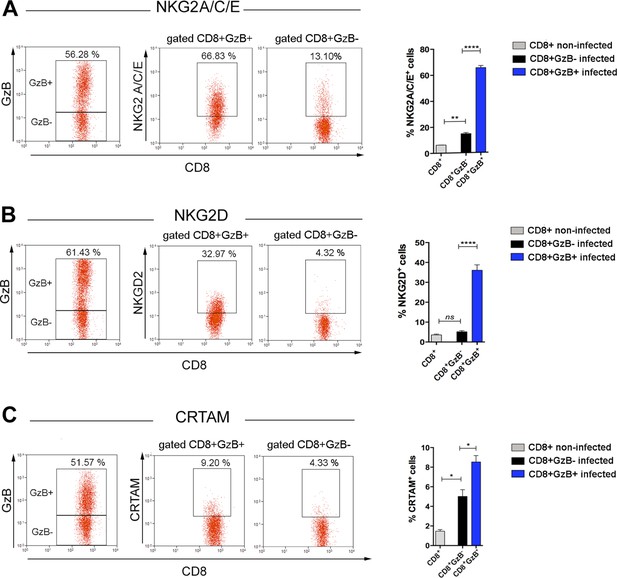
Expression of NKG2A/C/E, NKG2D, and CRTAM cytotoxic markers by CD8+GzB+ T cells.
Representative dot plots and gating strategies of spleen cells from B6 infected mice (left panels). Graphs on the right: bars represent mean values of cell frequency of each indicated cell subpopulation in individually analyzed mice; n=4; error bars = SEM; ns = non-significant; *p≤0.05; **p≤0.01; ****p≤0.0001 (two-tailed Student’s t-test). Data are representative of three independent experiments.
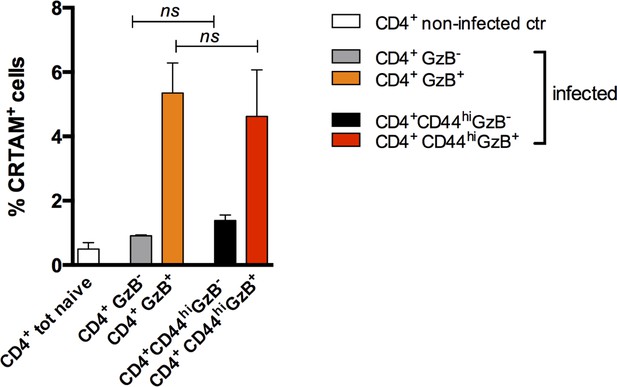
Frequency of CRTAM-expressing cells among splenic granzyme B-positive (GzB+) and GzB- CD4+ T cells, gating or not on CD44hi cells.
Bars represent mean values of the frequency of CRTAM+ cells among: CD4 naïve cells of non-infected (ctr) mice (white bar) and CD4+GzB- (gray bar), CD4+GzB+ (orange bar), CD4+CD44hiGzB- (black bar) or CD4+CD44hiGzB+ (red bars) of infected mice at day 14 post-infection (pi); Gating as shown in Figures 1A–B–3H and Figure 7—figure supplement 2. Mice were individually analyzed, n=4; error bars = SEM; ns = non-significant, Student’s t-test.
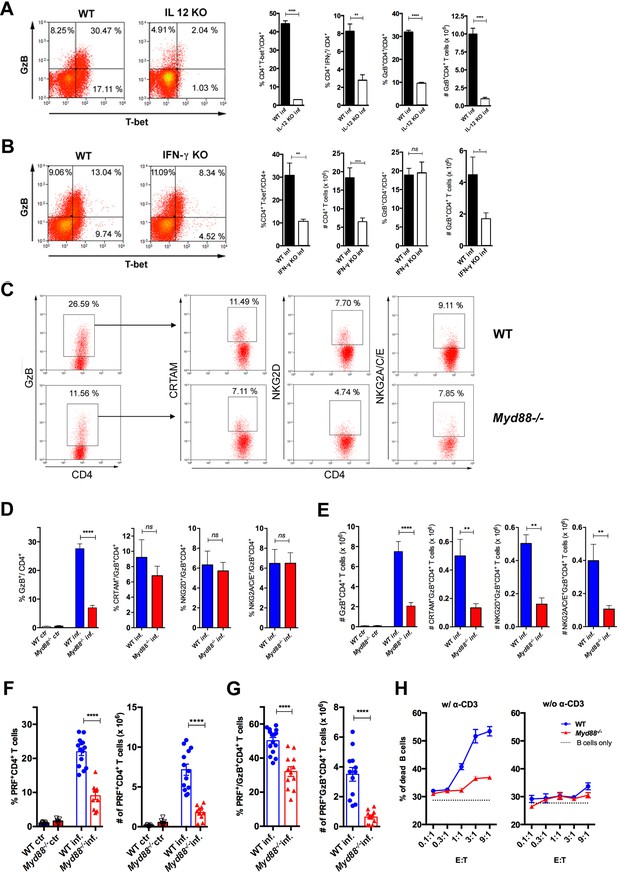
Cytotoxic CD4+ T cells (CD4CTLs) are severely reduced in Myd88-/- mice.
(A–B) Representative dot plots of granzyme B (GzB) and T-bet staining on gated CD4+ T splenocytes (left panels) and mean frequencies of T-bet+, IFN-γ+ and GzB+ CD4+ T cells and absolute numbers of GzB+CD4+ T cells (on the right) from (A) wild-type (B6) and Il12p40-/- mice on day 14 post-infection (pi) and (B) wild-type (WT) and Ifng-/- mice on day 11 pi. (C) Representative dot plots and gating strategy. (D) Mean frequencies and (E) absolute numbers of GzB+ or NKG2A/C/E+, NKG2D+ and CRTAM+ cells among GzB+CD4+ T cells in the spleen of WT (blue) and Myd88-/- (red) mice at day 13 pi. Bars represent mean values in each group; n=5 individually analyzed mice. Error bars = SEM; ns = non-significant; *p≤0.05; **p≤0.01; ****p≤0.0001 (Student’s t-test). Representative dot plots of GzB, NKG2A/C/E, NKG2D and CRTAM staining on CD4+ T cells from non-infected animals are shown in Figure 4—figure supplement 1B. (F–G) Frequencies (left) and absolute numbers (right) of perforin-positive (PRF+) cells among CD4+ T cells (F) and of PRF+ cells among GzB+CD4+ T cells (G) in the spleen of non-infected (ctr) or infected (inf.) WT (blue bars) and Myd88-/- (red bars) mice. GzB+CD4+ T cells were gated as shown in Figure 1A-D. Data in (F) and (G) are compiled from four independent experiments with n=3 animals in each group, each symbol represents an individual analyzed mice. ****p≤0.0001 (Student’s t-test). (H) Frequency of dead target cells (LPS-induced B cell blasts) in cytotoxic assay, after 14 hr of co-culture with CD4+CD44hi T cells sorted from WT (blue line) or Myd88-/- (red line) infected mice at day 13 pi. B cells were coated with anti-CD3 (left) or not (right). Mean of triplicates +/- SEM for each E:T ratio point are shown. Data are representative of three independent experiments.
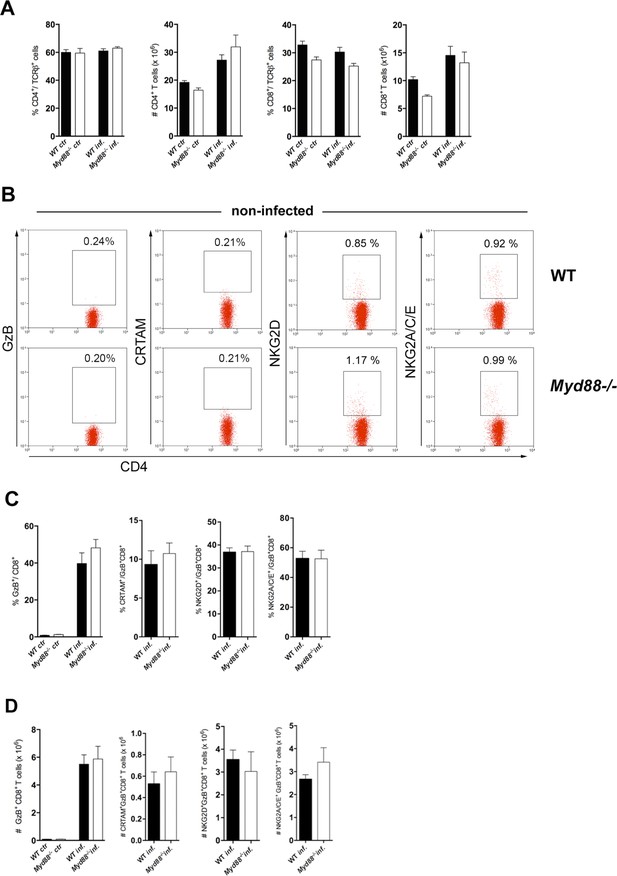
Equivalent frequencies and absolute numbers of total CD4+ and CD8+ T cells and equivalent frequencies and absolute numbers of CD8+ T cells expressing cytotoxic markers in the spleens of infected Myd88-/- and wild-type (WT) mice.
(A) Mean frequencies and absolute numbers of total CD4+ and CD8+ T cells in the spleens of non-infected (ctr) and infected (inf.) Myd88-/- and WT (B6) mice, at day 13 post-infection (pi). (B) Representative dot plots and gating strategies for granzyme B (GzB), CRTAM, NKG2D and NKG2A/C/E staining of CD4+ T splenocytes from non-infected (ctr) WT (B6) and Myd88-/- mice. Staining results obtained with infected mice are shown on Figure 4C–E. (C) Mean frequencies and (D) absolute numbers of GzB+, GzB+CRTAM+, GzB+NKG2D+ and GzB+NKG2A/C/E+ cells among CD8+ T cells in the spleen of WT and Myd88-/- infected mice at day 13 pi, gated as in Figure 3—figure supplement 4. Bars represent mean values in each group; error bars = SEM; n=5 individually analyzed mice. Results are representative of two independent experiments.
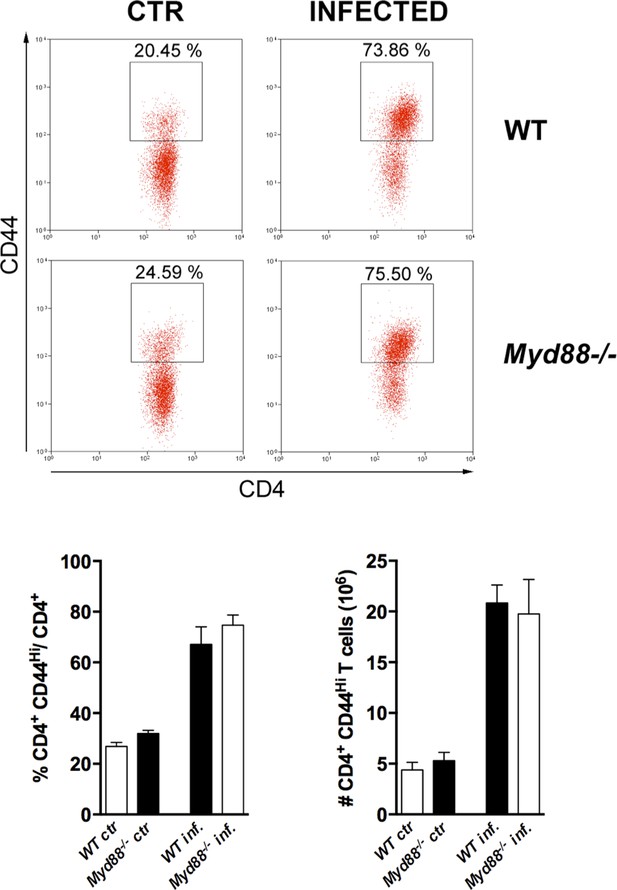
Equivalent frequency and absolute number of activated/memory (CD44hi) CD4+ T cells in wild-type (WT) and Myd88-/- mice infected with T. cruzi.
Representative dot plots (top) and mean frequencies and absolute numbers (bottom) of CD4+CD44hi T cells in the spleens of non-infected (ctr) and infected (inf.) WT (B6) and Myd88-/- mice, at day 14 post-infection (pi). Mice were individually analyzed; n=4; error bars = SEM. Representative of two independent experiments.
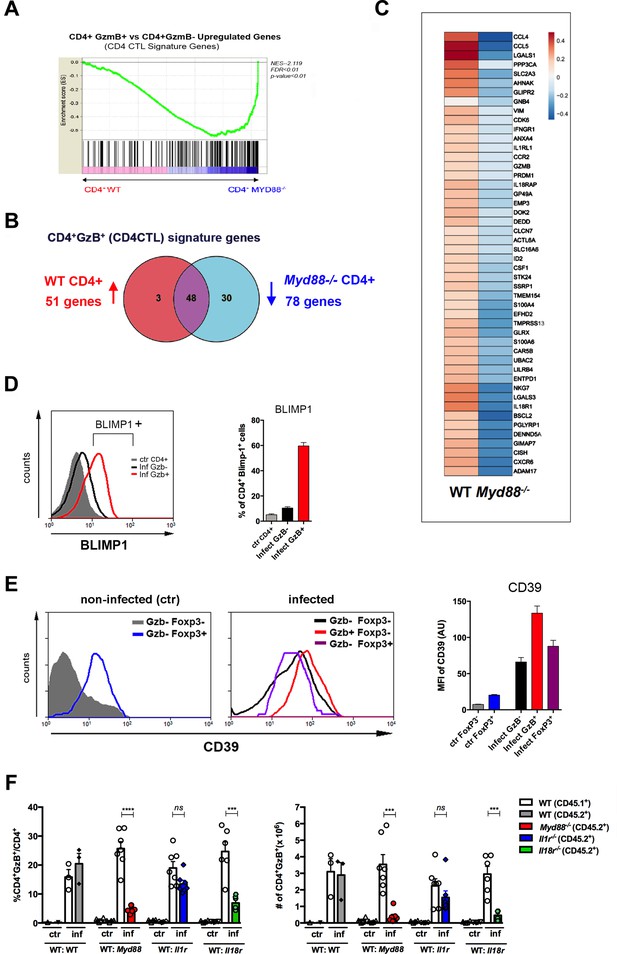
The cytotoxic CD4+ T cells (CD4CTL) gene program is absent in MyD88-deficient CD4+ T cells.
(A) GSEA performed using pre-ranked list of genes expressed in wild-type (WT) or Myd88-/- CD4+ T cells sorted from mixed bone marrow (mix-BM) chimeras using the CD4CTL gene signature previously described as gene set (Donnarumma et al., 2016). (B) Venn diagram of CD4CTL genes upregulated in WT and downregulated in Myd88-/- CD4+ T cells sorted from chimeric mice. (C) Heat map of normalized CD4CTL-signature gene expression in WT or Myd88-/- CD4+ T cells sorted from chimeric mice: values indicate mean Z-score, upregulated genes in red and downregulated genes in blue. (D) Representative histograms of Blimp-1 expression (right) and mean frequency of Blimp-1+ splenocytes (left) among total gated CD4+ T cells from non-infected mice (ctr, gray shaded area and bar) or among CD4+GzB+ (red line and bar) or CD4+GzB- (black line and bar) from WT infected mice at day 14 post-infection (pi). Individually analyzed mice, n=4. Error bars = SEM, data are representative of two independent experiments. (E) Representative histograms (left and central panel) and mean mean fluorescence intensity (MFI) values of CD39 expression (right panel) on gated FoxP3- (gray shaded area and bar) or FoxP3+ (blue line and bar) CD4+ T cells from non-infected (ctr) mice (left); GzB-FoxP3- (black line and bar), GzB-FoxP3+ (purple line and bar) or GzB+FoxP3- (red line and bar) CD4+ T cells from infected WT mice (center), at day 14 pi; individually analyzed mice (n=4), error bars = SEM; data are representative of four independent experiments. (F) Mean frequency (left) and absolute numbers (right) of GzB-expressing cells among CD45.1+ (B6.SJL WT) or CD45.2+ (B6 WT or KO) CD4+ T splenocytes in mix-BM chimeric mice, at day 14 pi; WT:WT control chimera group, n=3; other chimera groups, n=6 or 7. Error bars = SEM, ns = non-significant, ***p≤0.001,****p≤0.0001 (paired Student’s t-test). Gating strategy and representative dot plots are shown in Figure 5—figure supplement 1. Data are representative of three independent experiments.
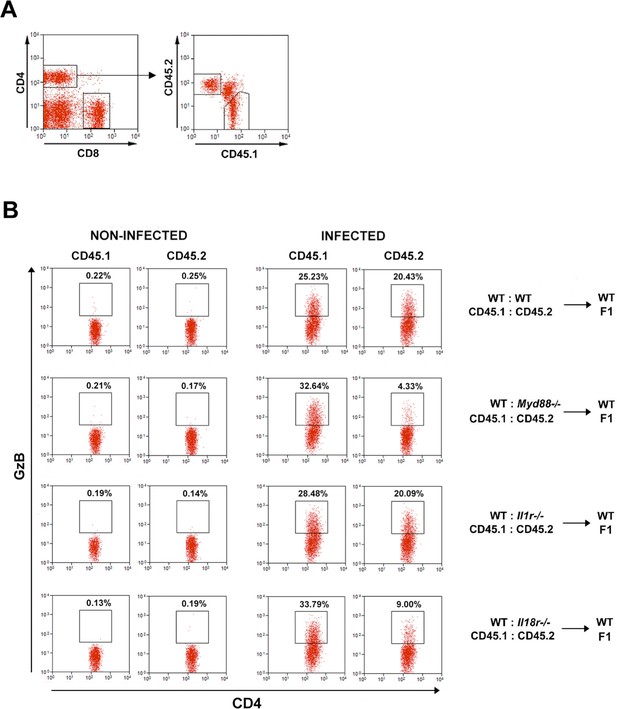
GzB expression in CD4+ T cells of mixed bone marrow (mixed-BM) chimeric mice.
Gating strategy and representative dot plots of (A) CD45.1+ and CD45.2+ cells (on the right) gated CD4+CD8- T lymphocytes (on the left) from the spleens of mixed-BM chimeric mice. (B) GzB expression in CD4+CD45.1+ (wild-type; WT) and CD4+CD45.2+ (WT or KO) T cells from the spleens of non-infected and infected chimeric mice at day 14 post-infection (pi), gated as in (A). Mean frequencies and absolute numbers of CD4+GzB+ in each group are shown in Figure 5F.
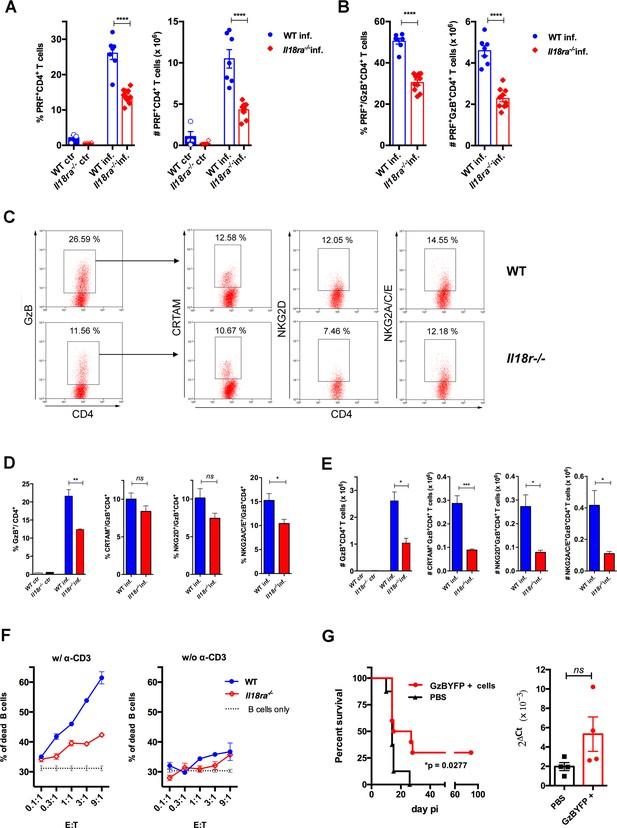
Cytotoxic CD4+ T cells (CD4CTLs) are severely reduced in Il18ra-/- mice.
(A and B) Frequencies and absolute numbers of perforin-positive (PRF+) cells among CD4+ T cells (A) and of PRF+ cells among GzB+CD4+ T cells (B) in wild-type (WT) (blue bars) and Il18ra-/- (red bars) spleens of non-infected (ctr) or infected (inf.) mice. Data are compiled from two independent experiments with n=4–5 animals in each group, each symbol represents an individual mouse. (C) Representative dot plots of GzB, CRTAM, NKG2D and NKG2A/C/E staining in WT and Il18ra-/- mice. (D) Mean frequencies and (E) absolute numbers of GzB+ or CRTAM+, NKG2D+ and NKG2A/C/E+ cells among CD4+ and CD4+GzB+ T cells, respectively. (A–E) CD4+ T cells gated as in Figure 1A–D; n=4 individually analyzed mice in each group; error bars = SEM, *p≤0.05, **p≤0.01, ***p≤0.001, ns = non-significant (Student’s t-test). Data are representative of two independent experiments. (F) Frequency of dead target cells (LPS-induced B cell blasts) in cytotoxic assay, after 14 hr of co-culture with CD4+CD44hi T cells sorted from WT (blue line) or Il18ra-/- (red line) infected mice at day 13 post-infection (pi). B cells were coated with anti-CD3 (left) or not (right). Mean of triplicates +/- SEM for each E:T ratio point are shown. (G) Survival curve (left) and parasite load in the heart (right) of infected Il18ra-/- mice, adoptively transferred with CD4+GzBYFP+ cells at day 7 pi (red curve and bar), or not (black curve and bar); time-line in Figure 6—figure supplement 1; n=10 mice in each group; p=0.0277 Log-rank (Mantel-Cox) test; survival data are compiled from two independent experiments; ns = non-significant (Student’s t-test).
Time-line of the adoptive transfer experiment shown on Figure 6G.
Description of the protocol is available in the Methods and materials section.
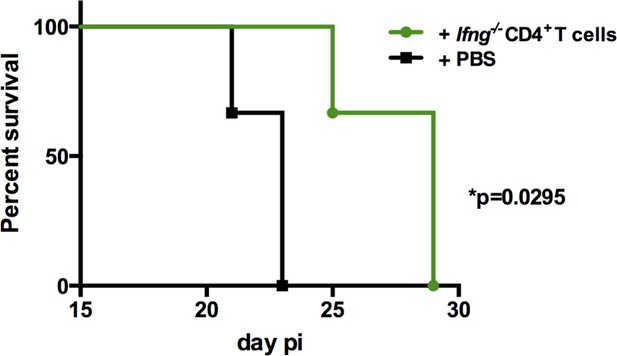
The adoptive transfer of Ifng-/- CD4+ T cells increased survival to infection.
Survival curve of Il18ra-/- mice, infected with 1 × 103 trypomastigotes of the Y strain and adoptively transferred with sorted (>98% pure) Ifng-/- CD4+ T cells (green curve) or not (phosphate-buffered saline [PBS] group, black curve); n=6 mice in each group; p=0.0295 Log-rank (Mantel-Cox) test.
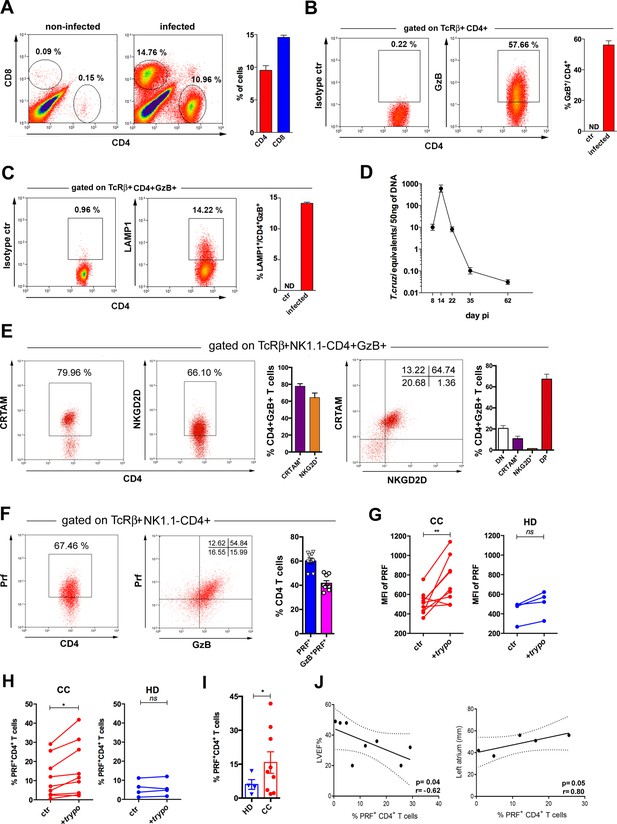
Cells with cytotoxic CD4+ T cell phenotype predominate in the cardiac tissue of infected mice and are at higher frequencies in patients with Chagas cardiomyopathy (CC).
(A) Representative dot plots (gated as in Figure 7—figure supplement 1A), and mean frequency of CD8+ and CD4+ T cells infiltrating the heart of B6 infected mice at day 14 post-infection (pi). (B and C) Representative dot plot and mean frequency of (B) granzyme B-positive (GzB+) and (C) LAMP-1+ (CD107a+) cells, among gated CD4+ T cells infiltrating the infected heart; ctr = non-infected, ND = non-detected. (D) Mean parasite load in the cardiac tissue at different time points pi, obtained by real-time PCR, of individually analyzed mice as described in Methods; data are representative of at least eight (A and B) or two (C and D) independent experiments. (E) Representative dot plots and mean frequency of CRTAM+ and NKG2D+ cells among gated intracardiac CD4+GzB+ T cells. DN and DP=CRTAM and NKG2D double negative and doulbe positive cells, respectively. Individually analyzed mice, n=4; error bars = SEM; data are representative of four independent experiments. (F) Representative dot plots and mean frequency of perforin-positive (PRF+) and GzB+PRF+ cells among intracardiac CD4+ T cells; data are compiled from two independent experiments with n=4 or 5 animals in each group, each symbol represents an individual analyzed mice. (G) Intensity of PRF staining (mean fluorescence intensity [MFI]) and (H) Frequency of PRF+ cells among CD4+ T cells in PBMCs from patients with chronic Chagas disease cardiomyopathy (CC; n=10) and healthy donors (HD; n=4) cultured overnight in medium alone (ctr) or in the presence of trypomastigote antigens (+trypo); *p≤0.05, **p≤0.01, ns = non-significant; data analyzed by paired Student’s t test. (I) Frequency of PRF+ cells among CD4+ T cells in the peripheral blood of patients with chronic Chagas disease cardiomyopathy (CC; n=10) and healthy donors (HD; n=4) cultured ON in the presence of trypomastigote antigens. Error bars = SEM; *p≤0.05, Student’s t test. Gating strategy and representative dot plots are shown in Figure 7—figure supplement 5. (J) Correlation analysis between the percentage of CD4+PRF+ T cells among CD4+ T cells in PBMCs from CC patients and left ventricular ejection fraction (LVEF) (left panel) or left atrium diameter (right panel); indicated p and r values were calculated using Pearson correlation test.
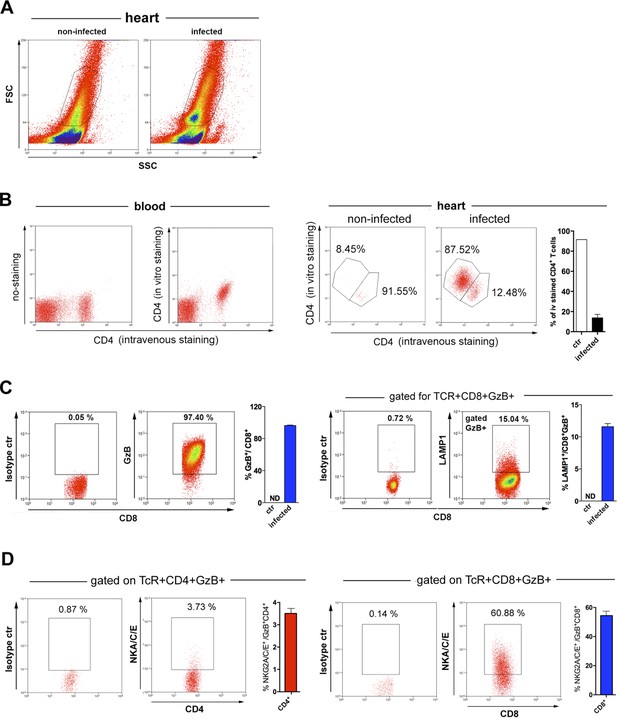
CD4+ and CD8+ T cells in the heart of mice infected with T. cruzi.
(A) Representative FSC × SSC dot plots and gating strategy of cells from the hearts of non-infected (left panel) and infected (right panel) wild-type (WT) mice, employed for data analysis shown on this figure and Figure 7. (B) Representative dot plots of blood samples from infected mice injected iv with anti-CD4-FITC mAb, 3 min before euthanasia, with no other staining (left panel) or after staining with anti-CD4-PECy7 mAb in vitro (right panel). Representative dot plots of cells from the heart of non-infected (ctr) and infected mice, injected iv with anti-CD4-FITC mAb and stained in vitro with anti-CD4-PECy7 (two panels on the right), and mean frequencies of intravenously stained CD4+ T cells in the hearts of non-infected (ctr) and infected mice. Mice were individually analyzed; n=3; error bars = SEM. Representative of two independent experiments. (C) Representative dot plots and gating strategy of isotype and granzyme B (GzB; left panels) or LAMP1 (right panels) staining and corresponding mean frequencies of GzB+CD8+ and LAMP1+CD8+ cells among gated CD8+ T cells infiltrating the infected myocardium at day 14 post-infection (pi). (D) Representative dot plots of isotype and NKG2A/C/E staining on gated CD4+GzB+ (left panels) or GzB+CD8+ (right panels) T cells and corresponding mean frequencies of NKG2A/C/E+ cells among GzB+CD4+ and GzB+CD8+ T cells infiltrating the infected myocardium at day 14 pi. Individually analyzed mice, n=4; error bars = SEM; ND = not detected. Data are representative of two independent experiments.
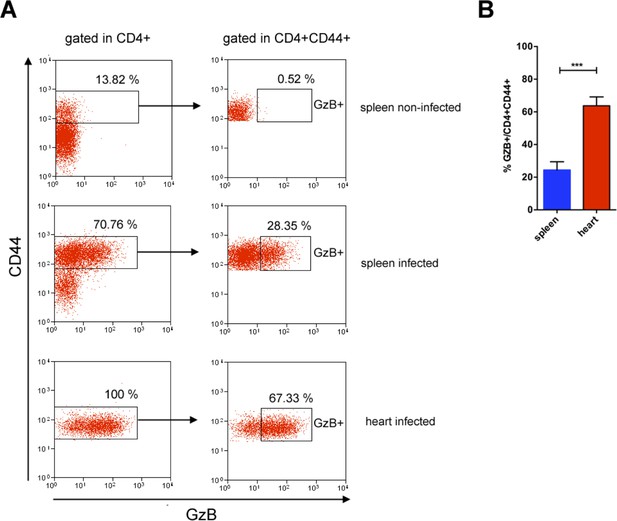
GzB+CD4+ T cells are enriched among activated/memory CD4+ T cells in the heart compared to the spleen of mice infected with T. cruzi.
(A) Representative dot plots of granzyme B (GzB) and CD44 staining of CD4+ T cells in the spleen and in the heart, gated as in Figures 1 and 7A. (B) Frequencies of GzB+ cells among activated/memory CD4+ T cells in the spleen and in the heart of individually analyzed infected mice; n=9; error bars = SEM, ***p≤0.001 Student’s t test. Data are compiled from three independent experiments.
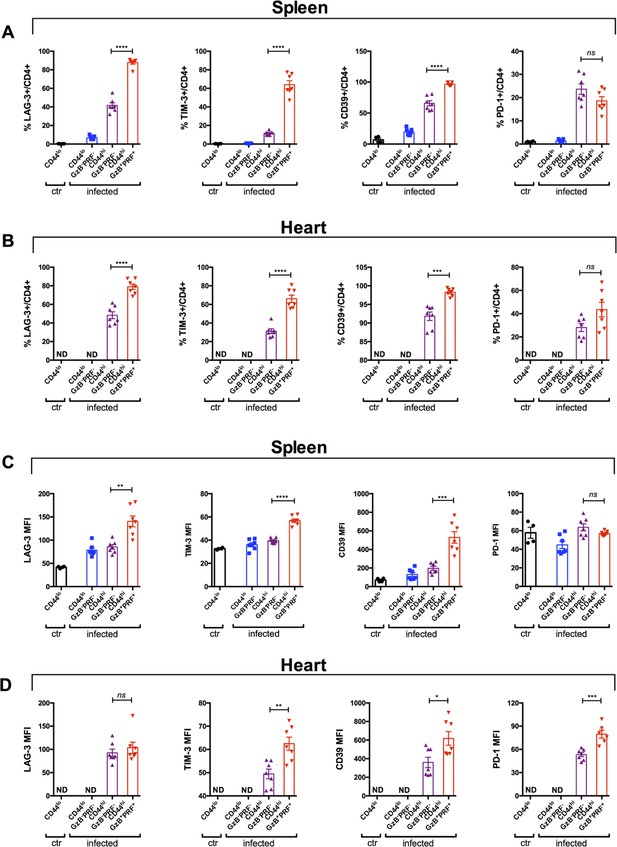
Cytotoxic CD4 +T cells express the higher levels of immunoregulatory molecules.
(A and B) Frequency of cells expressing. Lag-3, Tim-3, CD39 or PD-1 molecules among naïve cells form non-infected mice (CD44lo, ctr) and naïve cells (CD44loGzB-PRF-), activated non-cytotoxic (CD44hiGzB-PRF-) or activated cytotoxic (CD44hiGzB+PRF+) cells from infected mice, and (C and D) their respective levels of expression (mean fluorescence intensity, MFI), in the spleen and in the heart at day 14 post-infection (pi), as indicated. Bars represent mean frequency (A and B) or MFI (C and D) ± SEM of individually analyzed mice; n=7; ns = non-significant, *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001 Student’s t test; ND = not detected. Data are compiled from two independent experiments.
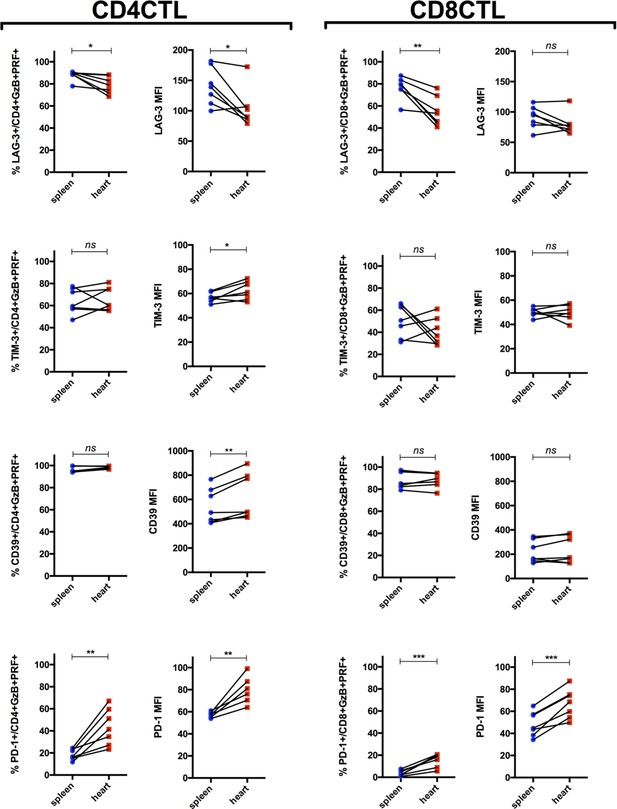
The expression of PD-1, CD39 and Tim-3, but not of Lag-3, is increased on cytotoxic CD4+ T cells (CD4CTLs) infiltrating the heart, compared to their levels in the spleen.
Graphs indicate the frequency of GZB+PRF+ T cells expressing Lag-3, Tim-3, CD39 and PD-1 among CD4+ and CD8+ T cells (CD4TLs on the left and CD8CTLs on the right), and their corresponding mean fluorescence intensity (MFI), in the spleen (blue dots) and in the heart (red squares), at day 14 post-infection (pi). Symbols represent individually analyzed mice; n=7; ns = non-significant, *p≤0.05, **p≤0.01, ***p≤0.001; paired Student’s t test. Data are compiled from two independent experiments.
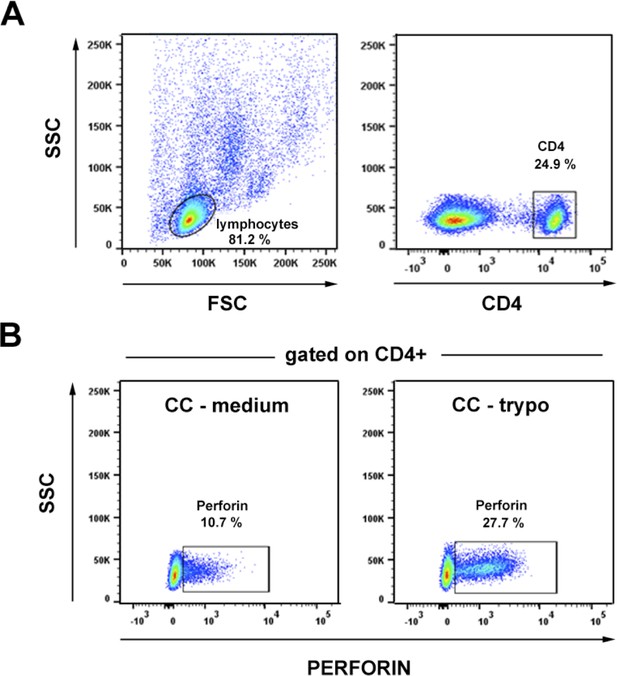
CD4+ T cells expressing perforin (PRF) are found in the circulation of patients suffering from chronic Chagas cardiomyopathy (CC).
(A) Dot plot showing the gating strategy for CD4+ T cell staining on PBMC. (B) Gating strategy and representative dot plots of perforin (PRF) staining, gated CD4+ T cells as in (A), from CC patient incubated with medium (on the left), or with trypomastigote antigens (trypo) (on the right). Frequencies of PRF+CD4+ T cells and mean fluorescence intensity (MFI) of PRF staining for CC patients and healthy donor controls are shown on Figure 7G–I.
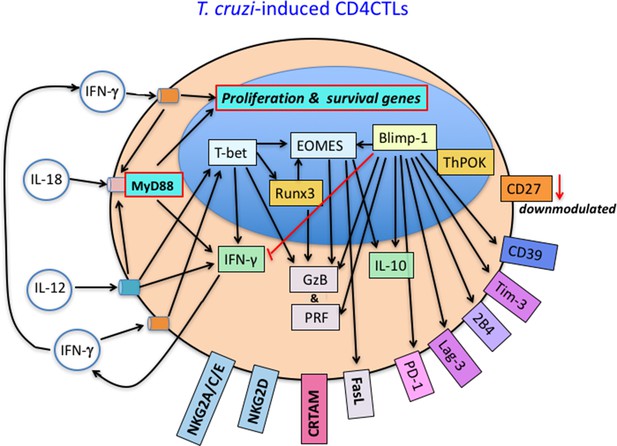
Predicted model for the generation of cytotoxic CD4+ T cells (CD4CTLs) in response to T. cruzi infection.
This cartoon summarizes our results and hypothesis and also contains information from previous published studies, cited in this work. IL-12 produced during infection acts on TCR-triggered CD4+ T cells, leading to T-bet expression, which promotes IFN-γ upregulation and induces Runx3d and Eomesodermin (Eomes) gene expression. In turn, IFN-γ- and IL-12-signaling induce IL-18R expression. T-bet, Eomes and Runx3d drive the expression of cytotoxic effector molecules, including granzyme B (GzB), perforin (PRF) and FasL. IL-10 expression can be induced by Blimp-1 and Eomes, although few IL-10+ CD4+GzB+ T cells were found in our infection model. As opposed to intraepithelial CD4CTLs of the gut, ThPOK expression is not downmodulated in splenic CD4+GzB+ T cells, until day 14 pi. Only around 18% of CD4+GzB+ T cells produce IFN-γ at the peak of the CD4+ T cell response (14 dpi), and IFN-γ is necessary for CD4+GzB+ T cell proliferation and/or survival. Most CD4+GzB+ T cells express Lag-3, Tim-3, PD1 and CD39 immunoregulatory molecules, possible under the control of Blimp-1, while CD27 is downmodulated. Notably, the expansion and/or survival of CD4+GzB+ T cells expressing CRTAM, NKG2D, and NKG2A/C/E cytotoxic markers depend on T-cell intrinsic IL-18R/MyD88 signaling.





