Chlamydomonas ARMC2/PF27 is an obligate cargo adapter for intraflagellar transport of radial spokes
Figures
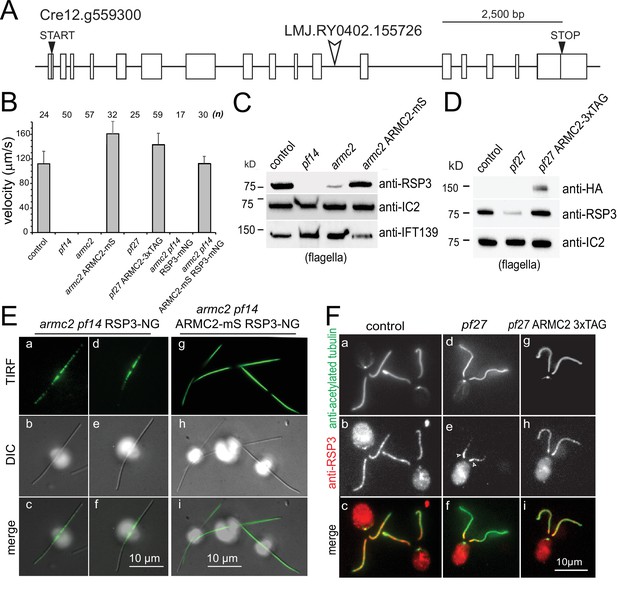
PF27 encodes ARMC2.
(A) Map of the ARMC2 gene. The open arrowhead indicates the position of the insertion in the CLiP mutant LMJ.RY0402.155726. (B) Average swimming velocity of the strains as indicated. The standard deviation and the number of cells analyzed are indicated. (C) Western blot analysis of isolated flagella of control, the RSP3 mutant pf14, armc2, and the armc2 ARMC2-mS rescue strain with antibodies to RSP3 and as loading controls, the outer arm dynein subunit IC2 and IFT139. Note accumulation of IFT139 in pf14 and armc2 as previously reported for paralyzed central pair mutants of Chlamydomonas (Lechtreck et al., 2013; see Figure 2—figure supplement 1B). (D) Western blot analysis of isolated flagella of control, pf27, and the pf27 ARMC2-3xTAG rescue strain with antibodies to RSP3, anti-HA, and anti-IC2, as a loading control. Anti-HA was used to document expression of ARMC2-3xTAG (the 3xTAG encompasses a triple HA tag). A representative Western blot of three biological replicates is shown. (E) DIC and TIRF imaging of live cells showing the distribution of RSP3-NG in the armc2 pf14 RSP3-NG mutant (a–f) and the armc2 pf14 ARMC2-mS RSP3-NG rescue strain (g–i). Bars = 10 µm. (F) Immunofluorescence staining of methanol-fixed control (a–c), pf27 (d–f), and pf27 ARMC2-3xTAG (g–i) cells stained with anti-acetylated-α-tubulin (a, d, g) to visualize flagella and affinity-purified anti-RSP3 (b, e, h); merged images are shown in the bottom row (c, f, i). Arrowheads in e, residual RSP3 near the proximal end of the pf27 flagella. The bright signal of the cell body likely results from unspecific binding of the anti-RSP3 antibody and chlorophyll autofluorescence. Bar = 10 µm.

Cloning, mass spectroscopy, and structure of Chlamydomonas ARMC2.
(A) Western blots of flagella isolated from control, pf27, and armc2 probed with antibodies against RSP3 or RSP23/NDK5; to test for equal loading, we used antibodies directed against the outer dynein subunit IC1. In pf27 and armc2, the radial spoke (RS) proteins migrated faster due to altered phosphorylation and were less abundant. To visualize RSP3 and NDK5 isoforms, we used a 6% acrylamide gel and let it run until the 50 kD marker reached the gel front. (B) In vivo epifluorescence imaging of pf14 RSP3-NG rescue cells and pf14 armc2 RSP3-NG cells. Bar = 10 µm. (C) Description of the ARMC2 expression construct with or without a tag. The 15.3 kB genomic construct was generated from ligating PCR-derived fragments of 4.6, 6.5, and 2.5 kB into the pGEM-T vector, along with the hygromycin resistance cassette (Hyg), using the indicated restriction sites. To insert the tags, an XhoI site was introduced into the 3’ fragment by fusing two PCR products. See Materials and methods for details. (D) Ni-NTA-chromatographic purification of ARMC2-3xTAG from whole cell extracts. The input, flow-through (FLOW), and eluate were assessed by Western blot analysis using anti-HA. The ~160 kD band enriched in the eluates (E) but absent in the flow-through was excised from a silver-stained gel and subjected to mass spectrometry. (E) Support of the 1053-residue ARMC2 sequence from mass spectrometry. Peptide sequences obtained by mass spectrometry of purified ARMC2-3xTAG and data mining are indicated in red letters. Yellow letters indicate the eight phosphorylation sites identified by phosphoproteomics of whole cell extracts (Wang et al., 2014). The encoding exons are marked in two shades of blue. (F) Predicted structure of ARMC2. Top: D, disordered regions. In the helical region the three armadillo repeats (ARM) are indicated in dark green. Bottom: The N-terminal ~400 residues of ARMC2 are predicted to be largely disordered (IUPred2, red line, score close to 1) and to assume a more structured configuration when binding to other proteins (blue line, ANCHOR2 score close to 1). The C-terminal region of ARMC2 is predicted to be largely ordered and encompasses three armadillo repeats (ARM).
armc2 has a paralyzed flagella phenotype.
Time lapse video of armc2 mutant cells. The video was recorded at 3 fps using phase contrast and a Nikon Eclipse 55i microscope equipped with a 40×/0.65 objective and a Zeiss AxioCam ERc5S. The timer displays seconds. The video is related to Figure 1B.
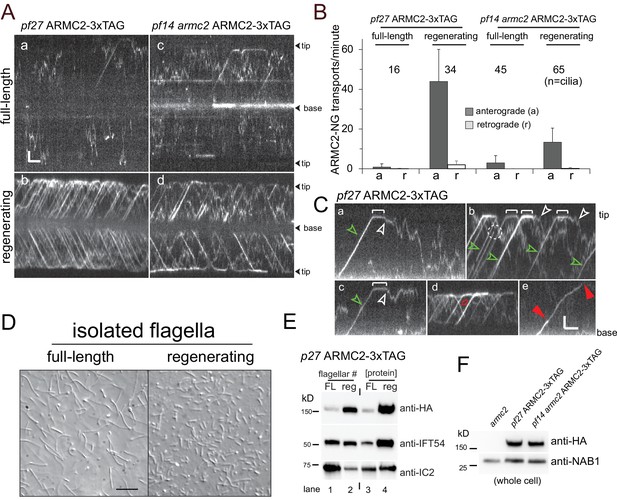
ARMC2-3xTAG is highly enriched in regenerating flagella.
(A) TIRF imaging of ARMC2-3xTAG in the pf27 background (a, b) and the pf14 armc2 double mutant background (c, d) in full-length (a, c) and in regenerating flagella (b, d). Bars = 2 s 2 µm. The flagellar tips and bases are indicated. (B) Bar graph showing the average frequencies (events/min/flagellum) of anterograde and retrograde transport of ARMC2-3xTAG in full-length and regenerating flagella of the pf27 ARMC2-3XTAG and the pf14 armc2 ARMC2-3xTAG strain. The standard deviation and the number of flagella analyzed are indicated. (C) Kymograms of ARMC2-3xTAG in late regenerating pf27 flagella. The white brackets in a–c mark the dwell time of individual ARMC2-3xTAG particles between arrival at the tip by anterograde IFT and the onset of diffusion (white arrowheads). Green arrowheads in a–c, anterograde transport of ARMC2-3xTAG, red open arrow in d, retrograde IFT of ARMC2-3xTAG; red arrowheads in e, stepwise bleaching of ARMC2-3xTAG indicating for the presence of two copies. In c, a single step bleaching event is marked by a dashed circle. Bars = 2 s and 2 µm. (D) DIC images of full-length and regenerating flagella of the pf27 ARMC2-3xTAG strain. Regenerating flagella were harvested ~22 min after deflagellation by a pH shock. Bar = 10 µm. (E) Western blot analysis of the full-length and regenerating flagella shown in C with the antibodies indicated. On the left side, an equal number of flagella were loaded and on the right side, approximately equal loading of protein was attempted. (F) Western blot comparing the presence of ARMC2-3xTAG in the pf27 ARMC2-3xTAG and the pf14 armc2 ARMC2-3xTAG strain. Antibodies to the cell body protein nucleic acid binding protein 1 (NAB1) were used as a loading control.
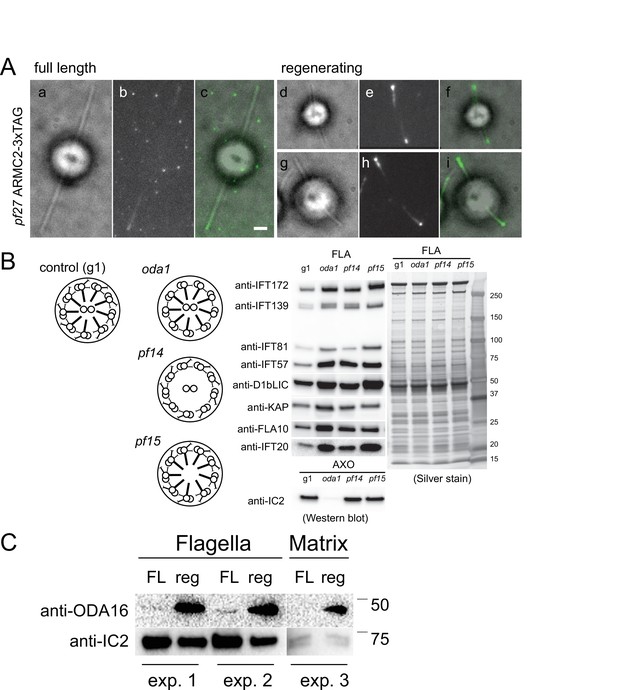
ARMC2-3xTAG accumulates at the tip of growing flagella.
(A) Bright field (a, d, g), TIRF (b, e, h), and merged images (c, f, i) of ARMC2-3xTAG in full-length (a–c) and regenerating (d–i) flagella of the pf27 ARMC2-3xTAG strain. Bar = 2 µm. (B) Schematic presentation, Western blot analysis and silver-stained gel of isolated flagella from control (g1), and the oda1, pf14, and pf15 mutants. Note accumulation of intraflagellar transport (IFT) proteins in the flagella these motility mutants. (C) Comparison of ODA16 levels in full-length (FL) and regenerating (reg) wild-type flagella from two independent biological replicates (experiments 1 and 2); from a third experiment (experiment 3) only the matrix fraction was available for analysis. For experiments 1 and 2, approximately equal number of flagella were loaded as apparent from the stronger IC2 bands in the FL samples.
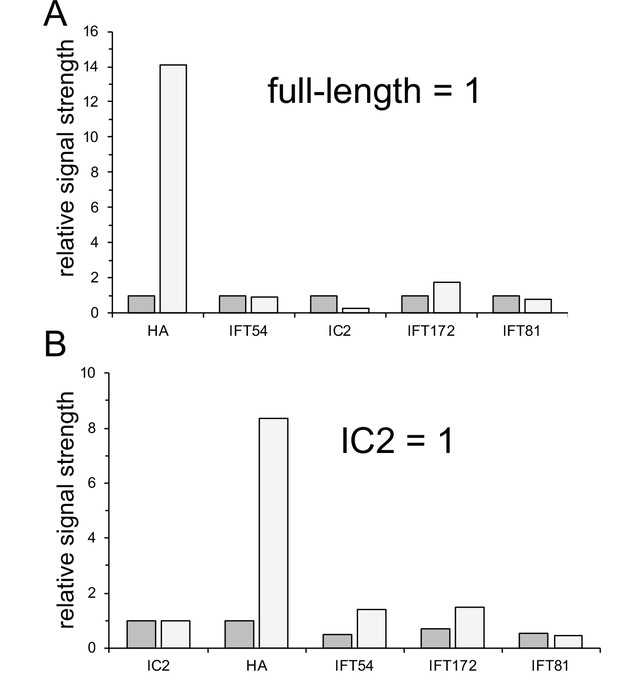
ARMC2-3xAG is enriched in growing flagella.
(A, B) Quantification of the Western blot shown in Figure 2E. (A) Quantification of lanes 1 and 2, in which equal number of flagella were loaded. To normalize, the signal obtained with the full-length flagella sample was set to one for each antibody. (B) Quantification of lanes 3 and 4, in which we attempted to load equal amounts of protein. The signals were normalized for the anti-IC2 signal, which was set to one for the full-length and regenerating flagellar sample. The analysis is based on N = 1 Western blots.
Anterograde intraflagellar transport (IFT) of ARMC2-3xTAG.
Video and corresponding kymogram showing transport of ARMC2-3xTAG by anterograde IFT to the flagellar tip, dwelling at the tip, and return to the ciliary base by diffusion. The video was recorded at 10 fps and the timer counts seconds. The video is related to Figure 2C.
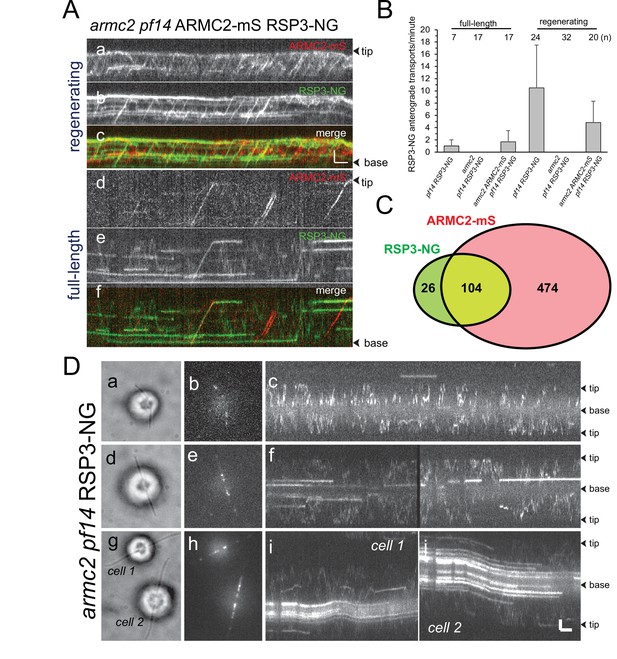
ARMC2-mS and RSP3-NG co-migrate by anterograde intraflagellar transport.
(A) Two-color TIRF imaging of a regenerating (a–c) and a full-length (d–f) flagellum of the armc2 pf14 ARMC2-mS RSP3-NG strain. Horizontal trajectories result from residuals unbleached RSP3-NG in the axoneme. Bars = 2 s 2 µm. (B) Bar graph showing the frequencies (events/min/flagellum) of RSP3-NG transports by anterograde IFT in full-length and regenerating flagella of the pf14 RSP3-NG, armc2 pf14 RSP3-NG, and armc2 pf14 ARMC2-mS RSP3-NG strains. The standard deviation and the number of flagella analyzed are indicated. (C) Venn diagram showing the distribution of anterograde ARMC2-mS and RSP3-NG transports; the overlap area represents the cotransports corresponding to 82% of all RSP3-NG and 18% of the ARMC2-mS transports. (D) IFT of RSP3-NG requires ARMC2. Analysis of RSP3-NG in armc2 mutant flagella. Brightfield (a, d, g) and TIRF (b, e, h) still images and corresponding kymograms (c, f, i, and j) of early (a–c), mid (g–f), and late stage (g–j) regenerating armc2 pf14 RSP3-NG cells. Bars = 2 s and 2 µm.
Cotransport of RSP3-NG and ARMC2-mS.
Movies and corresponding kymograms of a flagellum form armc2 pf14 ARMC2-mS RSP3-NG cells. Shown are the RSP3-NG and ARMC2 single channels and the merged image. An out-of-focus stretch of frames was deleted in the middle of the video as indicated by white bars in the kymograms. The video was recorded at 10 fps and the timer counts seconds. The video is related to Figure 3A.
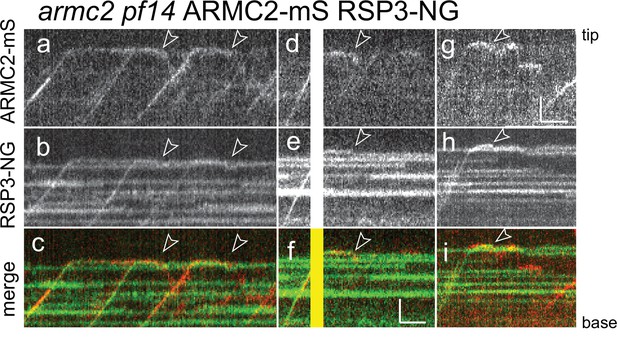
RSP3-NG ARMC2-mS complexes dissociate at the flagellar tip.
Kymograms from simultaneous imaging of the cargo adapter ARMC2-mS (a, d, g) and its cargo RSP3-NG (b, e, h); the merged images are shown in c, f, and i. The end of the dwell phase and concomitant onset of ARMC2-mS and RSP3-NG movements are marked with white arrowheads. The white/yellow frames in d–f result from overexposure due to the use of the bleaching laser pointed at the other flagellar tip of the cell. Bars = 2 s and 2 µm.
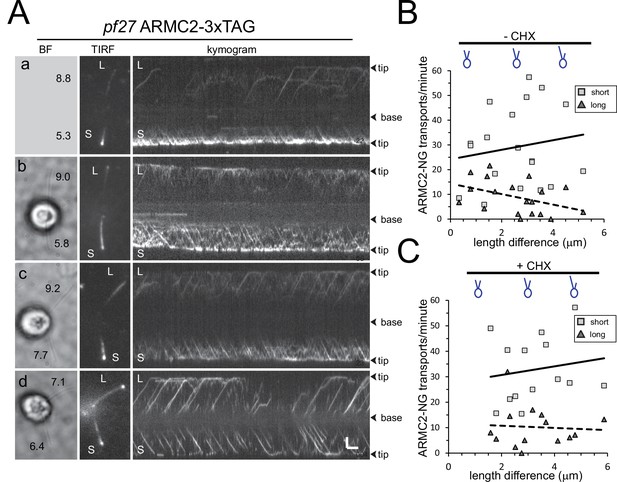
ARMC2-3xTAG transport is upregulated in short flagella.
(A) Gallery of brightfield (BF) and TIRF still images and the corresponding kymograms of long-short p27 ARMC2−3xTAG cells. No BF image was recorded for the cell shown in a. The length of the long (L) and short (S) flagella is indicated (in µm in a–d). Bars = 2 s and 2 µm. (B) Plot of the ARMC2-3xTAG transport frequency (events/min/flagellum) in the short (squares) and the long (triangles) flagella against the length difference between the two flagella. Trendlines, solid for the long and dashed for the short flagella, were added in Excel. (C) As B, but for cells treated with cycloheximide prior and during the experiment.
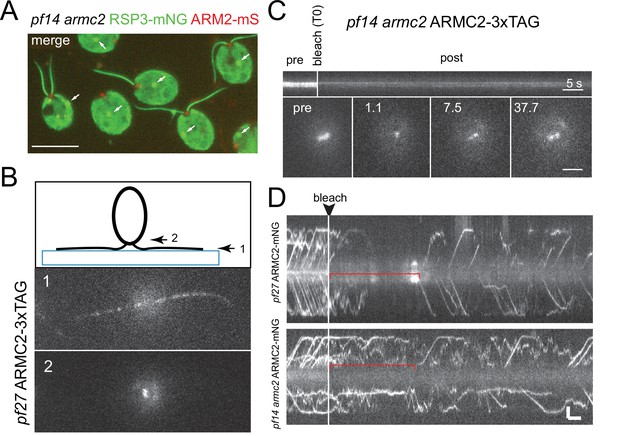
ARMC2-FP forms a pool near the basal bodies.
(A) Two-color epifluorescence image of the pf14 armc2 RSP3-NG ARMC2-mS strain. Note accumulation of ARMC2-mS (red) at the flagellar base. The bright signal of the cell body likely results from unspecific binding of the antibody and chlorophyll autofluorescence. The arrows mark the unspecific signal of the eyespot apparatus (arrows). Bar = 10 µm. (B) Focal series using flat-angle ‘TIRF’ illumination showing the flagella level (1) and the two ARMC2-3xTAG signals at the basal body level (2). The diagram illustrates the focal levels of the two images. (C) FRAP experiment using a focused laser beam to bleach one of the two ARMC2-3xTAG signals at the flagella base. Note slow and partial recovery of the signal. Statistical analysis was not performed as only three cells were analyzed. Bars = 5 s and 2 µm. (D) FRAP experiments to determine the dwell time of ARMC2-3xTAG in the basal body-associated pool. Regenerating flagella of the pf27 ARMC2-3xTAG and the pf14 armc2 ARMC2-3xTAG strains were analyzed. As in D, we used a focused laser beam to bleach the entire pool at the flagellar base and then analyzed the length of the gap before IFT of ARMC2-3xTAG resumed.
ARMC2-3xTAG transport in long-short cells.
Bright-field and TIRF video and the corresponding kymogram of a long-short pf27 ARMC2-3xTAG cell. Cells were sheared to generate long-zero cells and allowed to regrow missing flagella. The video was recorded at 10 fps and the timer counts seconds. The video is related to Figure 5.
ARMC2-3xTAG transport in long-short cells.
Bright-field and TIRF video and the corresponding kymogram of a long-short pf27 ARMC2-3xTAG cell. Cells were treated for 1 hr in CHX, sheared to generate long-zero cells and allowed to regrow missing flagella in the presence of CHX. The video was recorded at 10 fps and the timer counts seconds. Note that the bright-field video loops repeatedly. The video is related to Figure 5
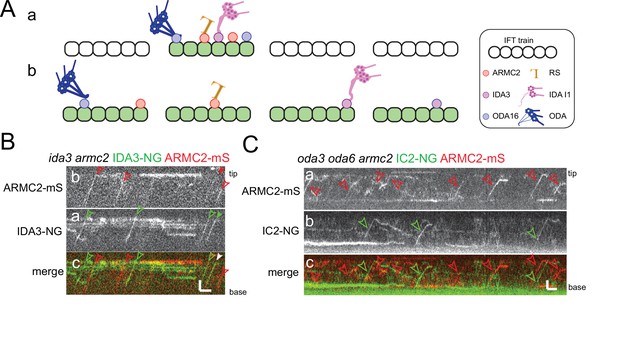
ARMC2-mS is transported independently of IDA3-NG and IC2-NG.
(A) Schematic presentation of two models for intraflagellar transport (IFT)-cargo interaction using radial spokes (RSs), outer dynein arms (ODAs), and I1 inner dynein arms (IDAs) and their adapters as examples. (a) Most cargoes use a specific subset of IFT trains, which have a high propensity to bind axonemal proteins, for example, because they are in a hypothetical open configuration. (b) All IFT trains are similarly capable of binding axonemal cargoes; thus, cargoes are stochastically distributed onto the trains. (B) Kymograms of two-color TIRF imaging of ARMC2-mS and IDA3-NG in an ida3 armc2 mutant cell. ARMC2-mS trajectories are marked with open red arrowheads and IDA3-NG trajectories with open green arrowheads. Filled arrowheads indicate a cotransport. Bars = 2 s and 2 µm. (C) Kymograms of two-color TIRF imaging using the oda3 oda6 armc2 IC2-NG ARMC2-mS strain. Trajectories of ARMC2-mS and IC2-NG transports are marked with red and green arrowheads, respectively. Bars = 2 s and 2 µm.
Tables
Frequency of cargo cotransport.
The table list the observed anterograde transports for ARMC2-mS, RSP3-NG, IDA3-NG, and IC2-NG in flagella of the corresponding double-mutant-double-rescue strains. For calculating the probability, by which an intraflagellar transport (IFT) train carries a cargo, we assumed an IFT frequency of 1/s. For cotransports, we calculated the observed probability (cotransports/total time) and compared it to the probability of cotransports occurring by chance as calculated by the following formula: P(cotransport−calculated) = P(cargo A) × P(cargo B).
| Strain | ARMC2-mS (n) | RSP3-NG (n) | Cotransports (n) | Time(s) | P(ARMC2-mS) | P(RSP3-NG) | P(cotransports-observed) | P(cotransport-calculated) | Cilia (n) |
|---|---|---|---|---|---|---|---|---|---|
| pf14 armc2RSP3-NG ARMC2-mS (full length) | 125 | 42 | 26 | 1504 | 0.083 | 0.028 | 0.017 | 0.0023 | 17 |
| pf14 armc2RSP3-NG ARMC2-mS (regenerating) | 578 | 130 | 104 | 1622 | 0.36 | 0.08 | 0.064 | 0.029 | 20 |
| ARMC2-mS (n) | IDA3-NG (n) | Cotrans-ports (n) | Time(s) | P(ARMC2-mS) | P(IDA3-NG) | P(cotransports-observed) | P(cotransport-calculated) | Cilia (n) | |
| ida3 armc2IDA3-NG ARMC2-mS (regenerating) | 243 | 106 | 20 | 905 | 0.27 | 0.12 | 0.022 | 0.03 | 35 |
| ARMC2-mS (n) | IC2-NG(n) | Cotrans-ports (n) | Total time(s) | P(ARMC2-mS) | P(IC2-NG) | P(cotransports-observed) | P(cotransport-calculated) | Cilia (n) | |
| oda3 oda armc2 ODA6-NG ARMC2-mS (regenerating) | 82 | 78 | 3 | 1575 | 0.052 | 0.049 | 0.0019 | 0.0026 | 20 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Chlamydomonas reinhardtii) | CC-1387, pf27, mt+ | Chlamydomonas Resource Center | RRID: SCR_014960 | |
| Genetic reagent (Chlamydomonas reinhardtii) | CC-613, pf14, mt- | Chlamydomonas Resource Center | RRID: SCR_014960 | |
| Genetic reagent (Chlamydomonas reinhardtii) | LMJ.RY0402.155726, armc2 mt- | Chlamydomonas Resource Center | RRID: SCR_014960 | |
| Genetic reagent (Chlamydomonas reinhardtii) | CC-2238 oda16 mt+ | Chlamydomonas Resource Center | RRID: SCR_014960 | |
| Genetic reagent (Chlamydomonas reinhardtii) | CC-5412, ida3:IDA3-NG, mt+ | Chlamydomonas Resource Center | RRID: SCR_014960 | |
| Transfected construct (Escherichia coli) | DH10B cells | New England BioLabs | – | Competent cells |
| Antibody | Rabbit anti-Sheep IgG (H + L) Secondary Antibody, HRP | Thermo Fisher | Catalog #: 31480. http://antibodyregistry.org/AB_228457 | WB 1:2000–5000 |
| Antibody | Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody | Thermo Fisher | Catalog #: 31432. http://antibodyregistry.org/AB_228302 | WB 1:2000–5000 |
| Antibody | IgG (H + L) Goat anti-Rat, HRP, Invitrogen | Thermo Fisher | Catalog #: 31470. http://antibodyregistry.org/AB_228356 | WB 1:2000–5000 |
| Antibody | Goat anti-mouse IgG (H + L) Alexa Fluor 488 (mouse polyclonal) | Invitrogen | Catalog #: A-11029.RRID: AB_2534088 | IF 1:800 |
| Antibody | Goat anti-rabbit IgG (H + L) Alexa Fluor 568 (rabbit polyclonal) | Invitrogen | Catalog #: 11,036.RRID: AB_10563566 | IF 1:800 |
| Antibody | Anti-HA, High Affinity (rat monoclonal, clone 3F10) | Roche/Sigma | Catalog #: 11867423001 | WB 1:800 |
| Recombinant DNA reagent | pGEMT-ARMC2(–3xTAG/mS) plus Hyg | This paper | Expression vector encompassing the up- and downstream flanking regions of the ARMC2 gene, optional mS or 3xTAG (NG-3xHA-6xHis) epitope tags and the aph7” selectable marker gene. Available from the corresponding authors. | |
| Sequence-based reagent | S1** | This paper | PCR Primer | CCGCCTGCACCCTTATCGCTGCCTCTGTCCCTCTTCC |
| Sequence-based reagent | AS2 | This paper | PCR Primer | CCTGTTCCGCACGCTGGTCTACCGTCTACC |
| Sequence-based reagent | S3* | This paper | PCR Primer | CGAGGCGGTGAGCGAGCACGTGTTCCGACTCATG |
| Sequence-based reagent | AS3* | This paper | PCR Primer | GCCTCACGGTACCGTGAGCACATGCATGGGTTTGC |
| Sequence-based reagent | S4 | This paper | PCR Primer | CGCAACCCCCGCTACTCTAACCTCGAGG |
| Sequence-based reagent | AS4Hind | This paper | PCR Primer | CAGAAGCTTGAAGCCCGAAAGCTGACGAAGTGGG |
| Sequence-based reagent | HindS6.1 | This paper | PCR Primer | GAGAAGCTTACCTACCTGGGTCTTGACATGCCCTGTCC |
| Sequence-based reagent | AS5Xho | This paper | PCR Primer | CCTCGAGCTCCGGCAACGCCTCCAGCTCC |
| Sequence-based reagent | XhoS7 | This paper | PCR Primer | CCTCGAGTAGGGGCCCTTGCTTAGGGAATTCAGGG |
| Sequence-based reagent | AS6 | This paper | PCR Primer | CTCGCTTTCACAACTCCAGGGTGCCCATGC |
| Sequence-based reagent | ida3f | This paper | PCR Primer | ATTTGGACGGAGCCTTGAC |
| Sequence-based reagent | ida3r | This paper | PCR Primer | TGTTTCGCACGCCTTCA |
| Chemical compound, drug | ProLong Gold Antifade Mountant | Thermo Fisher | Catalog #: P36934. RRID:SCR_015961 | Catalog number #: P36930 |





