Conformational dynamics and allosteric modulation of the SARS-CoV-2 spike
Figures
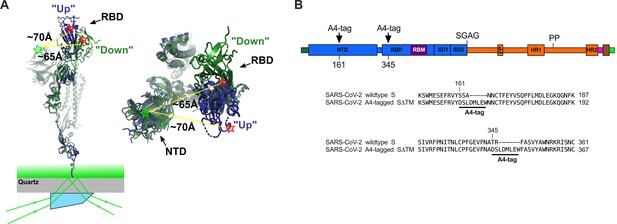
Single-molecule Förster resonance energy transfer (smFRET) imaging of the conformational dynamics of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) S ectodomain.
(A) (Left) SARS-CoV-2 SΔTM containing a single fluorescently labeled A4-tagged protomer within an otherwise untagged trimer was immobilized on a streptavidin‐coated quartz microscope slide by way of a C-terminal 8x-His-tag and biotin‐NiNTA. For clarity, only a monomer is depicted. Individual SΔTM trimers were visualized with prism-based TIRF microscopy using a 532 nm laser. Overlay of two S protomers with receptor-binding domains (RBD) in the ‘up’ (blue) and ‘down’ (green) conformations are shown with approximate positions of fluorophores indicated by green (LD550) and red (LD650) stars. (Right) Top view of the same S protomer overlay. The approximate distances between the sites of labeling are shown. Structures adapted from PDB 6VSB. (B) Domain organization of the SARS-CoV-2 SΔTM construct used for smFRET experiments, indicating the sites of A4 tag insertion. The S1 and S2 subunits are in blue and orange, respectively. Additional domains and features are as follows, ordered from N- to C-terminus: signal peptide, dark green; NTD, N-terminal domain; RBD and RBM, receptor-binding domain and motif (purple), respectively; SD1, subunit domain 1; SD2, subunit domain 2; SGAG, furin cleavage site mutation; FP, fusion peptide; HR1 and HR2, heptad repeat 1 and 2, respectively; PP, diproline mutations; T4 fibritin trimerization motif (foldon), magenta; TEV protease cleavage site, brown; 8x-His-tag, green. (Bottom) Amino acid sequence alignments indicating sites of A4 tag insertions in SΔTM. A4 peptide sequences (DSLDMLEW) are underlined. Fluorophores get attached to the serine amino acid within the A4 peptide.
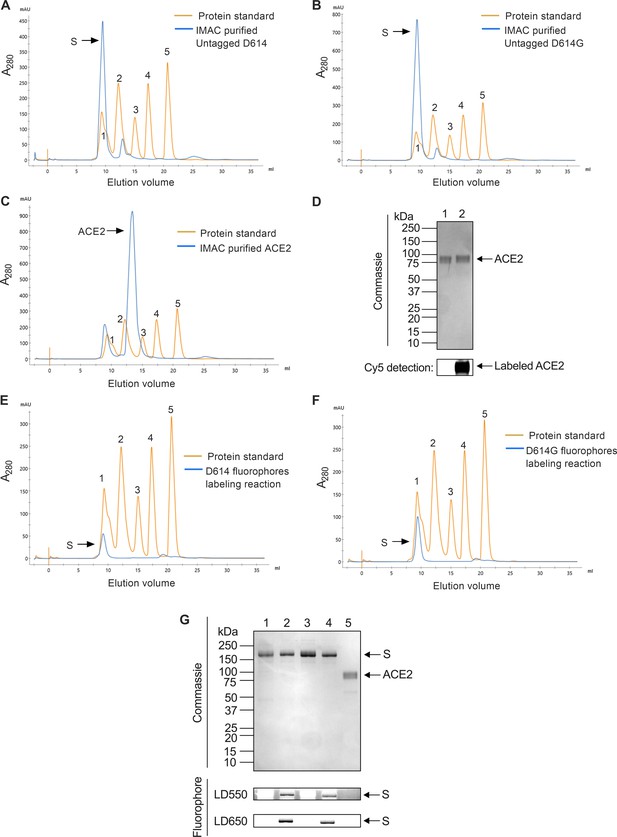
Purification and labeling of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) SΔTM and soluble angiotensin-converting enzyme 2 (ACE2).
(A) Chromatogram from size exclusion chromatography (SEC) purification of SΔTM D614 (orange) as described in Materials and methods, with a gel filtration standard (Bio-Rad) overlaid. The 670, 158, 44, 17, and 1.35 kDa molecular weight references are numbered as 1, 2, 3, 4, and 5, respectively. (B) The same SEC data for SΔTM D614G and (C) ACE2. (D) Cy5 labeling of purified ACE2 was analyzed by denaturing PAGE. Representative gel showing gel Coomassie staining (top) and Cy5 fluorescence (bottom) with unlabeled and labeled ACE2 in lanes 1 and 2, respectively. (E) SEC purification, displayed as in (A), for LD550/650-labeled A4-tagged SΔTM D614 and (F) SΔTM D614G hetero-trimers. (G) Denaturing PAGE analysis of purified unlabeled and labeled proteins. Labeling was performed as described in Materials and methods. Coomassie staining (top) and in-gel fluorescence detection are as indicated. Lane 1: SΔTM D614; lane 2: LD550/650-labeled SΔTM D614; lane 3: SΔTM D614G; LD550/650-labeled SΔTM D614G; and lane 5: unlabeled ACE2. Raw data is provided in Figure 1—figure supplement 1—source data 1.
-
Figure 1—figure supplement 1—source data 1
Numeric chromatography data from purification of SΔTM and angiotensin-converting enzyme 2 (ACE2), and original Western blot images.
- https://cdn.elifesciences.org/articles/75433/elife-75433-fig1-figsupp1-data1-v2.zip
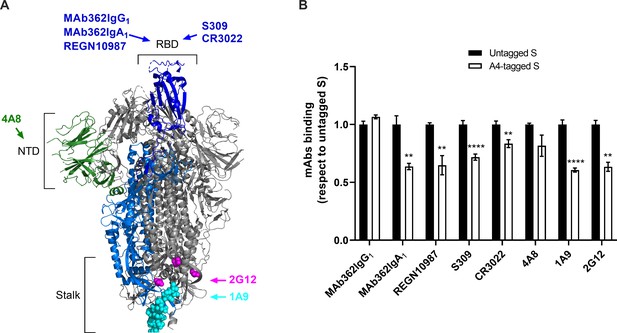
Characterization of SΔTM containing A4 peptide tags for site-specific fluorophore attachment.
(A) Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) S domains and regions recognized by the monoclonal antibodies (mAbs) used in this study are indicated. An S protomer with its receptor-binding domain (RBD) in the up conformation is depicted in colors: RBD, blue; N-terminal domain (NTD), dark green; cyan and magenta spheres, stalk epitopes for 1A9 (Zheng et al., 2020) and 2G12 (Williams et al., 2021), respectively. The remainder of the protomer is shown in light blue. The remaining two protomers with their RBDs in the down conformation are shown in gray. Structure adapted from PDB 6VSB. (B) ELISA indicating mAb binding to A4-tagged SΔTM spikes as described in Materials and methods. Results from three independent experiments with two technical replicates each, using both untagged (white bars) or A4-tagged (black bars) SΔTM D614 spikes and the indicated mAbs are shown. Statistical significance was evaluated as described in Materials and methods and p-values < 0.05 were considered significant. Significance values are indicated as **p < 0.01 and ****p < 0.0001. Raw data is provided in Figure 1—figure supplement 2—source data 1.
-
Figure 1—figure supplement 2—source data 1
Numeric angiotensin-converting enzyme 2 (ACE2)-bound fraction data.
- https://cdn.elifesciences.org/articles/75433/elife-75433-fig1-figsupp2-data1-v2.zip
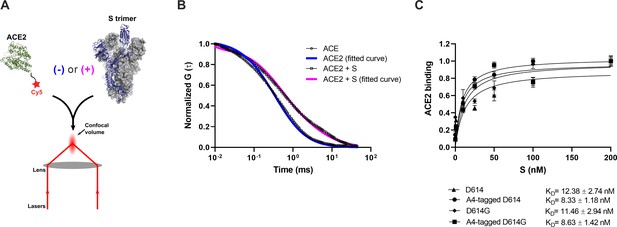
Verification of angiotensin-converting enzyme 2 (ACE2)-binding to A4-tagged SΔTM trimers using fluorescence correlation spectroscopy (FCS).
(A) Cy5-labeled ACE2 was incubated in the absence or presence of untagged or A4-tagged SΔTM spikes. The diffusion of Cy5-ACE2 was evaluated by FCS using a 647 nm laser as indicated in Materials and methods. (B) Representative normalized autocorrelation curves for Cy5-ACE2 in the absence (circles) or presence (squares) of SΔTM, and the corresponding fits are shown in blue or magenta, respectively. The shift in the autocorrelation to longer timescales seen in the presence of SΔTM reflects the slower diffusion resulting from the larger size of the complex. (C) Cy5-ACE2 (100 nM) was incubated with different concentrations of the indicated SΔTM spikes and the resulting mixture was evaluated by FCS as described in Materials and methods. Dissociation constants (KD) determined from fitting the titration are indicated for the different SΔTM constructs. Data are presented as the mean ± standard deviation from three independent measurements. Raw data is provided in Figure 2—source data 1.
-
Figure 2—source data 1
Numeric autocorrelation and bound angiotensin-converting enzyme 2 (ACE2) fraction data for panels B and C.
- https://cdn.elifesciences.org/articles/75433/elife-75433-fig2-data1-v2.zip
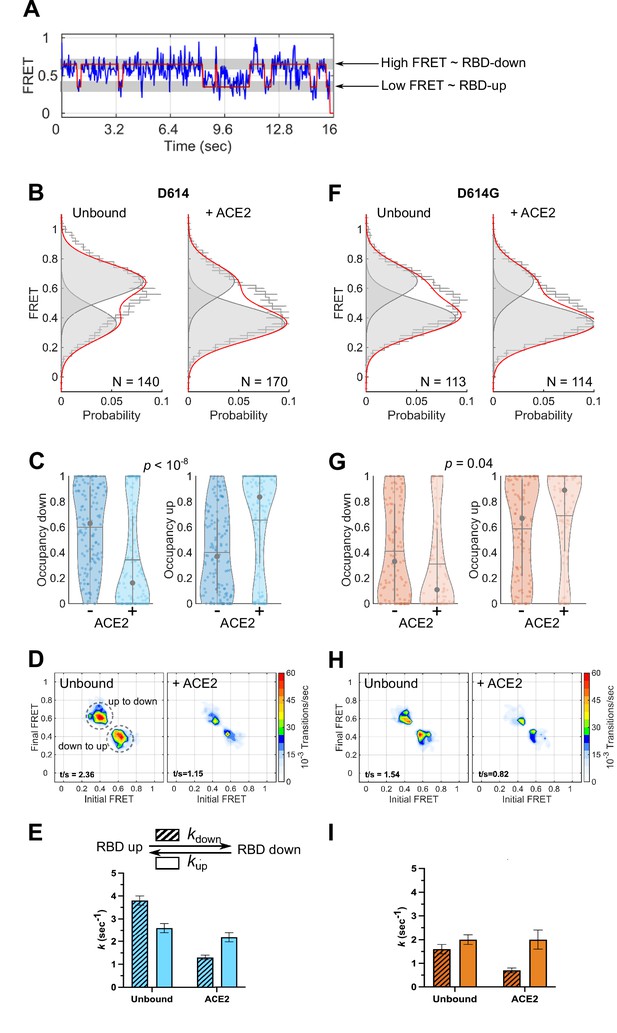
Angiotensin-converting enzyme 2 (ACE2)-binding modulates the receptor-binding domain (RBD) conformation of SΔTM D614 and D614G.
(A) Representative single-molecule Förster resonance energy transfer (smFRET) trajectory acquired from an individual SΔTM trimer (blue). Idealization resulting from Hidden Markov modeling (HMM) analysis is overlaid (red). The high-FRET (0.65) and low-FRET (0.35) states correspond to the RBD-down and RBD-up conformations, respectively, as indicated. Bulk fluorescence lifetime and anisotropy measurements supported the interpretation of changes in FRET as arising due to conformational transitions that reposition the fluorophores and are presented in Table 1. (B) (Left) FRET histogram for unbound SΔTM D614 overlaid with the sum of two Gaussian distributions centered at 0.65 and 0.35 FRET (sum, red; single distributions, gray) generated from the results of HMM analysis. FRET histograms are presented as the mean ± standard error determined from three technical replicates. The total number of smFRET traces used in the HMM analysis is shown (N). (Right) The same data for the ACE2-bound SΔTM D614 spike. (C) Violin plots indicating the distribution of occupancies in the 0.65-FRET (RBD-down) and 0.35-FRET (RBD-up) states seen for the smFRET traces analyzed. For each plot the gray circles and horizontal lines indicate the median and mean occupancy, respectively. The vertical gray lines extend to the 25th and 75th quantiles. The statistical significance of the differences in occupancies seen for the unbound and ACE2-bound SΔTM D614 trimers was evaluated with a one-way ANOVA (p-value is indicated). (D) Transition density plots (TDPs) for (left) unbound and (right) ACE2-bound SΔTM D614 indicating the frequency of observed FRET transitions determined through HMM analysis. The assignment of the two transitions is indicated on the left-hand TDP. (E) (Top) Kinetic scheme defining the rates of transition between FRET states. (Bottom) Rates of transition for unbound and ACE2-bound SΔTM D614 determined from HMM analysis of the individual smFRET traces. Rate constants are presented as the mean ± standard error determined from the same populations of smFRET traces used to construct the FRET histograms. (F) FRET histograms for the unbound and ACE2-bound SΔTM D614G spike, displayed as in (B). (G) Violin plots indicating FRET state occupancies for the SΔTM D614G spike, displayed as in (C). (H) TDPs for the SΔTM D614G spike, displayed as in (D). (I) Rate constants for the unbound and ACE2-bound SΔTM D614G spike, displayed as in (E). Numeric data are provided in Figure 3—source data 1.
-
Figure 3—source data 1
Matlab figure files containing numeric data for Förster resonance energy transfer (FRET) histograms, violin plots, and transition density plots (TDPs), and numeric kinetics data.
- https://cdn.elifesciences.org/articles/75433/elife-75433-fig3-data1-v2.zip
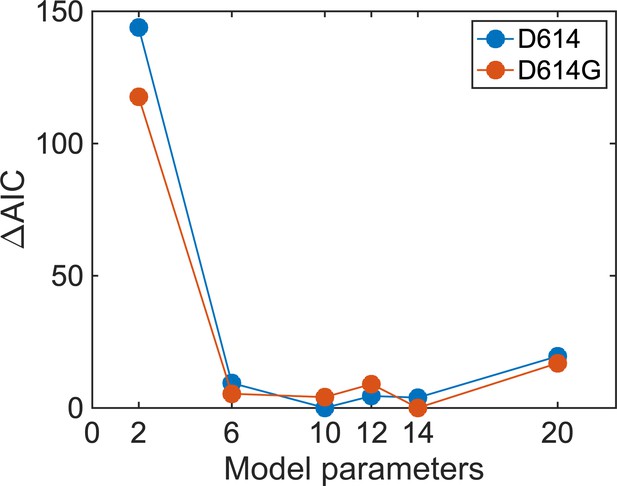
Selection of a model for analysis of single-molecule Förster resonance energy transfer (smFRET) trajectories.
The Akaike information criterion ( AIC) values were calculated from the maximized log-likelihoods per trace determined using the maximum point likelihood (MPL) algorithm. Model fitness was evaluated using both the SΔTM D614 and D614G data sets, which yielded equivalent results. A circular model with two non-zero FRET states and a 0-FRET state (three states in total with six rate constants) was selected as the simplest model that adequately represented the data. AIC values were not further reduced, indicating no improvement in model fitness, upon consideration of models with greater numbers of parameters.
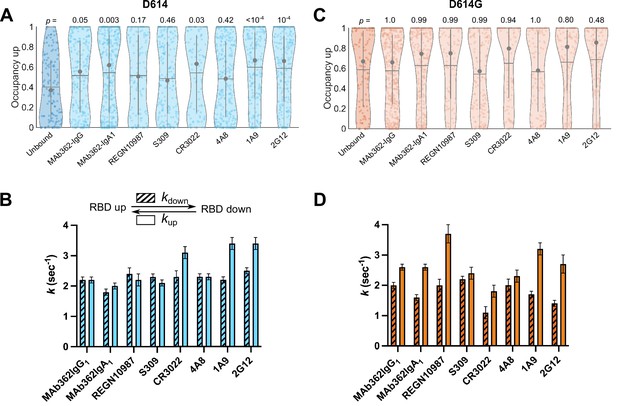
Antibodies directly and allosterically modulate SΔTM receptor-binding domain (RBD) conformation.
(A) The RBD-up conformation occupancy (low-Förster resonance energy transfer [FRET] state) determined through Hidden Markov modeling (HMM) analysis for SΔTM D614 in the absence or presence of the indicated monoclonal antibodies (mAbs). Occupancy data are presented as violin plots as in Figure 3. The indicated p-values were determined by comparing mAb-bound to unbound SΔTM through one-way ANOVA. (B) (Top) Kinetic scheme defining the rates of transition between RBD-up and -down conformations. (Bottom) Rates of transition for SΔTM D614 in the presence of mAbs determined through HMM analysis of the single-molecule FRET (smFRET) traces. Rate constants are presented as mean ± standard error determined from three technical replicates. (C) RBD-up conformation occupancy data for SΔTM D614G displayed as in (A). (D) Kinetic data for SΔTM D614G displayed as in (B). Corresponding FRET histograms for each mAb-bound SΔTM trimer is shown in Figure 4—figure supplement 1. Data are shown numerically in Tables 2 and 3 and provided in Figure 4—source data 1.
-
Figure 4—source data 1
Matlab figure files containing numeric data for violin plots and numeric kinetics data.
- https://cdn.elifesciences.org/articles/75433/elife-75433-fig4-data1-v2.zip
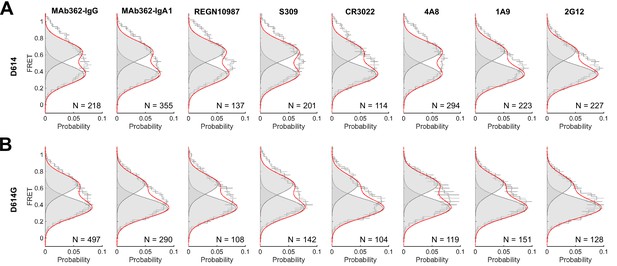
Förster resonance energy transfer (FRET) histograms for SΔTM in the presence of monoclonal antibodies (mAbs).
Data are shown for both the D614 and D614G spikes in the presence of the indicated mAbs and are presented exactly as in Figure 3. Raw data are provided in Figure 4—figure supplement 1—source data 1.
-
Figure 4—figure supplement 1—source data 1
Matlab figure files contains numeric Förster resonance energy transfer (FRET) histogram data.
- https://cdn.elifesciences.org/articles/75433/elife-75433-fig4-figsupp1-data1-v2.zip
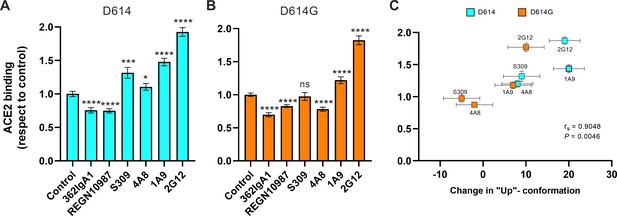
Allosteric modulation of the receptor-binding domain (RBD) position promotes angiotensin-converting enzyme 2 (ACE2) binding.
(A) Binding of ACE2 by (A) SΔTM D614 or (B) D614G spikes pre-incubated with the indicated monoclonal antibodies (mAbs) was measured by fluorescence correlation spectroscopy (FCS) as described in Materials and methods. Data are presented as the average of two independent experiments, each consisting of 20–25 10 s acquisitions. Statistical significance was evaluated through a two-tailed, unpaired Mann-Whitney test as indicated in Materials and methods. p-Values < 0.05 were considered significant and significance values are indicated as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (C) The change in the RBD-up conformation of SΔTM spikes pre-incubated with the indicated mAbs exhibited a positive correlation with the binding of ACE2 determined through FCS. Statistical significance (p = 0.0046) was found when Spearman test was performed with the 95% level of confidence (α = 0.05). Raw data are provided in Figure 5—source data 1.
-
Figure 5—source data 1
Numeric angiotensin-converting enzyme 2 (ACE2)-bound fraction data, and numeric change in receptor-binding domain (RBD)-up conformation data.
- https://cdn.elifesciences.org/articles/75433/elife-75433-fig5-data1-v2.zip
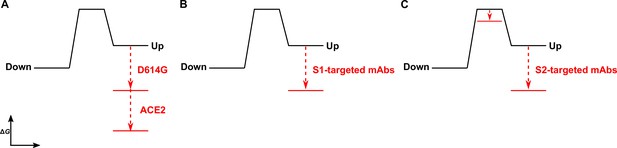
The D614G mutation and ligands modulate the S energetic landscape.
(A) The D614G mutation and angiotensin-converting enzyme 2 (ACE2) have additive effects on the thermodynamic stabilization of the receptor-binding domain (RBD)-up conformation. (B) The predominant effect of monoclonal antibodies (mAbs) that target the S1 domain, either the RBD (MAb362, REGN10987, S309, CR3022) or N-terminal domain (NTD) (4A8), is to stabilize the RBD-up conformation. (C) mAbs that target the S2 domain have a more complex allosteric effect, resulting in stabilization of the RBD-up conformation coupled to reduction in the activation energy for transition from the RBD-down to the -up conformation.
Tables
Fluorescence lifetime and anisotropy measurements.
Data are presented as the mean ± standard error determined from three technical replicates. ACE2, angiotensin-converting enzyme 2; ND, not determined.
| Lifetime (ns) | Anisotropy | |
|---|---|---|
| LD550 | 1.167 ± 0.004 | 0.143 ± 0.002 |
| SΔTM-LD550 | 1.463 ± 0.006 | 0.101 ± 0.003 |
| SΔTM-LD550 + ACE2 | 1.523 ± 0.008 | 0.093 ± 0.004 |
| LD650 | ND | 0.140 ± 0.001 |
| SΔTM-LD650 | ND | 0.258 ± 0.004 |
| SΔTM-LD650 + ACE2 | ND | 0.247 ± 0.003 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Cricetulus griseus) | ExpiCHO-S | Gibco, Thermo Fisher Scientific (Waltham, MA) | Cat. No. A29127 | |
| Cell line (Homo sapiens) | Expi293F | Gibco, Thermo Fisher Scientific (Waltham, MA) | Cat. No. A14527 | |
| Antibody | MAb362-IgG1(Human monoclonal) | PMID:32826914 | ELISA (600 nM),FCS (600 nM), smFRET (see Materials and methods section ‘smFRET imaging and data analysis’). | |
| Antibody | MAb362-IgA1(Human monoclonal) | PMID:32826914 | ELISA (600 nM),FCS (600 nM), smFRET (see Materials and methods section ‘smFRET imaging and data analysis’). | |
| Antibody | REGN10987(Mouse monoclonal) | This work and PMID:32540901 | See Materials and methods section ‘Antibodies’.ELISA (600 nM),FCS (600 nM), smFRET (see Materials and methods section ‘smFRET imaging and data analysis’). | |
| Antibody | S309(Human monoclonal) | This work andPMID:32422645 | See Materials and methods section ‘Antibodies’.ELISA (600 nM),FCS (600 nM), smFRET (see Materials and methods section ‘smFRET imaging and data analysis’). | |
| Antibody | CR3022(Human monoclonal) | This work and PMID:16796401 | See Materials and methods section ‘Antibodies’.ELISA (600 nM),FCS (600 nM), smFRET (see Materials and methods section ‘smFRET imaging and data analysis’). | |
| Antibody | 2G12(Human monoclonal) | This work | See Materials and methods section ‘Antibodies’.ELISA (600 nM),FCS (600 nM), smFRET (see Materials and methods section ‘smFRET imaging and data analysis’). | |
| Antibody | 4A8(Human monoclonal) | BioVision (Milpitas, CA) | Cat. No. A2269-100 | ELISA (600 nM),FCS (600 nM), smFRET (see Materials and methods section ‘smFRET imaging and data analysis’). |
| Antibody | 1A9(Mouse monoclonal) | GeneTex (Irvine, CA) | Cat. No. GTX632604 | ELISA (600 nM),FCS (600 nM),SmFRET: see Materials and methods section ‘smFRET imaging and data analysis’,WB (1:2000). |
| Antibody | Anti-6x-His-tag(Rabbit polyclonal) | Invitrogen (Waltham, MA) | Cat. No. PA1-983B | WB (1:2000). |
| Antibody | HRP-conjugated anti-mouse IgG Fc(Rabbit polyclonal) | Invitrogen (Waltham, MA) | Cat. No. 31455 | ELISA (1:5000), WB (1:5000). |
| Antibody | HRP-conjugated anti-human IgG Fc(Goat polyclonal) | Invitrogen (Waltham, MA) | Cat. No. A18823 | ELISA (1:10,000), WB (1:10,000). |
| Antibody | HRP-conjugated anti-human kappa(Goat polyclonal) | SouthernBiotech (Birmingham, AL) | Cat. No. 2060–05 | ELISA (1:4000). |
| Antibody | HRP-conjugated anti-rabbit IgG(Goat polyclonal) | Abcam (Cambridge, UK) | ab205718 | WB (1:50,000). |
| Recombinant DNA reagent | pcDNA3.1_ REGN10987-heavy_chain | This work and PMID:32540901 | See Materials and methods section ‘Antibodies’. | |
| Recombinant DNA reagent | pcDNA3.1_ REGN10987-light_chain | This work and PMID:32540901 | See Materials and methods section ‘Antibodies’. | |
| Recombinant DNA reagent | pcDNA3.1_S309-heavy_chain | This work andPMID:32422645 | See Materials and methods section ‘Antibodies’. | |
| Recombinant DNA reagent | pcDNA3.1_S309-light_chain | This work andPMID:32422645 | See Materials and methods section ‘Antibodies’. | |
| Recombinant DNA reagent | pcDNA3.1_CR3022-heavy_chain | This work and PMID:16796401 | See Materials and methods section ‘Antibodies’. | |
| Recombinant DNA reagent | pcDNA3.1_CR3022-light_chain | This work and PMID:16796401 | See Materials and methods section ‘Antibodies’. | |
| Recombinant DNA reagent | Plasmid_2G12-heavy_chain | Peter D Kwong laboratory | ||
| Recombinant DNA reagent | Plasmid_2G12-light_chain | Peter D Kwong laboratory | ||
| Recombinant DNA reagent | pcDNA3.1 SARS-CoV-2 SΔTM (plasmid) | This work.GenScript (Piscataway, NJ) | See Materials and methods section ‘Plasmids and site-directed mutagenesis’. | |
| Recombinant DNA reagent | pcDNA3.1 SARS-CoV-2 SΔTM 161/345A4 double-tagged (plasmid) | This work | See Materials and methods section ‘Plasmids and site-directed mutagenesis’. | |
| Recombinant DNA reagent | pcDNA3.1 SARS-CoV-2 SΔTM D614G (plasmid) | This work | See Materials and methods section ‘Plasmids and site-directed mutagenesis’. | |
| Recombinant DNA reagent | pcDNA3.1 SARS-CoV-2 SΔTM D614G 161/345A4 double-tagged (plasmid) | This work | See Materials and methods section ‘Plasmids and site-directed mutagenesis’. | |
| Recombinant DNA reagent | pCAGGS-ACE2-his (plasmid) | PMID:32991842 | Addgene No. 158089 (Watertown, MA) | |
| Peptide, recombinant protein | SARS-CoV-2 SΔTM | This work | See Materials and methods section ‘Protein expression and purification’. | |
| Peptide, recombinant protein | SARS-CoV-2 SΔTM 161/345A4 double-tagged | This work | See Materials and methods section ‘Protein expression and purification’. | |
| Peptide, recombinant protein | SARS-CoV-2 SΔTM D614G | This work | See Materials and methods section ‘Protein expression and purification’. | |
| Peptide, recombinant protein | SARS-CoV-2 SΔTM D614G 161/345A4 double-tagged | This work | See Materials and methods section ‘Protein expression and purification’. | |
| Peptide, recombinant protein | ACE2 | This work | See Materials and methods section ‘Protein expression and purification’. | |
| Peptide, recombinant protein | Acyl carrier protein synthase (AcpS) | PMID:31952255 | ||
| Commercial assay, kit | Q5 Site-Directed Mutagenesis Kit | New England Biolabs(Ipswich, MA) | Cat. No. E0554S | |
| Commercial assay, kit | Coomassie Plus (Bradford) Assay Kit | Thermo Fisher Scientific (Waltham, MA) | Cat. No. 23,236 | |
| Commercial assay, kit | SuperSignal West Pico PLUS Chemiluminescent Substrate | Thermo Scientific (Waltham, MA) | Cat. No. 34580 | |
| Commercial assay, kit | 1-Step Ultra TBM-ELISA | Thermo Scientific (Waltham, MA) | Cat. No. 34028 | |
| Chemical compound, drug | Cy5-conjugated n-hydroxysuccinimide ester | Cytiva (Marlborough, MA) | Cat. No. PA15100 | |
| Chemical compound, drug | Coenzyme A (CoA)-conjugated LD550 fluorophore | Lumidyne Technologies (New York, NY), | Cat. No. LD550-CS | |
| Chemical compound, drug | Coenzyme A (CoA)-conjugated LD650 fluorophore | Lumidyne Technologies (New York, NY), | Cat. No. LD650-CS | |
| Software, algorithm | Micromanager | PMID:25606571micro-manager.org | v2.0 | |
| Software, algorithm | SPARTAN | https://www.scottcblanchardlab.com/software and PMID:26878382 | Version 3.7 | |
| Software, algorithm | Matlab (Mathworks, Natick, MA) | Mathworks (Natick, MA) | Version R2018b | |
| Software, algorithm | Maximum point likelihood algorithm | PMID:11023897 | ||
| Software, algorithm | GraphPad Prism | GraphPad Software(San Diego, CA) | Version 9.2.0 | |
| Software, algorithm | PyMOL software | The PyMOL Molecular Graphic System, Schrödinger Inc(New York, NY) | Version 2.0.7 | |
| Other | Pierce Protein G Agarose | Thermo Fisher Scientific (Waltham, MA) | Cat. No. 20398 | |
| Other | Ni-NTA Agarose | Invitrogen (Waltham, MA) | Cat. No. R901-15 | |
| Other | Superdex 200 Increase 10/300 GL column | GE Healthcare (Chicago, IL) | Cat. No. 28990944 | |
| Other | Gel Filtration Standard | Bio-Rad (Hercules, CA) | Cat. No. 1511901 | |
| Other | Synergy H1 microplate reader | BioTek (Winooski, VT) | ||
| Other | Typhoon 9,410variable mode imager | GE Amersham Biosciences (Amersham, UK) | ||
| Other | CorTector SX100 instrument | LightEdge Technologies(Beijing, China) | ||
| Other | QuantaMaster 400 fluorimeter | Horiba (Kyoto, Japan) |
Förster resonance energy transfer (FRET)-state occupancies and rate constants for SΔTM D614.
Data are presented at mean ± standard error determined from the total population of traces analyzed.
| SARS-CoV-2 spikes D614 | FRET-state occupancies (%) | Rate constants (s–1) | ||
|---|---|---|---|---|
| Low-FRET (0.35)RBD-up conformation | High-FRET (0.65)RBD-down conformation | kdown(Low to high FRET) | kup(High to low FRET) | |
| Unbound | 40 ± 3 | 60 ± 3 | 3.8 ± 0.2 | 2.6 ± 0.2 |
| + ACE2 | 66 ± 3 | 34 ± 3 | 1.3 ± 0.1 | 2.2 ± 0.2 |
| + MAb362IgG1 | 52 ± 2 | 48 ± 2 | 2.2 ± 0.1 | 2.2 ± 0.1 |
| + MAb362IgA1 | 54 ± 2 | 46 ± 2 | 1.8 ± 0.1 | 2.0 ± 0.1 |
| + REGN10987 | 52 ± 3 | 48 ± 3 | 2.4 ± 0.2 | 2.2 ± 0.2 |
| + S309 | 49 ± 3 | 51 ± 3 | 2.3 ± 0.1 | 2.1 ± 0.1 |
| + CR3022 | 54 ± 4 | 46 ± 4 | 2.3 ± 0.2 | 3.1 ± 0.2 |
| + 4A8 | 48 ± 2 | 52 ± 2 | 2.3 ± 0.1 | 2.3 ± 0.1 |
| + 1A9 | 60 ± 2 | 40 ± 2 | 2.2 ± 0.1 | 3.4 ± 0.2 |
| + 2G12 | 59 ± 2 | 41 ± 2 | 2.5 ± 0.1 | 3.4 ± 0.2 |
Förster resonance energy transfer (FRET)-state occupancies and rate constants for SΔTM D614G.
Data are presented at mean ± standard error determined from the total population of traces analyzed.
| SARS-CoV-2 spikes D614G | FRET-state occupancies (%) | Rate constants (s–1) | ||
|---|---|---|---|---|
| Low-FRET (0.35)RBD-up conformation | High-FRET (0.65)RBD-down conformation | kdown(Low to high FRET) | kup(High to low FRET) | |
| Unbound | 59 ± 3 | 41 ± 3 | 1.6 ± 0.2 | 2.0 ± 0.2 |
| + ACE2 | 69 ± 4 | 31 ± 4 | 0.7 ± 0.1 | 2.0 ± 0.4 |
| + MAb362IgG1 | 58 ± 2 | 42 ± 2 | 2.0 ± 0.1 | 2.6 ± 0.1 |
| + MAb362IgA1 | 63 ± 2 | 37 ± 2 | 1.6 ± 0.1 | 2.6 ± 0.1 |
| + REGN10987 | 63 ± 4 | 37 ± 4 | 2.0 ± 0.2 | 3.7 ± 0.3 |
| + S309 | 54 ± 3 | 46 ± 3 | 2.2 ± 0.1 | 2.4 ± 0.2 |
| + CR3022 | 65 ± 4 | 35 ± 4 | 1.1 ± 0.2 | 1.8 ± 0.2 |
| + 4A8 | 57 ± 3 | 43 ± 3 | 2.0 ± 0.2 | 2.3 ± 0.2 |
| + 1A9 | 66 ± 3 | 34 ± 3 | 1.7 ± 0.1 | 3.2 ± 0.2 |
| + 2G12 | 69 ± 3 | 31 ± 3 | 1.4 ± 0.1 | 2.7 ± 0.3 |






