Visualizing molecules of functional human profilin
Figures
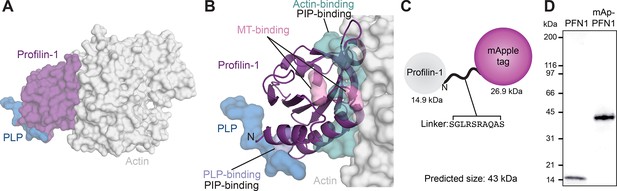
Strategy for fluorescently tagging and purifying profilin-1 (PFN1).
(A) View of profilin (PFN1; purple) with an actin monomer (gray) and poly-L-proline (PLP; blue), from PDB: 2BTF and 2PAV. (B) Profilin residues that contact actin (teal), microtubules (MT) (M114 and G118; pink), PLP (Y6D; blue) or phosphoinositide (PIP) lipids. (C) Position of genetically encoded tag and linking sequence (SGLRSRAQAS) on PFN1. (D) Coomassie-stained SDS-PAGE gel of PFN1 and mApple-PFN1 (mAp-PFN1). Source file contains uncropped gel in (D).
-
Figure 1—source data 1
Full SDS-PAGE gel and gel filtration trace.
- https://cdn.elifesciences.org/articles/76485/elife-76485-fig1-data1-v2.zip
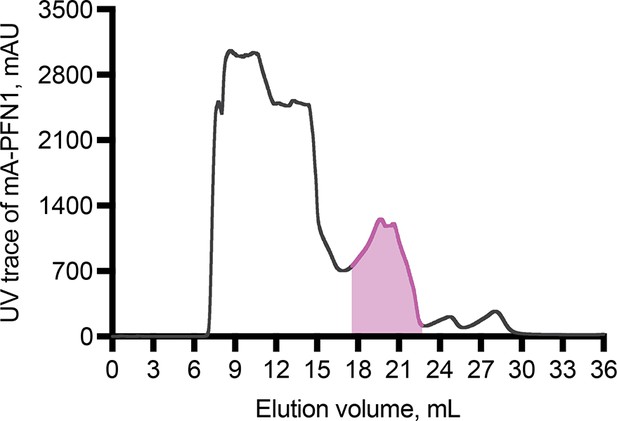
Gel filtration trace of mApple-profilin-1 (mAp-PFN1).
Trace of mAp-PFN1 (pink) elution from gel filtration column. Source file contains values plotted to obtain the UV trace.
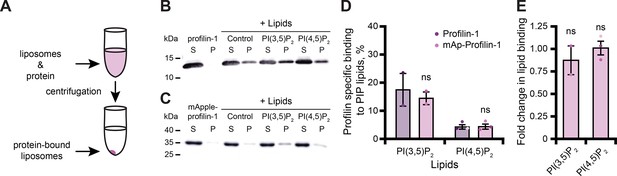
Tagged profilin binds phosphoinositide (PIP) lipids.
(A) Schematic of liposome pelleting assay. (B) Blot of supernatant and pellet samples from 1 µM profilin-1 (PFN1) in buffer, PS control, or 0.33 mM PI(3,5)P2 or PI(4,5)P2. Blots were probed with anti-PFN1 primary antibody (1:5000; SantaCruz 137235, clone B-10) paired with goat anti-mouse:IRDye 800CW secondary (1:10,000; LI-COR Biosciences 926–32210). (C) Blot for 1 µM mApple-profilin-1 (mAp-PFN1). Assay as in (B). (D) Band densitometry after background subtraction. (E) Fold change in binding normalized to PFN1. Shaded values are independent data points (n=2–3). Error bars, SE. Not significant by Student’s t-test, ns. Full blots in Figure 2—figure supplement 1. Figure 2—source data 1 contains uncropped blots and quantification values for (D and E).
-
Figure 2—source data 1
Full blots associated with PFN1 lipid binding experiments.
- https://cdn.elifesciences.org/articles/76485/elife-76485-fig2-data1-v2.zip
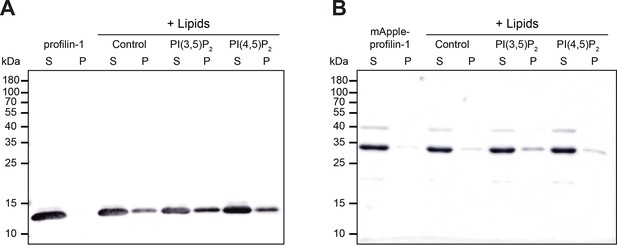
Full blots associated with tagged profilin-1 (PFN1) binding phosphoinositide (PIP) lipids.
(A) Supernatant and pellet samples from liposome pelleting assays in Figure 2B. (B) Blot from assay in (A) with mApple-PFN1 (mAp-PFN1). Blots were probed with anti-PFN1 antibody (1:5000; SantaCruz 137235, clone B-10) paired with goat anti-mouse:IRDye 800CW (1:10,000; LI-COR Biosciences 926–32210). Source file contains uncropped blot images.
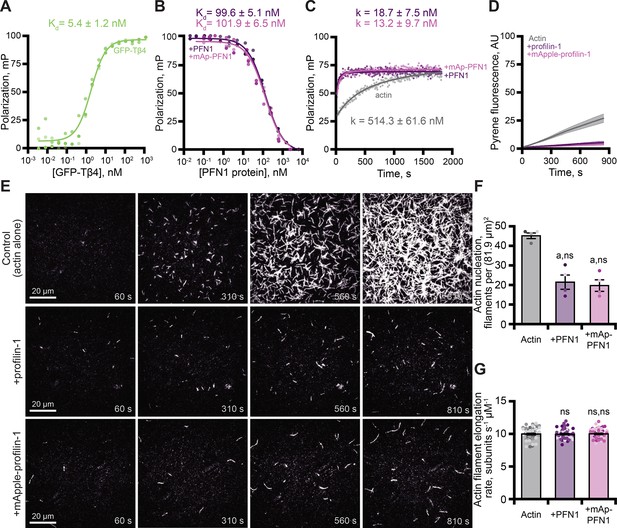
Tagged profilin binds actin monomers and stimulates nucleotide exchange.
(A) Fluorescence polarization of 10 nM actin (unlabeled) with concentrations of GFP-thymosin β4 (GFP-Tβ4). (B) Competitive polarization of 10 nM GFP-Tβ4, 10 nM actin, and concentrations of profilin-1 (PFN1; purple) or mApple-PFN1 (mAp-PFN1; pink). (C) Kinetics of 500 nM ATP-ATTO-488–2 µM actin in the presence of MEI buffer (gray), 1 µM PFN1, or mAp-PFN1. Dots represent time resolved means from n=3 replicates. (D) Actin assembly via bulk fluorescence. Reactions contain: 2 µM actin (5% pyrene-labeled), and 3 µM PFN1 or mAp-PFN1. Shaded values are SE from n=3 assays. (E) Time lapse total internal reflection fluorescence (TIRF) of 1 µM actin (20% Oregon Green-labeled, 0.6 nM biotin-actin) in buffer (control), or 3 µM PFN1 or mAp-PFN1. Scale bars, 20 µm. See Figure 3—video 1 and Figure 4—video 2. (F) Actin filament nucleation 100 s following initiation from movies as in (E), n=4 fields of view. (G) Actin filament elongation rates from movies as in (E) (n=51 filaments per condition). Shaded dots are single data points. Error bars, SE. Statistics, one-way ANOVA with Bartlett’s correction: ns, not different; (A) p<0.05 from control. No difference found for mAp-PFN1 to PFN1. Source file contains quantification values for (A–D and F and G).
-
Figure 3—source data 1
Polarization readings for binding assays associataed with Figure 3, Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/76485/elife-76485-fig3-data1-v2.zip
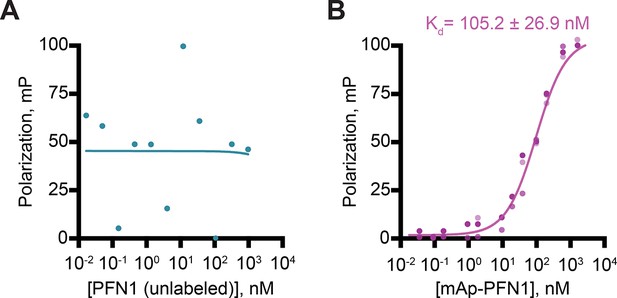
mApple-profilin-1 (mAp-PFN1) binds Oregon Green (OG)-actin monomers and is suitable for fluorescence-based binding assays.
(A) Fluorescence polarization of 10 nM OG-actin mixed with concentrations of profilin-1 (PFN1). PFN1 did not elicit a change in polarization (n=1). (B) Polarization of 10 nM actin (unlabeled) with mAp-PFN1 bound actin equivalent to PFN1 (Figure 3B; n=3). Source file contains quantification values.
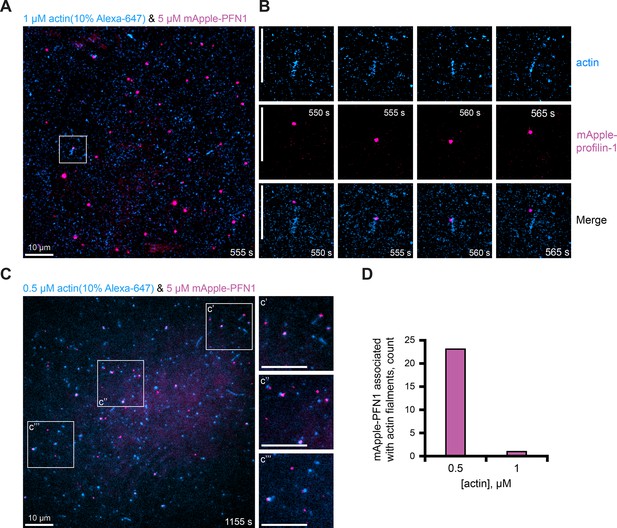
Localization of mApple-profilin-1 (mAp-PFN1) with actin filaments in vitro.
(A) Image and montage from a total internal reflection fluorescence (TIRF) assay containing: 1 µM actin (10% Alexa-647-labeled; 0.6 nM biotin-actin; blue), and 5 µM mAp-PFN1 (pink). (B) Magnified view of (A). (C) Image of 0.5 µM actin (10% Alexa-647-labeled; 0.6 nM biotin-actin; blue) and 5 µM mAp-PFN1. (D) Quantification of mAp-PFN1 with actin. Scale bars, 10 µm. Source file contains quantification values for (D).
Total internal reflection fluorescence (TIRF) movie of profilin-1 (PFN1) or mApple-PFN1 (mAp-PFN1) on actin assembly.
Reaction contains: 1 µM actin (20% OGoOregon gGreen-labeled; 0.6 nM biotin-actin) with buffer (control) or 3 µM PFN1 or mAp-PFN1. Playback, 10 fps. Scale bars, 10 µm.
TIRF movie of mAp-PFN1 on actin assembly.Reaction contains: 1 µM actin (20% OG-labeled; 0.6 nM biotin-actin) (cyan) and 3 µM mAp-PFN1 (pink).
Box corresponds to Figure 3—figure supplement 2A Figure S4A inset. Playback, 10 fps. Scale bars, 10 µm.
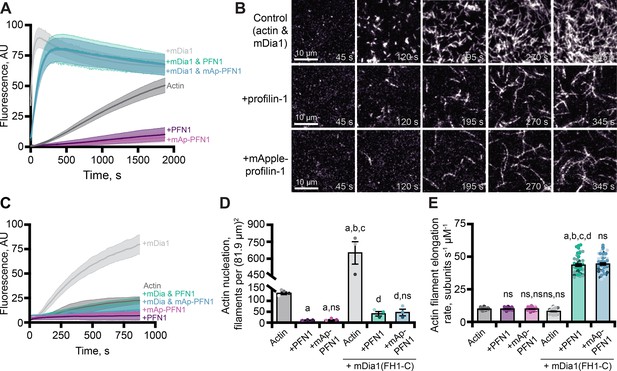
Effects of mApple-profilin-1 (mAp-PFN1) on formin-mediated actin assembly.
(A) Bulk actin assembly: 2 µM actin (5% pyrene-labeled), 25 nM mDia1(FH1-C), and 5 µM profilin-1 (PFN1) or mApple-PFN1 (mAp-PFN1). Shaded values are SE from n=2 assays. (B) total internal reflection fluorescence (TIRF) of 1 µM actin (10% Alexa-647-labeled, 0.6 nM biotin-actin), 25 nM mDia1(FH1-C), and 5 µM PFN1 or mAp-PFN1. Scale bars, 10 µm. See Figure 4—video 1, Figure 4—video 2 . (C) Actin fluorescence from TIRF videos. Shading indicates SE from n = 3 videos. (D) Mean nucleation 100 s after initiation, from n=4 fields of view. (E) Actin filament elongation rates from movies as in (B) (n=51 total filaments per condition). Shaded dots are individual data points. Error bars, SE. Statistics, one-way ANOVA with Bartlett’s correction: ns, not different; (A) p<0.05 from control; (B) p<0.05 from PFN1; (C) p<0.05 from mAp-PFN1; (D) p<0.05 from actin and mDia1 control. No difference was found for mAp-PFN1 to PFN1. Source file contains quantification values for (C–E).
-
Figure 4—source data 1
Full views, nucleation, and elognation rate values for experiments exploring the effects of mAp-PFN1 in formin-based actin assembly assays.
- https://cdn.elifesciences.org/articles/76485/elife-76485-fig4-data1-v2.zip
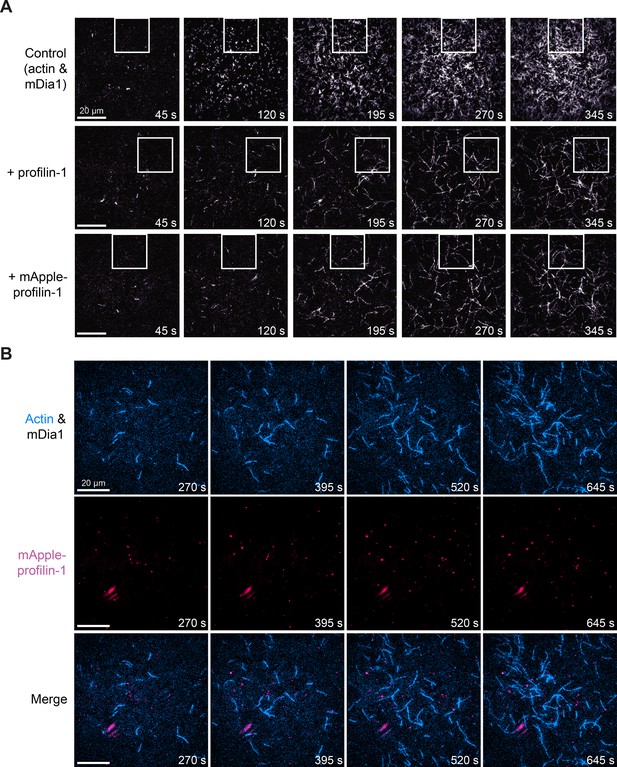
Full views of mApple-profilin-1 (mAp-PFN1) on formin-mediated actin assembly.
(A) Full views formin-mediated actin polymerization montage in Figure 4B (white boxes). (B) Multi-color total internal reflection fluorescence (TIRF) montage containing: 1 µM actin (10% Alexa-647-labeled; 0.6 nM biotin-actin), 25 nM mDia1(FH1-C) and 5 µM mAp-PFN1. Scale bars, 20 µm.
TIRF movie of PFN1 or mAp-PFN1 on formin-mediated actin assembly.
Reaction contains: 1 µM actin monomers (10% Alexa-647-labeled; 0.6 nM biotin-actin) and 5 µM PFN1 or mAp-PFN1. Box corresponds to montage in Figure 4B and Figure 4—figure supplement 1A. Playback, 10 fps. Scale bars, 10 µm.
Total internal reflection fluorescence (TIRF) video of mApple-profilin-1 (mAp-PFN1) in formin-mediated actin assembly assay.
Reaction contains: 1 µM actin (10% Alexa-647-labeled; 0.6 nM biotin-actin; cyan) and 5 µM mAp-PFN1 (pink). Playback, 10 fps. Scale bars, 10 µm.
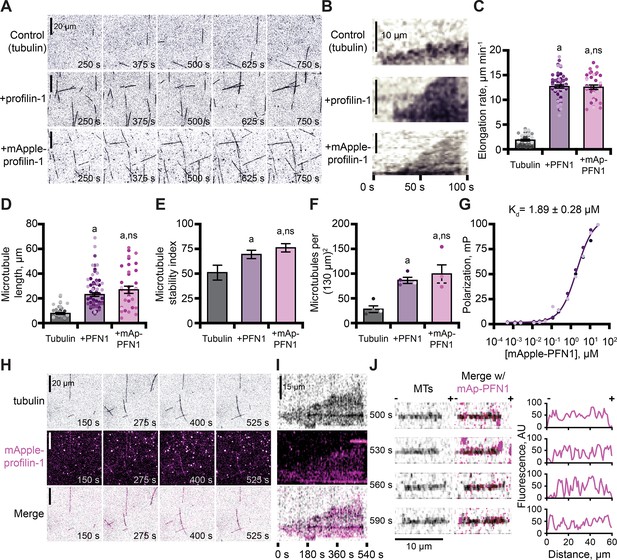
Profilin binds tubulin dimers and associates with the microtubule lattice.
(A) Total internal reflection fluorescence (TIRF) images of reactions of biotinylated GMP-CPP seeds, 10 µM free tubulin (5% HiLyte-488 labeled), and buffer (control) or 5 µM profilin-1 (PFN1) or mApple-PFN1 (mAp-PFN1). Scale bars, 20 µm. See Figure 5—video 1. (B) Kymographs display dynamics as in (A). Scale bars: length, 10 µm; time, 100 s. (C) Microtubule polymerization (35–58 microtubules from n=3 experiments). (D) Microtubule length (n=35–58 microtubules from n=3 experiments). (E) Stability index: rescue/catastrophe frequency (n=18–46 microtubules from n=3 experiments). (F) Number of microtubules (n=4 experiments). Shaded dots are single data points. Error bars, SE. (G) Polarization of 10 nM tubulin (unlabeled) and mAp-PFN1. (H) TIRF as in (A), but visualizing tubulin (black) and mAp-PFN1 (pink). Scale bars, 20 µm. See Figure 5—video 2. (I) Kymographs display dynamics in (H). Scale bars: length, 15 µm; time, 540 s. (J) Montage of a microtubule (black) and mAp-PFN1 (pink) merged, with the intensity profiles of mAp-PFN1 along the microtubule lattice. See Figure 5—video 3. Scale bar, 10 µm. + and -, microtubule polarity. Statistics, one-way ANOVA with Bartlett’s correction: (A) p<0.05 from control. No difference was found for mAp-PFN1 to PFN1. Source file contains quantification values for (C–G and J).
-
Figure 5—source data 1
Full views of TIRF movies and values for measured parameters associated with Figure 5.
- https://cdn.elifesciences.org/articles/76485/elife-76485-fig5-data1-v2.zip
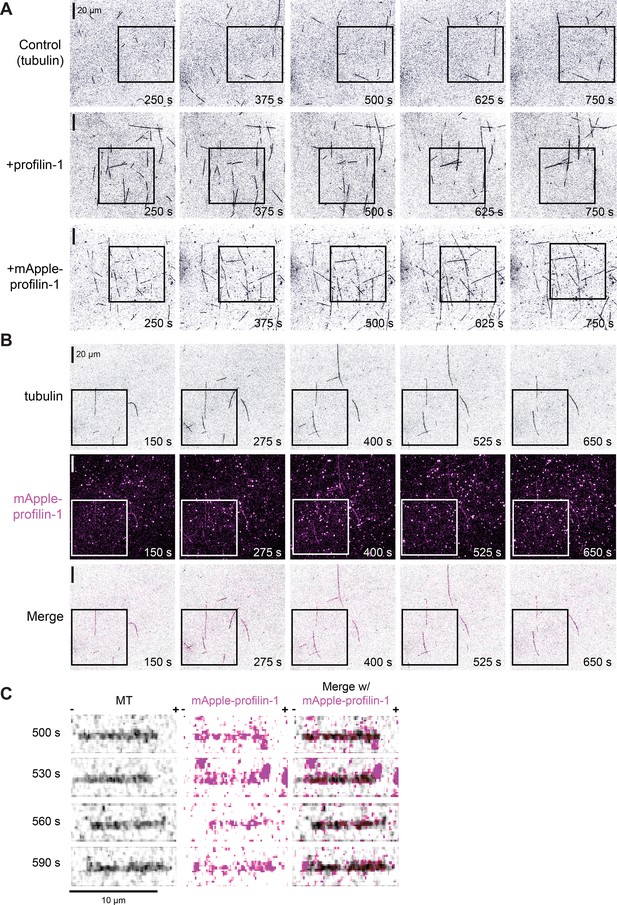
Additional views of the effects of mApple-profilin-1 (mAp-PFN1) on microtubule dynamics.
(A) Full views of montages present in Figure 5A (black boxes). (B) Full view of multi-color time lapse total internal reflection fluorescence montage present in Figure 5H (black boxes). Scale bars, 20 µm. (C) Montage of a microtubule (black), mAp-PFN1 (pink) and merge from Figure 5I. Scale bar, 10 µm. + and - indicate microtubule polarity.
TIRF movie of PFN1 or mAp-PFN1 on microtubules.
Reaction contains: 647-biotinylated-GMP-CCP microtubule seeds (not shown), 10 µM tubulin (5% HiLyte 488) in buffer or 5 µM profilin or mAp-PFN1. Box corresponds to Figure 5A and Figure 5—figure supplement 1A. Playback, 10 fps. Scale bars, 20 µm.
TIRF movie of mAp-PFN1 on microtubules.
Reaction contains: 647- biotinylated-GMP-CCP microtubule seeds (not shown), 10 µM free tubulin (5% HiLyte 488) (black) and 5 µM mAp-PFN1 (pink). Box corresponds to montage inset from Figure 5H and Figure 5—figure supplement 1B. Playback 10 fps. Scale bars, 20 µm.
mAp-PFN1 transiently associates with the microtubule lattice.
Reaction contains: 647-biotinylated-GMP-CCP microtubule seeds (not shown), 10 µM free tubulin (5% HiLyte-488); (black) and 5 µM mAp-PFN1 (pink). +and -, microtubule polarity. Playback, 10 fps. Scale bar, 10 µm.
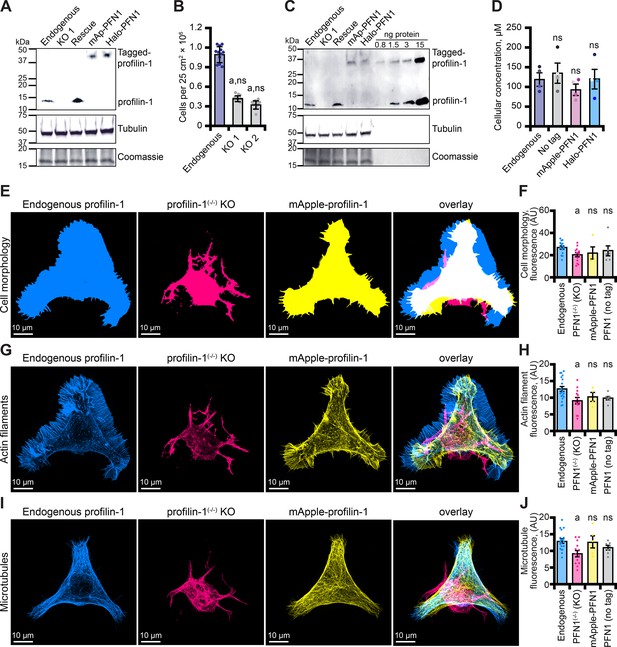
Tagged versions of profilin rescue protein levels and cell-based phenotypes in profilin-1 (PFN1) deficient cells.
(A) Blot confirming knockout and rescue of PFN1 with tag-free PFN1, mApple-PFN1 (mAp-PFN1) or Halo-PFN1 plasmids. Extracts prepared from endogenous PFN1(+/+), PFN1(-/-), and PFN1(-/-) transfected with tag-free (rescue), mAp-PFN1 or Halo-PFN1 plasmids. Blot probed with anti-PFN1 antibody (1:3500; SantaCruz 137235, clone B-10) paired with goat anti-mouse:IRDye 800CW secondary (1:5000; LI-COR Biosciences 926–32210), and anti-α-tubulin (1:10,000; Abcam 18251) paired with donkey anti-rabbit 926–68073 secondary (1:20,000). Coomassie stained membrane used as loading control. Full blot in Figure 6—figure supplement 1A-C. (B) PFN1(+/+) cells proliferate significantly better than PFN1-deficient cell lines. (C) Blot probed as in (A) with known quantities of purified PFN1 and mAp-PFN1. Full blot in Figure 6—figure supplement 1D-F. (D) PFN1 levels in neuroblastoma (N2a) cells. Shaded dots are data points from n=4 experiments. Error bars, SE. Maximum intensity images and quantification of (E–F) cell morphology and (G–H) fluorescence of phalloidin-stained actin filaments or (I–J) microtubules from N2a cells expressing endogenous PFN1 (blue), PFN1(-/-) (pink), PFN1(-/-) transfected with mAp-PFN1 (yellow) or tag-free PFN1 plasmids (Figure 6—figure supplement 1G). Cells were plated on micropatterns and stained with anti-α-tubulin antibody (1:100; Abcam 18251) paired with donkey anti-rabbit conjugated to AlexaFluor-647 (1:100; Life Technologies A31573). Scale bars, 10 µm. Shaded dots are individual cells (n=4–15 cells) from n=3 coverslips. Error bars, SE. Statistics, one-way ANOVA with Bartlett’s correction: ns, not different from PFN1(+/+); (A) p<0.05 from PFN1(+/+). No significant difference was found for mAp-PFN1, Halo-PFN1 or tag-less PFN1 to endogenous PFN1(+/+). Source file contains uncropped blots and quantification values for (B, D, F, H, and J).
-
Figure 6—source data 1
Full blots and additional cell views associated with Figure 6.
- https://cdn.elifesciences.org/articles/76485/elife-76485-fig6-data1-v2.zip

Full blots used to determine profilin-1 (PFN1) levels in neuroblastoma-2a (N2a) cells.
Blots confirming knockout or rescue of PFN1 as in Figure 6A. (A) Blots were probed with anti-PFN1 antibody (1:3500; SantaCruz 137235, clone B-10) paired with goat anti-mouse:IRDye 800CW secondary antibody (1:5000; LI-COR Biosciences 926–32210) and (B) anti-α-tubulin primary (1:10,000; Abcam 18251) paired with donkey anti-rabbit:IRDye 680RD (1:20,000; LI-COR Biosciences 926–68073). (C) Coomassie stained membrane from (A). (D) Example blot used to determine PFN1 concentration N2a cells (n=4 total). Blot probed as in (A–C) for (D) PFN1 and (E) α-tubulin, and (F) Coomassie stained. (G) Morphology measurements related to Figure 6 for tag-free PFN1. Source file contains uncropped blots.
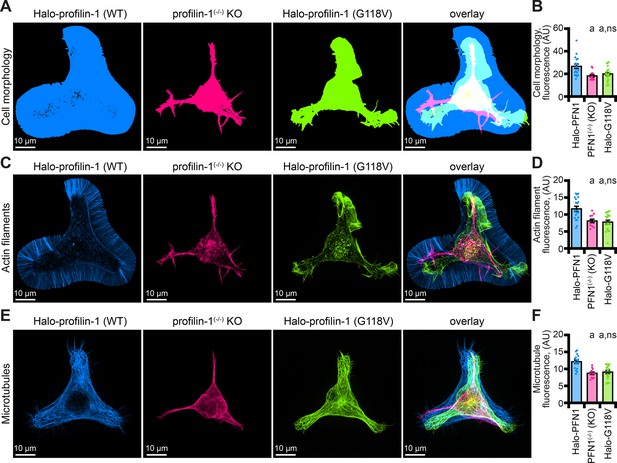
The profilin-1(G118V) amyotrophic lateral sclerosis (ALS) variant does not rescue morphology or cytoskeletal phenotypes present in profilin-1 (PFN1) deficient cells.
(A–B) Maximum intensity images and quantification of cell morphology and (C–D) fluorescence of phalloidin-stained actin filaments (E–F) or microtubules from neuroblastoma-2a PFN1(-/-) transfected with Halo-PFN1 (blue), PFN1(-/-) (pink), or PFN1(-/-) cells transfected with Halo-PFN1(G118V; lime). Cells plated and stained as in Figure 6I–J. Scale bars, 10 µm. Shaded dots are individual cells (n=16–25 cells) from at least n=3 coverslips. Error bars, SE. Statistics, one-way ANOVA with Bartlett’s correction: ns, not different from PFN1(-/-) expressing Halo-PFN1 (control); (A) p<0.05 from control. Source file contains quantification values for (B, D, and F).
-
Figure 7—source data 1
Source values for cell morphology, and total fluorescence of actin filaments and microtubules.
- https://cdn.elifesciences.org/articles/76485/elife-76485-fig7-data1-v2.zip
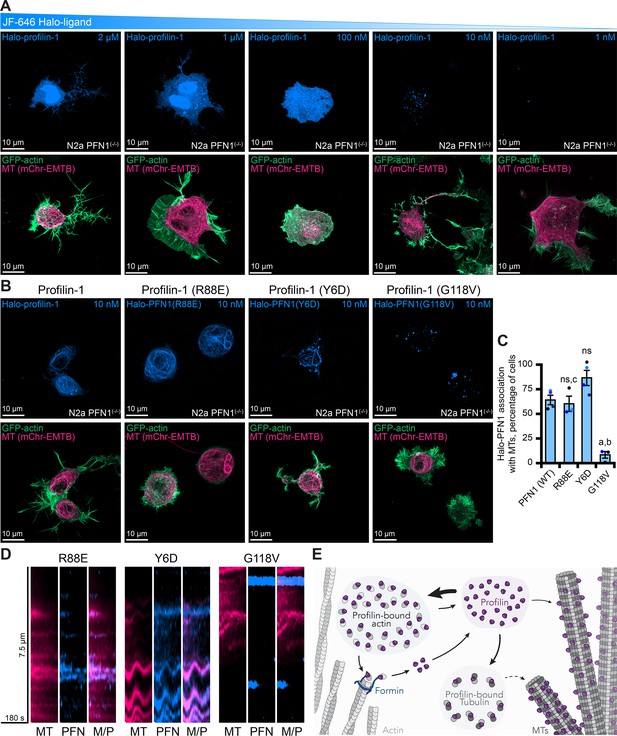
Live-cell visualization of individual molecules of profilin-1 (PFN1).
(A) Maximum intensity projections of PFN1(-/-) expressing markers for actin (green), microtubules (pink) or Halo-PFN1 (blue; top) visualized with JF-646 and imaged as in Figure 6. Titration of JF-646 to illuminate PFN1 molecules. (B) PFN1(-/-) transfected with Halo-PFN1 plasmids (wild-type, R88E, Y6D, and G118V) as in (A). Scale bars, 10 µm. (C) Halo-PFN1-microtubule overlap in cells from (B). Overlap analysis was performed in n=3–4 experiments. Averages from 10 to 25 cells per experiment. Statistics, one-way ANOVA with Bartlett’s correction: ns, not different from PFN1(-/-) expressing Halo-PFN1 (control); (A) p<0.05 from control; (B) p<0.05 from Halo-PFN1(R88E) or Halo-PFN1(Y6D); (C) p<0.05 from Halo-PFN1(G118V). (D) Kymographs display microtubule dynamics (see Figure 8—figure supplement 2 and Figure 8—video 1). (E) Model of PFN1 distribution in cells. Source file contains quantification values for (C).
-
Figure 8—source data 1
Halo-profilin microtubule localization counts.
- https://cdn.elifesciences.org/articles/76485/elife-76485-fig8-data1-v2.zip
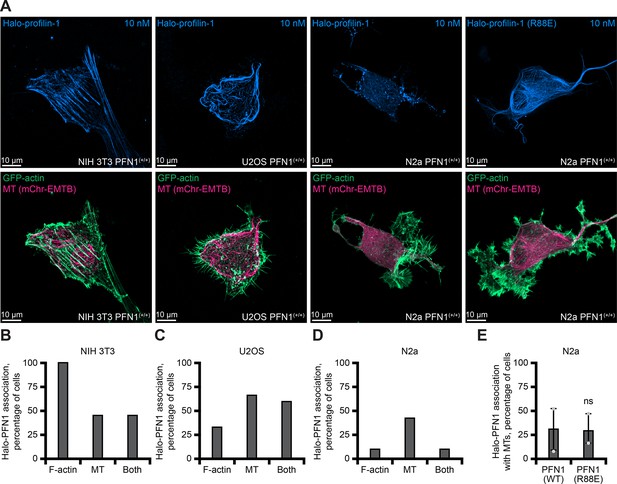
Localization of Halo-profilin-1 (Halo-PFN1) in different cell types.
(A) Maximum intensity images of different PFN1(+/+) cells transiently expressing GFP-actin (green), a marker for microtubules (EMTB-2× mCherry; pink), and either Halo-PFN1 (Halo-PFN1) or Halo-PFN1(R88E) (light blue), visualized with 10 nM JF-646 ligand. Scale bars, 10 µm. (B–D) Quantification of Halo-PFN1-microtubule overlap from different cell types from (A) (n=15–49 cells). (E) No difference was found Halo-PFN1 or Halo-PFN1(R88E)-microtubule colocalization (n=42–46 cells). Source file contains quantification values for (B–E).
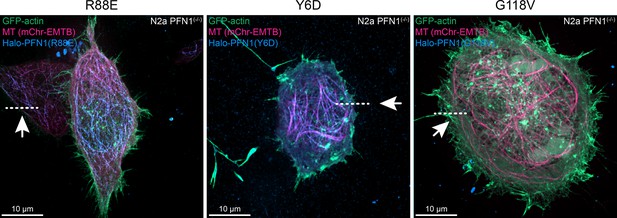
Live cell localization of constructs of Halo-profilin-1 (Halo-PFN1).
View of kymograph line drawn on cells expressing Halo-PFN1 mutants in Figure 8D. Scale bars, 10 µm.
Halo-profilin-1 (Halo-PFN1) dynamics in live neuroblastoma-2aN2a cells.
4D-spinning disk confocal movie of cells transiently expressing markers for actin (GFP-actin; cyan), microtubules (EMTB; yellow), and Halo-PFN1 plasmids labeled with 10 nM JF-646 (magenta). Playback, 10 fps. Scale bar, 10 µm.





