Structural basis for an unprecedented enzymatic alkylation in cylindrocyclophane biosynthesis
Figures
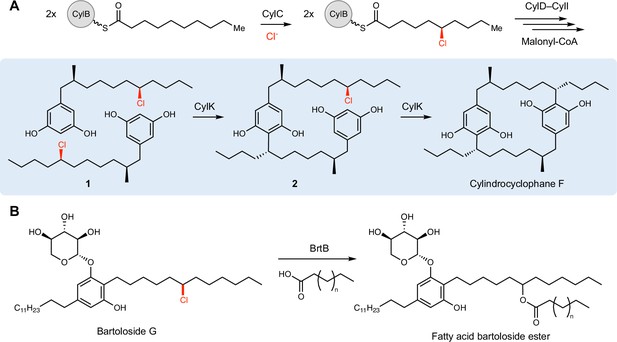
CylK and related enzymes use alkyl chloride substrates as biosynthetic intermediates.
(A) The halogenase CylC generates an alkyl chloride substrate for CylK, which catalyzes two stepwise Friedel–Crafts alkylations to construct a paracyclophane macrocycle in cylindrocyclophane biosynthesis. This involves an intermolecular reaction between two resorcinol-containing alkyl chloride substrates (1) to generate intermediate (2), followed by an intramolecular alkylation to afford cylindrocyclophane F. (B) The related enzyme BrtB (30% amino acid identity, 46% similarity) catalyzes an analogous but chemically distinct C–O bond-forming event between chlorinated bartolosides and fatty acid nucleophiles.

The CylK X-ray crystal structure reveals a distinct arrangement of two protein domains.
(A) An overall view of the structure of CylK. In this image and throughout, calcium ions are shown as blue spheres, magnesium ions as dark gray spheres, and chloride ions as green spheres. The N-terminal domain is depicted as a light gray ribbon diagram, and the C-terminal domain is shown as a pink ribbon diagram. (B) The N-terminal domain contains a right-handed parallel β-roll stabilized by three Ca2+ ions. The structure is capped by a three-strand antiparallel β-sheet and buttressed by additional helical secondary structures. Insets show the Ca2+ ion coordination environment within the β-roll. (C) A top-down view of the seven-bladed β-propeller C-terminal domain. The propeller blades are numbered from the N-terminal end of the domain and colored by blade. The inset shows a representative view of the Ca2+ coordination environment. A similar binding site exists at each blade junction. (D) Cavity mapping analysis (Ho and Gruswitz, 2008) of CylK reveals two cavities large enough to accommodate the alkyl resorcinol substrates.

Topology diagram of the N-terminal CylK β-roll domain, residues 7–251.
Ca2+ ions are shown as blue circles, residues implicated in catalysis are highlighted in green ovals, Ca2+-coordinating residues are shown in black, and Ca2+-coordinating residues that follow canonical repeat-in-toxin (RTX) motifs are shown in blue italics.

Comparison of the N-terminal CylK β-roll domain to similar C-terminal repeat-in-toxin (RTX) domains in other enzymes.
(A) The N-terminal domain of CylK (residues 7–251). (B) The C-terminal region of RTX-motif enzyme psychrophilic alkaline metalloprotease (PAP) that structurally aligns with the β-roll of CylK (residues 326–461) (PDB ID 1OMJ). (C) Some CylK Ca2+-coordinating loops (yellow, residues 131–152) extend beyond the tight RTX-motif turn (green, PAP residues 330–335, 348–353, and 366–371). (D) The Ca2+-binding β-roll motif in PAP is located far away from the active site of the enzyme (box) and is likely not involved in catalysis (Ravaud et al., 2003). In all panels, Ca2+ ions are represented as blue spheres. Mg2+ (CylK) and Zn2+ (PAP) ions are shown as gray spheres.
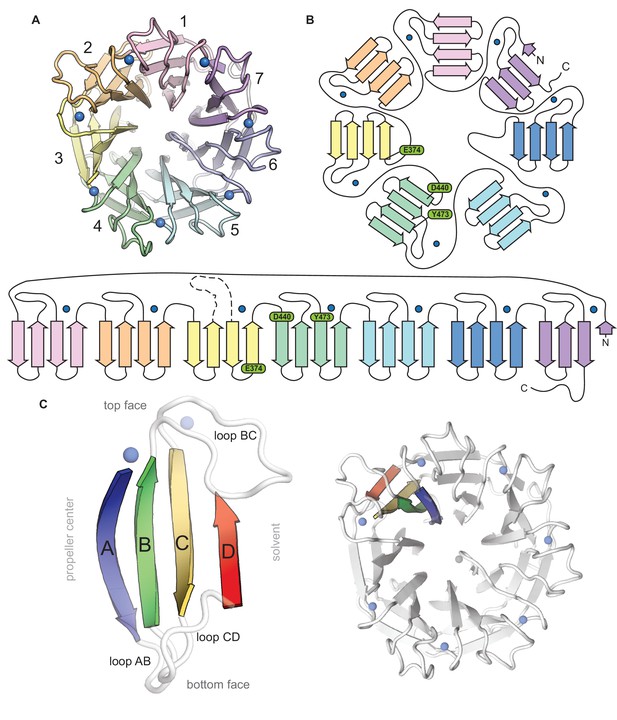
Topological and structural features of the C-terminal CylK β-propeller domain.
(A) Top-down view of the C-terminal β-propeller domain (residues 252–662). (B) Topology diagrams of the CylK β-propeller domain. Blades are colored as in (A), Ca2+ ions shown as blue circles. Residues implicated in catalysis are shown in green. (C) Example of a propeller blade (left) and the orientation of the blade within the β-propeller fold (right). β-strands within each blade are lettered A–D, with A corresponding to the innermost strand, closest to the central tunnel. Loops are labeled to indicate the strands they connect. For example, loop BC connects blade B and C within the blade while loop DA connects strand D of one blade to strand A of the next blade.
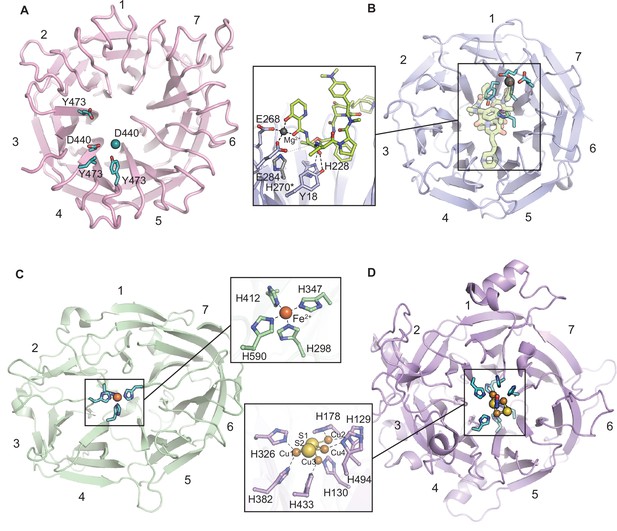
Comparison of the proposed CylK active site location to those of other characterized seven-bladed β-propeller enzymes.
Panels show a top-down ribbon diagram of β-propeller domains. Residues implicated in catalysis are shown as cyan sticks. (A) The C-terminal domain of CylK. Br- ions are shown as teal spheres, and Ca2+ ions are shown as transparent blue spheres. (B) Virginiamycin B lyase (VrgB) complexed with the substrate analog, quinupristin (lime green sticks) (PDB ID 2Z2P). Mg2+ ions are shown as gray spheres. *H270 modeled as in PDB ID 2Z2O. (C) Carotenoid cleavage dioxygenase (CCD) (PDB ID 3NPE). Fe2+ ions are shown as orange spheres. (D) Nitrous oxide reductase (NOR) complexed with N2O (blue and red sticks) (PDB ID 3SBR) and the active site Cu-S cluster. Cu ions are shown as orange spheres, and sulfur atoms are shown as yellow spheres.

Solvent accessibility and properties of the cavity located in between the C-terminal β-propeller domain and the N-terminal domain in CylK.
The walls of this cavity contain (A) charged (red/blue) and polar (purple) residues and (B) hydrophobic patches (red), ideal for interaction with the amphipathic resorcinol.
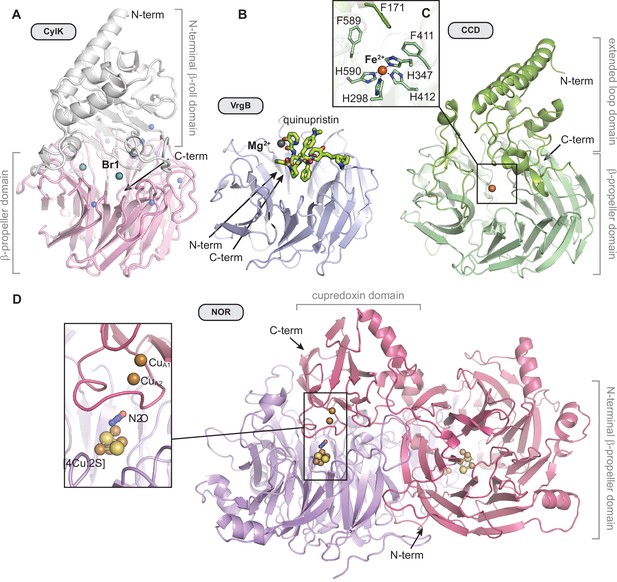
A comparison of the overall structures of CylK and selected seven-bladed β-propeller enzymes.
(A) CylK is a fusion of a Ca2+-binding β-roll fold and a β-propeller with the active site located at the domain interface. (B) Virginiamycin B lyase (VrgB) is a single-domain enzyme in which the active site is located on the top face of the propeller. Quinupristin substrate analog shown as green sticks (PDB ID 2Z2P). (C) The extended loops of carotenoid cleavage dioxygenase (CCD) form a helical domain on the top face of the propeller, oriented similarly to the N-terminal domain of CylK. The active site, delineated by the location of the iron cofactor (orange sphere), is located at the interface of these two domains (PDB ID 3NPE). (D) Dimeric nitrous oxide reductase (NOR) contains an active site at the interface between a C-terminal cupredoxin electron transfer domain (CuA) contributed by the other monomer and the β-propeller catalytic domain (inset) (PDB ID 3SBR). Unless otherwise noted, ions are shown as spheres and colored as in Figure 2—figure supplement 2.

The active site of CylK is located at the domain interface.
(A) Selected amino acids in CylK near cavities 1 and 2 subjected to mutant scanning are shown in stick format. Side chains shown in yellow near cavity 1 correspond to sites found to be essential for activity. (B) Non-native substrate pair used in CylK activity assays and product of the alkylation reaction (SNAc = N-acetylcysteamine). (C) Screen of mutant activity to locate the CylK active site. Product formation was measured by liquid chromatography-mass spectrometry (LC-MS), error bars represent the standard deviation from the mean of three biological replicates.
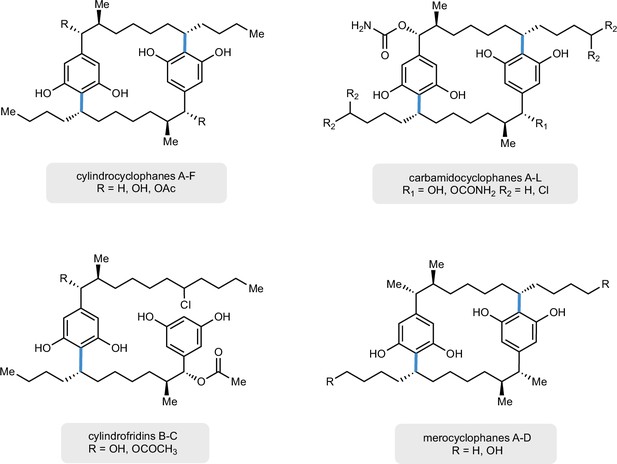
Structurally related, cylindrocyclophane-like natural products produced by cyanobacteria with related biosynthetic gene clusters and CylK homologs, namely, merocyclophanes (Nostoc sp. UIC 10110, MerH), cylindrofridins (Cylindrospermum stagnale PCC 7417, CylK), and carbamidocyclophanes (Nostoc sp. CAVN2, CabK).
The bonds highlighted in blue are hypothesized to be constructed by their respective CylK.
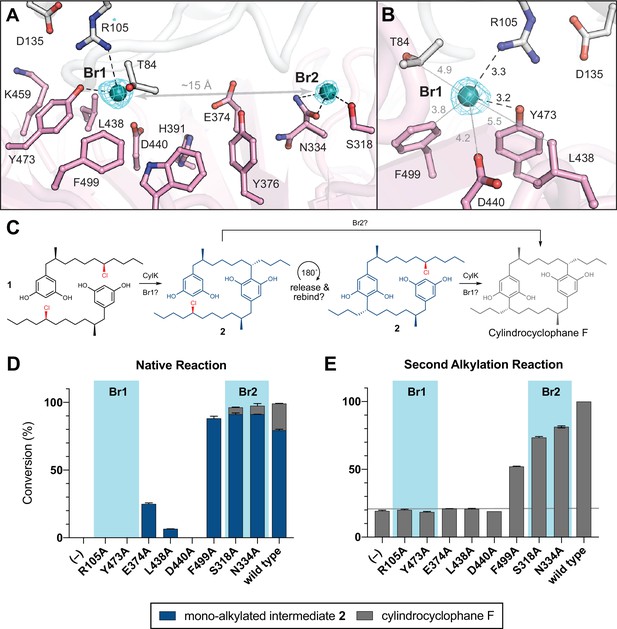
NaBr soak and mutagenesis of bromide-binding residues suggest a single site for catalysis.
(A) A soak of NaBr into CylK crystals reveals two strong peaks in the anomalous difference electron density map (cyan mesh, contoured at 5.0 σ) within cavity 1. Bromide ion 1 (Br1) and 2 (Br2) are shown as teal spheres, and selected amino acids in the vicinity of each site are shown in stick format. The C-terminal domain is colored white, and the N-terminal domain is colored pink. (B) Alternate view of Br1 with anomalous difference electron density map, polar contacts shown as black dashed lines, and other distances shown as gray lines. All distances measured in angstroms. (C) Proposed alkylation scheme with potential roles for two bromide-binding sites in paracyclophane formation, or invoking release and rebinding of monoalkylated intermediate 2 for alkylation at a single bromide site. (D) End point activity at 1 hr of select mutants performing the native reaction with substrate 1, highlighting the residues associated with Br1 or Br2. Product formation was quantified by high-performance liquid chromatography (HPLC), error bars represent the standard deviation from the mean of two biological replicates. (E) End point activity at 22 hr of select mutants performing the second alkylation reaction with intermediate 2.
-
Figure 4—source data 1
Western blot of Strep-tagged CylK mutants (from left to right): R105A, R105K, S318A, N334A, E374A, L438A; and serial dilutions of purified wild-type CylK: 2.0, 0.5, and 0.1 µM.
- https://cdn.elifesciences.org/articles/75761/elife-75761-fig4-data1-v2.zip
-
Figure 4—source data 2
Western blot of Strep-tagged CylK mutants (from left to right): D440A, D440N, Y473A, Y473F, F499A, wild-type; and serial dilutions of purified wild-type CylK: 2.0, 0.5, and 0.1 µM.
- https://cdn.elifesciences.org/articles/75761/elife-75761-fig4-data2-v2.zip
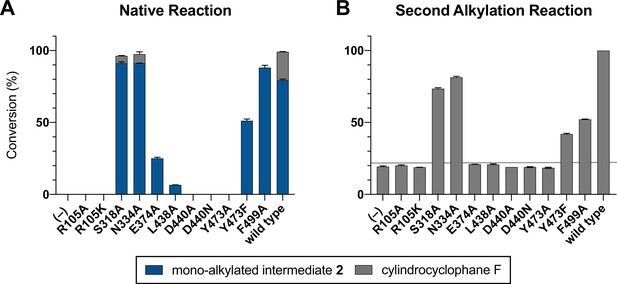
Mutagenesis of active site cleft.
(A) End point activity at 1 hr of select mutants performing the native reaction with substrate 1. Product formation was quantified by high-performance liquid chromatography (HPLC), error bars represent the standard deviation from the mean of two biological replicates. (B) End point activity at 22 hr of select mutants performing the second alkylation reaction with intermediate 2.

Western blotting of Strep-tagged CylK mutants from soluble lysate with anti-Strep-horseradish peroxidase (HRP, IBA) in order to ensure mutant enzyme expression and solubility.
Serial dilution of purified CylK-Strep included to visualize protein concentration range.
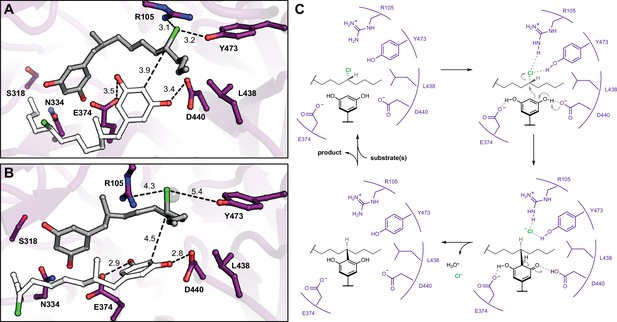
Molecular dynamics simulations reveal roles for key catalytic residues and enable a mechanistic proposal.
(A) An energy-minimized docking model of CylK in complex with two chlorinated alkylresorcinol molecules in cavity 1 before and (B) after unrestrained molecular dynamics simulation. This analysis shows that both substrates can be accommodated while maintaining contact with one another and essential catalytic residues. Substrate molecules and selected amino acid side chains shown in stick format. (C) Proposed mechanism for a single cycle of the CylK-catalyzed Friedel–Crafts alkylation with key residues illustrated.
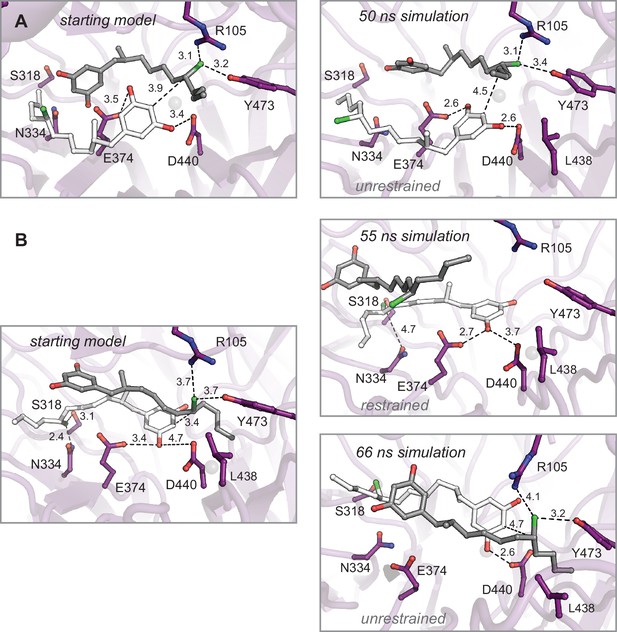
Molecular dynamics simulations of enzyme–substrate complexes manually docked into interdomain cavity support active site assignment in this region of CylK.
(A) An energy-minimized starting model before (left) and after restrained molecular dynamics simulation (right). The complex was generated using Br1 to place one equivalent of substrate 1 (gray). This complex also maximizes the interaction between the resorcinol of a second substrate molecule (white) and amino acids D440 and E374 in the propeller domain. Results from an unrestrained simulation are shown in the main text (Figure 4D and E). (B) Restrained (top right) and unrestrained (bottom right) simulations resulting from a starting model (left) that prioritized Br2 site interactions. Select residues shown as dark purple sticks, distances shown as dashed lines and measured in angstroms.

Molecular dynamics simulations of intermediate product 2 complexes manually docked into interdomain cavity reveal a halide-binding pocket involving R105 and Y473.
Energy-minimized starting models before (left) and after simulation (right). (A) Restrained and (B) unrestrained simulations of the intermediate product complex. (C) Restrained and (D) unrestrained simulations of the intermediate product complex in an alternate starting conformation. Select residues shown as dark purple sticks, product intermediate shown as light purple sticks, and distances shown as dashed lines and measured in angstroms.
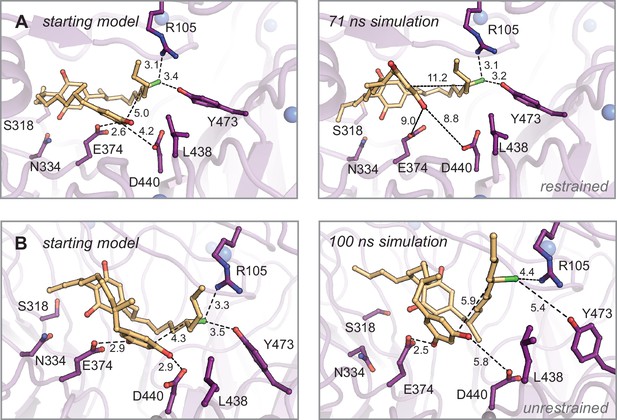
Molecular dynamics simulations of monoalkylated intermediate 2 positioned for the second alkylation reaction.
Energy-minimized starting models before (left) and after simulation (right). (A) Restrained and (B) unrestrained simulations of (A). Select residues shown as dark purple sticks, monoalkylated intermediate 2 shown as orange sticks, and distances shown as dashed lines and measured in angstroms.

Comparison of bound alkyl halides and their respective enzyme active sites.
(A) 5′-Chloro-5′-deoxyadenosine (5’-ClDA) and methionine (Met) products bound in the active site of wild-type SalL SAM-dependent chlorinase (PDB ID 2Q6I). The alkyl chloride forms apparent polar interactions (dashed lines) with the backbone amides of Gly 131 and Tyr 130; and nonpolar interactions (solid lines) with Tyr 72 and Thr 75. (B) 5′-fluoro-5′-deoxyadenosine (5’-FDA) and Met products bound in the active site of wild-type FlA SAM-dependent fluorinase (PDB ID 1RQR). The alkyl fluoride forms apparent polar interactions with the backbone amides of Ser 158 and Tyr 157, and the side chain of Ser 158; and nonpolar interactions with Phe 156 and Leu 17. (C) 1,2-Dichloroethane (DCE) substrate bound in the active side of wild-type haloalkane dehalogenase (PDB ID 2DHC). The alkyl chloride forms apparent polar interactions with Trp 125, 175, and the backbone amide of Pro 223; and nonpolar interactions with Phe 172 and Val 226. Asp 124 is the nucleophilic residue that displaces chloride. (D) Bromide ion product analog bound in the active site of CylK. The Br- ion forms polar interactions with Arg 105 and Tyr 473; and nonpolar interactions with Thr 84 and Phe 499. Select amino acid residues are represented in stick format, bromide ions are shown as teal spheres, calcium ions are shown as blue spheres, alkyl chloride in green, and alkyl fluoride in light aqua.

CylK homologs with partner CylC halogenases contain the proposed alkyl chloride-activating residues.
Maximum-likelihood phylogenetic tree of protein sequences homologous to CylK and BrtB that contain both N- and C-terminal domains (>24% amino acid identity), highlighting the key conserved catalytic residues, clustered halogenases, and associated monoalkylresorcinol-forming enzymes. Dialkylresorcinol-forming enzymes (DarA/B-like) were not found except as expected with BrtB and the bartoloside-producing cluster, indicated as an unfilled green circle. Bootstrap values are shaded based on the legend. Natural products associated with select CylK homologs are displayed; the bonds highlighted in blue are constructed by their respective CylK.
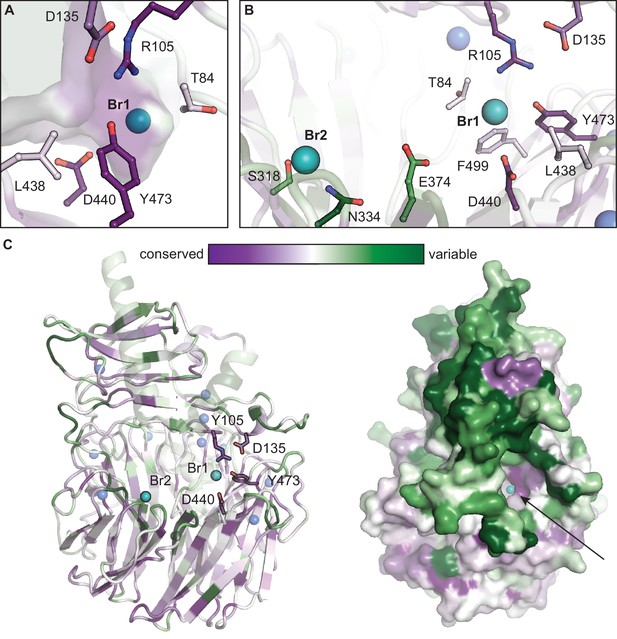
Conservation mapping analysis of functionally annotated CylK homologs.
Representations of sequence conservation generated in ConSurf (Ashkenazy et al., 2016) using a subset of cyanobacterial CylK homologs that have R105 and Y473 (48 unique sequences). Of those homologous enzymes, 37 contain a partner CylC halogenase in the genome of the encoding organism (~80% are co-localized with CylK). (A) The halide-binding pocket in CylK homologs with R105 and Y473. (B) Residues surrounding Br1 are generally conserved in this subset, while Leu 438, Glu 374, and other active site residues are not highly conserved, potentially suggesting a diversity of substrates used by these candidate enzymes. Residues surrounding Br2 are variable. (C) Ribbon and surface diagrams of overall sequence conservation in this subset of homologs. Selected amino acid residues are represented in stick format, bromide ions are shown as teal spheres, and calcium ions are shown as blue spheres. Arrow denotes the opening of the interdomain cavity proposed to contain the active site of CylK.
Tables
Data collection and refinement statistics for Tb-soaked C. licheniforme CylK structures.
| Tb anomalous | Native (final model) | |
|---|---|---|
| Data collection | ||
| Space group | C 2 2 21 | C 2 2 21 |
| Wavelength (Å) | 1.6314 | 0.97872 |
| Cell dimensions | ||
| a, b, c (Å) | 94.265, 139.286, 100.381 | 93.667, 138.613, 99.438 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50–2.38 (2.42–2.38) | 50.0–1.68 (1.71–1.68) |
| Rmerge | 0.218 (0.670) | 0.101 (1.326) |
| Rpim | 0.041 (0.192) | 0.026 (0.346) |
| I / σI | 25.2 (3.8) | 29.3 (2.15) |
| CC1/2 | 1.010 (0.906) | 0.999 (0.780) |
| Completeness (%) | 99.5 (99.9) | 99.8 (99.3) |
| Redundancy | 29.5 (11.4) | 14.8 (14.3) |
| Refinement | ||
| Resolution (Å) | 42.41–1.68 | |
| No. reflections | 68,476 | |
| Rwork/Rfree | 0.1556/0.1894 | |
| No. atoms | 5,292 | |
| Protein | 4,903 | |
| Ligand/ion | 50 | |
| Water | 339 | |
| B-factors (Å2) | ||
| Protein | 18.20 | |
| Ligand/ion | 28.54 | |
| Water | 25.82 | |
| RMS deviations | ||
| Bond lengths (Å) | 0.016 | |
| Bond angles (°) | 1.921 | |
| Molprobity clashscore | 2.29 (99th percentile) | |
| Rotamer outliers (%) | 0.78 | |
| Ramachandran favored (%) | 96.33 | |
| Ramachandran allowed (%) | 3.51 | |
| Ramachandran outlier *(%) | 0.16 | |
| PDB accession code | 7RON |
-
*
Values in parentheses are for highest-resolution shell.
Selected structural homologs of the N-terminal domain of CylK obtained by a structural comparison to other proteins in the PDB (Gáspári, 2020).
| Rank | PDB ID | Functional description | Z-score | RMSD | No. residues aligned | Total residues |
|---|---|---|---|---|---|---|
| 1 | 1OMJ-A | Psychrophilic alkaline metalloprotease (PAP); serralysin family | 11.2 | 6.8 | 135 | 456 |
| 2 | 2Z8Z-A | MIS38 lipase | 11.1 | 8.5 | 116 | 616 |
| 3 | 6SUS-A | RTX domain of blocks IV and V of adenylate cyclase toxin | 11.0 | 3.1 | 120 | 258 |
| 4 | 2QUA-A | LipA, extracellular lipase from Serratia marcescens | 10.6 | 2.3 | 110 | 615 |
| 5 | 2ML1-A | AlgE6R1, Mannuronan C5-epimerase | 10.6 | 3.0 | 122 | 153 |
Selected structural homologs of the C-terminal domain of CylK obtained by a structural comparison to other proteins in the PDB (Gáspári, 2020).
| Rank | PDB ID | Functional description | Z-score | RMSD | No. residues aligned | Total residues |
|---|---|---|---|---|---|---|
| 1 | 1SQ9-A | Antiviral protein Ski8 | 31.2 | 2.4 | 286 | 378 |
| 2 | 4H5J-B | Guanine nucleotide-exchange factor Sec12 | 31.1 | 2.9 | 300 | 347 |
| 3 | 5TF2-A | Prolactin regulatory element-binding protein | 31.0 | 2.7 | 291 | 338 |
| 4 | 4J0X-B | Ribosomal RNA-processing protein 9 | 30.9 | 2.4 | 290 | 366 |
| 5 | 2Z2O-C | Virginiamycin B lyase | 30.5 | 2.4 | 286 | 299 |
Conservation of CylK residues selected for mutagenesis experiments.
| Position | Residue(CylK, C. licheniforme) | Mutant | Residue (CabK) | Residue(CylK, C. stagnale) | Residue(MerH) | Residue(BrtB) |
|---|---|---|---|---|---|---|
| 78 | Lys | Glu | Lys | Lys | Lys | Lys |
| 96 | Asp | Ala | Asp | Asp | Asp | Asp |
| 105 | Arg | Ala, Lys | Arg | Arg | Arg | Arg |
| 473 | Tyr | Ala, | Tyr | Tyr | Tyr | Tyr |
| 265 | Ser | Ala | Ser | Ser | Ala | Ala |
| 266 | Ser | Ala | Ser | Ser | Ser | Thr |
| 267 | Thr | Ala | Thr | Thr | Thr | Ala |
| 324 | Ser | Ala | Ser | Ser | Ser | Ser |
| 377 | Lys | Ala | Arg | Lys | Arg | Gly |
| 379 | Thr | Ala | Thr | Thr | Thr | Ala |
| 502 | Asp | Ala | Asp | Asp | Asp | Asp |
| 504 | Glu | Ala | Glu | Glu | Glu | Ala |
| 558 | Leu | Ala | Leu | Leu | Leu | Met |
| 559 | Asp | Ala | Asp | Asp | Asp | Gly |
| 562 | Asp | Ala | His | Asp | Glu | Val |
| 616 | Thr | Ala | Thr | Thr | Thr | Gly |
| 618 | Thr | Ala | Thr | Thr | Ser | Ser |
| 374 | Glu | Ala | Glu | Glu | Glu | Arg |
| 438 | Leu | Ala | Leu | Leu | Leu | Leu |
| 440 | Asp | Ala, Asn | Asp | Asp | Asp | Glu |
| 499 | Phe | Ala | Phe | Phe | Phe | Tyr |
| 318 | Ser | Ala | Ser | Ser | Thr | Ile |
| 334 | Asn | Ala | Asn | Asn | Asn | Trp |
Data collection and refinement statistics for Br-soaked C. licheniforme CylK structures.
| Native | Br anomalous | |
|---|---|---|
| Data collection | ||
| Space group | C 2 2 21 | C 2 2 21 |
| Wavelength (Å) | 0.97872 | 0.9184 |
| Cell dimensions | ||
| a, b, c (Å) | 93.470, 138.656, 99.511 | 93.557, 138.726, 99.706 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0–1.52 (1.55–1.52) | 50–1.54 (1.57–1.54) |
| Rmerge | 0.076 (0.703) | 0.725 (4.755) |
| Rpim | 0.029 (0.295) | 0.149 (1.140) |
| I / σI | 23.3 (2.14) | 24.0 (2.06) |
| CC1/2 | 0.990 (0.843) | 0.978 (0.124) |
| Completeness (%) | 99.7 (98.9) | 100.0 (100.0) |
| Redundancy | 7.0 (5.5) | 24.2 (16.9) |
| Refinement | ||
| Resolution (Å) | 46.78–1.52 | |
| No. reflections | 89,122 | |
| Rwork / Rfree | 0.1693/0.1978 | |
| No. atoms | 5437 | |
| Protein | 4989 | |
| Ligand/ion | 45 | |
| Water | 403 | |
| B-factors (Å2) | ||
| Protein | 16.68 | |
| Ligand/ion | 23.03 | |
| Water | 23.77 | |
| RMS deviations | ||
| Bond lengths (Å) | 0.013 | |
| Bond angles (°) | 1.823 | |
| Molprobity clashscore | 1.64 (99th percentile) | |
| Rotamer outliers (%) | 0.76 | |
| Ramachandran favored (%) | 96.41 | |
| Ramachandran allowed (%) | 3.59 | |
| Ramachandran outlier (%) | 0.00 | |
| PDB accession code | 7ROO |
-
*Values in parentheses are for highest-resolution shell.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Cylindrospermum licheniforme ATCC 29412) | CylK | GenBank | ARU81125.1 | |
| Strain, strain background (Escherichia coli) | BL21(DE3) | Invitrogen | C6000-03 | |
| Strain, strain background (E. coli) | BL21 Gold CodonPlus (DE3) RIL | Agilent | 230245 | |
| Peptide, recombinant protein | Strep-Tactin HRP, anti-Strep-HRP (Streptomyces avidinii) | IBA Life Sciences | 2-1502-001 | (1:25,000) |
| Recombinant DNA reagent | pET-His6-Sumo-CylK-Strep (plasmid) | Schultz et al., 2019 | ||
| Recombinant DNA reagent | pPR-IBA1-CylK (plasmid) | Nakamura et al., 2017 | ||
| Recombinant DNA reagent | pPR-IBA1-CylK K78E (plasmid) | This work, constructed by GENEWIZ | Replaced AAG with GAG at position 232 | |
| Recombinant DNA reagent | pPR-IBA1-CylK D96A (plasmid) | This work, constructed by GENEWIZ | Replaced GAT with GCT at position 286 | |
| Recombinant DNA reagent | pPR-IBA1-CylK R105A (plasmid) | This work, constructed by GENEWIZ | Replaced CGT with GCC at position 313 | |
| Recombinant DNA reagent | pPR-IBA1-CylK Y473A (plasmid) | This work, constructed by GENEWIZ | Replaced TAT with GCT at position 1,417 | |
| Recombinant DNA reagent | pPR-IBA1-CylK S265A (plasmid) | This work, constructed by GENEWIZ | Replaced AGT with GCC at position 793 | |
| Recombinant DNA reagent | pPR-IBA1-CylK S266A (plasmid) | This work, constructed by GENEWIZ | Replaced TCC with GCC at position 796 | |
| Recombinant DNA reagent | pPR-IBA1-CylK T267A (plasmid) | This work, constructed by GENEWIZ | Replaced ACC with GCT at position 799 | |
| Recombinant DNA reagent | pPR-IBA1-CylK S324A (plasmid) | This work, constructed by GENEWIZ | Replaced AGT with GCT at position 970 | |
| Recombinant DNA reagent | pPR-IBA1-CylK K377A (plasmid) | This work, constructed by GENEWIZ | Replaced AAA with GCC at position 1129 | |
| Recombinant DNA reagent | pPR-IBA1-CylK T379A (plasmid) | This work, constructed by GENEWIZ | Replaced ACC with GCT at position 1135 | |
| Recombinant DNA reagent | pPR-IBA1-CylK D502A (plasmid) | This work, constructed by GENEWIZ | Replaced GAC with GCC at position 1504 | |
| Recombinant DNA reagent | pPR-IBA1-CylK E504A (plasmid) | This work, constructed by GENEWIZ | Replaced GAA with GCC at position 1510 | |
| Recombinant DNA reagent | pPR-IBA1-CylK L558A (plasmid) | This work, constructed by GENEWIZ | Replaced TTA with GCT at position 1672 | |
| Recombinant DNA reagent | pPR-IBA1-CylK D559A (plasmid) | This work, constructed by GENEWIZ | Replaced GAT with GCT at position 1675 | |
| Recombinant DNA reagent | pPR-IBA1-CylK D562A (plasmid) | This work, constructed by GENEWIZ | Replaced GAT with GCC at position 1684 | |
| Recombinant DNA reagent | pPR-IBA1-CylK T616A (plasmid) | This work, constructed by GENEWIZ | Replaced ACG with GCT at position 1846 | |
| Recombinant DNA reagent | pPR-IBA1-CylK T618A (plasmid) | This work, constructed by GENEWIZ | Replaced ACT with GCT at position 1852 | |
| Recombinant DNA reagent | pPR-IBA1-CylK D440A (plasmid) | This work, constructed by GENEWIZ | Replaced GAC with GCC at position 1318 | |
| Recombinant DNA reagent | pPR-IBA1-CylK L438A (plasmid) | This work, constructed by GENEWIZ | Replaced CTT with GCT at position 1312 | |
| Recombinant DNA reagent | pPR-IBA1-CylK E374A (plasmid) | This work, constructed by GENEWIZ | Replaced GAA with GCT at position 1120 | |
| Recombinant DNA reagent | pPR-IBA1-CylK F499A (plasmid) | This work, constructed by GENEWIZ | Replaced TTT with GCC at position 1495 | |
| Recombinant DNA reagent | pPR-IBA1-CylK N334A (plasmid) | This work, constructed by GENEWIZ | Replaced AAC with GCC at position 1000 | |
| Recombinant DNA reagent | pPR-IBA1-CylK S318A (plasmid) | This work, constructed by GENEWIZ | Replaced AGC with GCC at position 952 | |
| Recombinant DNA reagent | pPR-IBA1-CylK R105K (plasmid) | This work, constructed by GENEWIZ | Replaced CGT with AAG at position 313 | |
| Recombinant DNA reagent | pPR-IBA1-CylK Y473F (plasmid) | This work, constructed by GENEWIZ | Replaced TAT with TTT at position 1417 | |
| Recombinant DNA reagent | pPR-IBA1-CylK D440N (plasmid) | This work, constructed by GENEWIZ | Replaced GAC with AAC at position 1318 | |
| Software, algorithm | HKL2000 | Otwinowski and Minor, 1997 | http://www.hkl-xray.com/hkl-2000; RRID:SCR_015547 | |
| Software, algorithm | Phaser | McCoy et al., 2007 | https://www.phenix-online.org/documentation/reference/phaser.html; RRID:SCR_014219 | |
| Software, algorithm | Phenix | Liebschner et al., 2019 | https://www.phenix-online.org/; RRID:SCR_014224 | |
| Software, algorithm | Coot | Emsley and Cowtan, 2004 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/; RRID:SCR_014222 | |
| Software, algorithm | CCP4 (FFT, CAD) | Winn et al., 2011 | http://www.ccp4.ac.uk/; RRID:SCR_007255 | |
| Software, algorithm | Refmac5 | Vagin et al., 2004 | http://www.ccp4.ac.uk/html/refmac5/description.html; RRID:SCR_014225 | |
| Software, algorithm | PyMOL | The PyMOL Molecular Graphics System, version 2.0 Schrödinger, LLC | http://www.pymol.org/; RRID:SCR_000305 |
Additional files
-
Supplementary file 1
Catalog of 715 protein sequences with homology to CylK and/or BrtB from the IMG-JGI database.
- https://cdn.elifesciences.org/articles/75761/elife-75761-supp1-v2.xlsx
-
Supplementary file 2
Annotated table of putative CylK homologs that contain N-terminal fusions or are encoded by organisms with CylC- or CylI-like enzymes.
- https://cdn.elifesciences.org/articles/75761/elife-75761-supp2-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/75761/elife-75761-transrepform1-v2.pdf





