Cross-species analysis of LZTR1 loss-of-function mutants demonstrates dependency to RIT1 orthologs
Figures
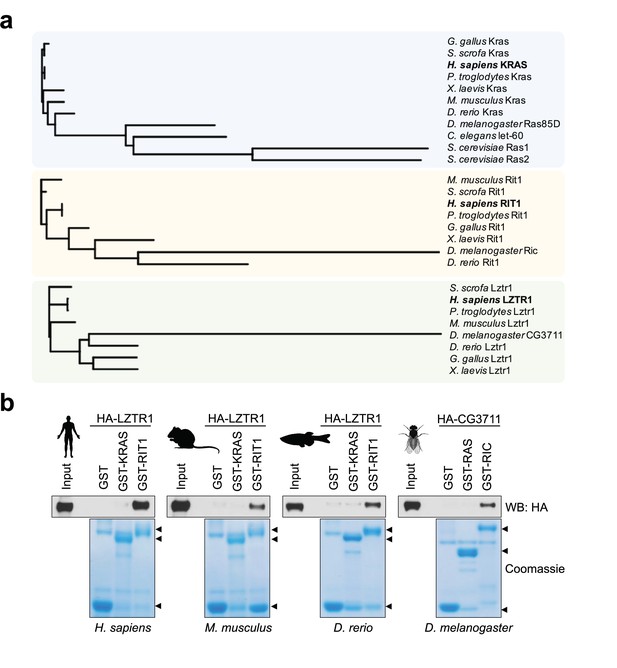
Evolutionary analysis of LZTR1 and the RAS proteins KRAS and RIT1.
(a) Phylogenetic trees of KRAS, RIT1, and LZTR1 orthologs based on multiple protein sequence alignments performed with Clustal Omega (Figure 1—source data 1). Orthologs were searched in the following model organisms: chimpanzee (Pan troglodytes), pig (Sus scrofa), chicken (Gallus gallus), mouse (Mus musculus), African clawed frog (Xenopus laevis), zebrafish (Danio rerio), fruit fly (Drosophila melanogaster), roundworm (Caenorhabditis elegans), and budding yeast (Saccharomyces cerevisiae). (b) Pull-down assays using GST-tagged KRAS and RIT1 or their mouse, zebrafish, or fruit fly orthologs produced recombinantly in bacteria. Recombinant proteins were incubated with lysates from HEK-293T cells expressing their corresponding species’ HA-tagged LZTR1 ortholog. Representative results from three biological replicates.
-
Figure 1—source data 1
Raw data for Figure 1.
- https://cdn.elifesciences.org/articles/76495/elife-76495-fig1-data1-v2.zip
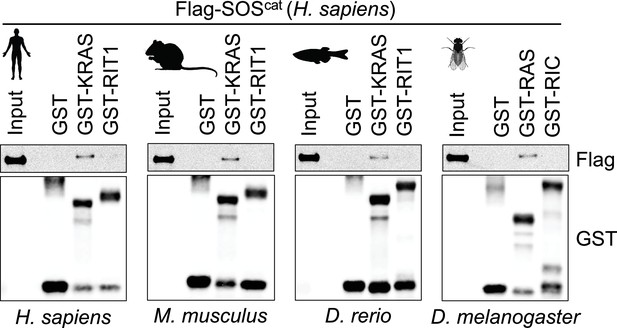
Interaction between the human SOScat domain and the different KRAS orthologs using pull-down assays demonstrates that the proteins are properly folded and GDP loaded in our assay.
Note that SOScat domain does not interact with RIT1 orthologs. Representative results from two biological replicates.
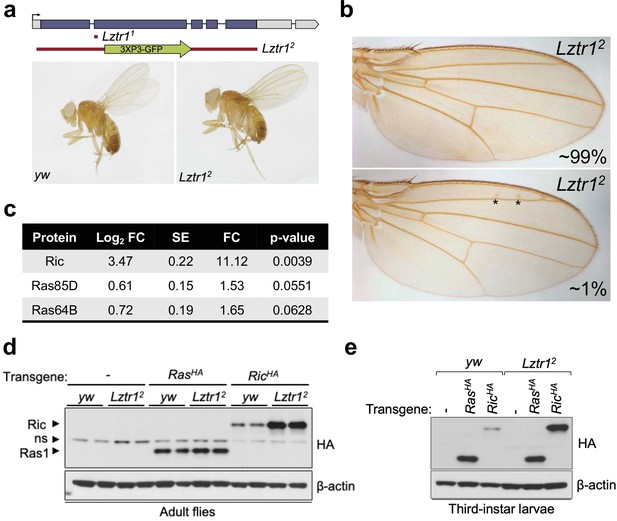
Drosophila Lztr1 regulates Ric stability.
(a) A schematic representation of the Drosophila CG3711/Lztr1 gene locus is shown. Coding exons are represented in blue. Two CRISPR/Cas9-mediated approaches were used to isolate the Lztr1 loss-of-function alleles, Lztr11 and Lztr12. (b) Wing vein patterning is not affected in Lztr1 null flies (upper panel), with very few individuals exhibiting small ectopic veinlets in two or three points (asterisks; lower panel) (n = 209 flies). (c) Estimated normalized protein abundance expressed as mean log2 fold change in Lztr12 vs yw (control) comparison. Corresponding single-protein standard error (SE) and t-test p-values are listed. (d) Immunoblot analysis of protein extracts isolated from the indicated transgenic adult flies in a yw or Lztr12 background. ns: non-specific band. (e) Same as panel (d) with protein extracts isolated from third-instar larvae. Representative results from three biological replicates.
-
Figure 2—source data 1
Raw data for Figure 2.
- https://cdn.elifesciences.org/articles/76495/elife-76495-fig2-data1-v2.zip
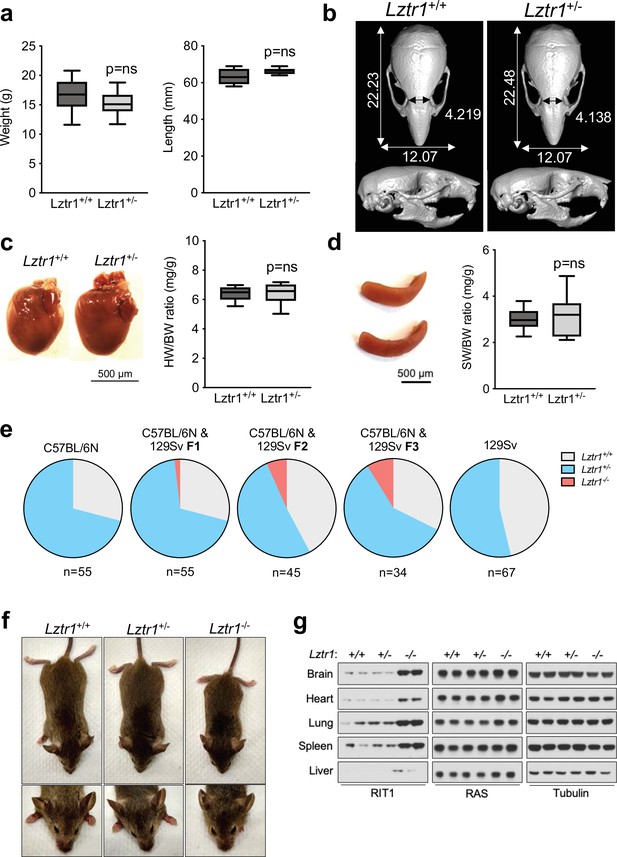
Lztr1 is haplosufficient in mice and its null phenotype can be modified by strain background.
(a) Weight (left panel) and length (right panel) of 4-week-old male Lztr1 wild type (n = 18) and heterozygous mutant (n = 20) mice. (b) Representative µCT imaging of the skull of an 8-week-old male Lztr1 wild type and heterozygous mutant mouse. The indicated values show the average measurement (mm) of length, width, and inner intercanthal distance in Lztr1 wild type (n = 5) and heterozygous mutant (n = 5) mice. Mann-Whitney p values were not significant for all the measurements. (c) Heart weight was similar between 8-week-old male Lztr1 wild type (n = 8) and heterozygous mutant (n = 6) mice, as assessed by heart to body weight ratio (HW/BW). Mann-Whitney test p value was not significant. (d) Same as panel (c), for the spleen of these mice. (e) Pie charts indicate the percentage of obtained genotypes upon weaning (21 days of age) the offspring of Lztr1 heterozygous mutant intercrosses. Each pie chart represents a different strain background and/or mixed background. (f) Representative image of female littermates with the indicated Lztr1 genotypes (C57BL/6N-129Sv F3 background). Note the decreased size, round skull, and proptosis of the homozygous Lztr1 knockout mouse. (g) Immunoblot analysis of RIT1, RAS, and Tubulin proteins isolated from the indicated tissues of Lztr1 wild type, heterozygous, and homozygous mice (C57BL/6N-129Sv F3 background). Protein lysates from two different mice were used for each genotype.
-
Figure 3—source data 1
Raw data for Figure 3.
- https://cdn.elifesciences.org/articles/76495/elife-76495-fig3-data1-v2.zip
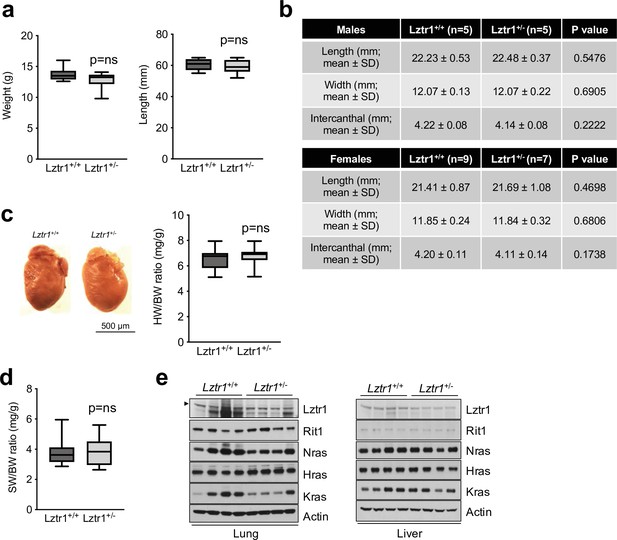
Phenotyping results of Lztr1 heterozygous knockout female mice.
(a) Weight (left panel) and length (right panel) of 4-week-old female Lztr1 wild type (n = 16) and heterozygous knockout (n = 24) mice. (b) µCT measurements of the skull of 8-week-old male and female Lztr1 wild type and heterozygous knockout mice. The indicated values show the average measurement (mm) of length, width, and inner intercanthal distance, as well as the standard deviation (SD). Mann-Whitney p values are indicated and were not significant for any of the measurements. (c) Heart weight was similar between 8-week-old female Lztr1 wild type (n = 9) and heterozygous mutant (n = 7) mice, as assessed by heart to body weight ratio (HW/BW). Mann-Whitney test p value was not significant. (d) Same as panel c, for the spleen of these mice. (e) Immunoblot analysis of lung and liver protein extracts isolated from Lztr1 wild type (n = 4) and heterozygous knockout (n = 4) mice. Arrowhead indicates Lztr1 band.
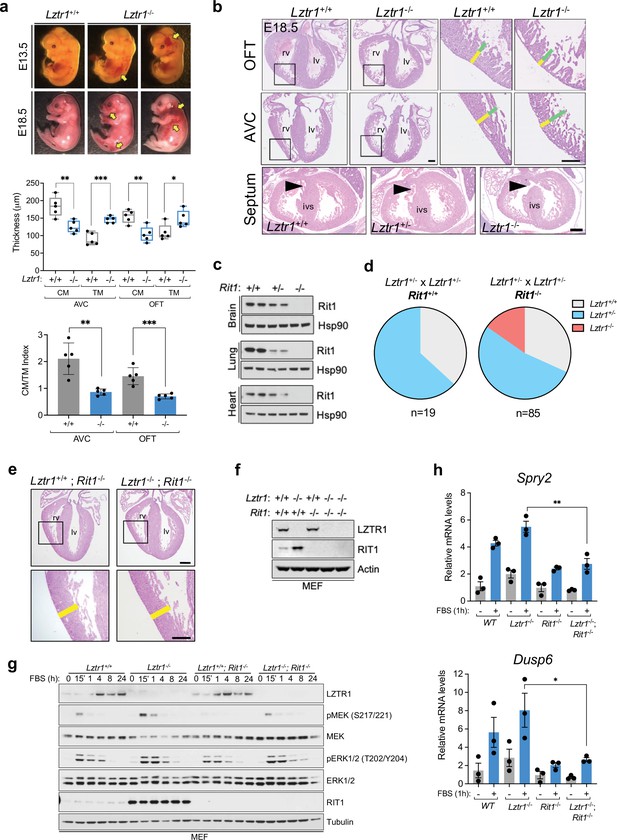
Lethality in Lztr1 knockout mutants as a result of Rit1-dependent cardiovascular defects.
(a) Gross morphology of Lztr1+/+ (wild type) and Lztr1-/- (knockout) embryos at E13.5 and E18.5. Note the presence of extensive hemorrhage in knockout embryos (yellow arrows). (b) Histological characterization of E18.5 hearts shows highly penetrant defects in ventricular wall thickness at both outflow tract (OFT) and atrioventricular canal (AVC) levels, as well as septal defects (arrows) in ~20% Lztr1-/- embryos. Quantification of ventricular wall thickness in both CM (yellow) and TM (green) and CM/TM index are shown in the graphs (n = 5). Rv: right ventricle; lv: left ventricle; ivs: interventricular septum; CM: compact myocardium; TM: trabecular myocardium. p Values were calculated using Student’s t-test. (c) Immunoblot analysis of different tissues isolated from Rit1+/+ (wild type), Rit1+/- (heterozygous), and Rit1-/- (knockout) adult mice. (d) Percentage of Lztr1 genotypes obtained upon weaning (21 days of age) the offspring of Lztr1 heterozygous mutant intercrosses in either a Rit1+/+ (n = 19) or Rit1-/- (n = 85) background. All these mice were maintained in a C57BL/6 N background. (e) Histological analysis of the heart of Lztr1; Rit1 double knockout E18.5 embryos (n = 3). (f) Immunoblot analysis of lysates isolated from primary MEF with the indicated genotypes. (g) Primary MEF derived from wild type, Lztr1 knockout, Rit1 knockout, and Lztr1; Rit1 double knockout were starved overnight and stimulated with 10% FBS during the indicated times. Protein lysates were immunoblotted as indicated. Immunoblots represent a representative result from two biological replicates. (h) Quantitative PCR analysis of Spry2 and Dusp6 mRNA levels in primary MEF with the indicated genotypes stimulated for 1 hr with 10% FBS (n = 3 biological replicates). p Values were calculated using Student’s t-test. p values: * (p < 0.05); ** (p < 0.01); *** (p < 0.005).
-
Figure 4—source data 1
Raw data for Figure 4 and Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/76495/elife-76495-fig4-data1-v2.zip
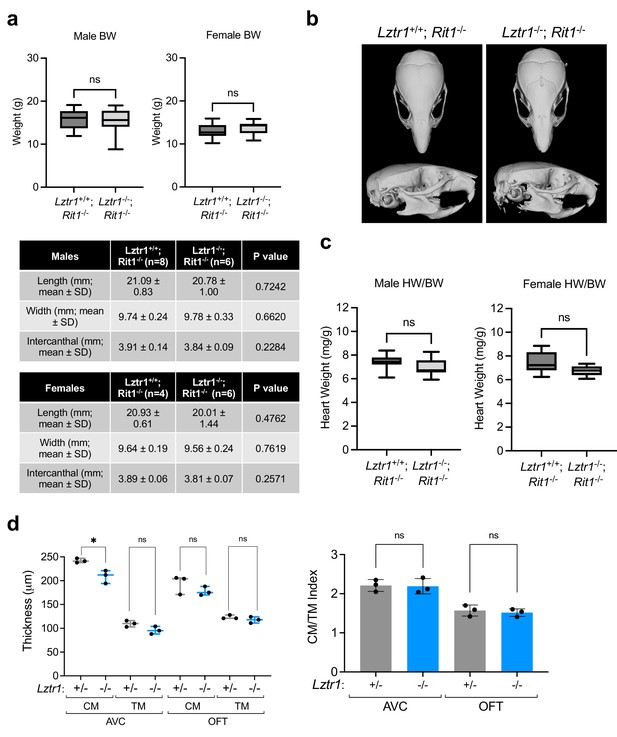
Phenotyping results of Lztr1/Rit1 double knockout mice and embryos.
(a) Weight of 4-week-old male (left) female (right) Rit1 knockout (male: n = 14; female: n = 13) and Lztr1/Rit1 double knockout (male: n = 24; female: n = 14) mice. (b) µCT measurements of the skull of eight-week-old male and female Rit1 knockout and Lztr1/Rit1 double knockout mice. The indicated values show the average measurement (mm) of length, width, and inner intercanthal distance, as well as the standard deviation (SD). Mann-Whitney p values are indicated and were not significant for any of the measurements. (c) Heart weight was similar between eight-week-old male (left) and female (right) Rit1 knockout (male: n = 11; female: n = 8) and Lztr1/Rit1 double knockout (male: n = 12; female: n = 11) mice, as assessed by heart to body weight ratio (HW/BW). Mann-Whitney test p value was not significant. (d) Quantification of ventricular wall thickness in both CM and TM and CM/TM index are shown in the graphs (n = 3). Lztr1 genotype for embryos is indicated and were all in the Rit1-/- background. CM: compact myocardium; TM: trabecular myocardium. p Values were calculated using Student’s t-test. p values: ns (p > 0.05); * (p < 0.05).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | BL21(DE3) | NEB | C2527H | |
| Genetic reagent (D. melanogaster) | Lztr11 | This paper | CG3711/Lztr1 knockout | |
| Genetic reagent (D. melanogaster) | Lztr12 | This paper | CG3711/Lztr1 knockout | |
| Genetic reagent (D. melanogaster) | RasHA | This paper | Transgene of HA-Ras at attP-86Fb landing site | |
| Genetic reagent (D. melanogaster) | RicHA | This paper | Transgene of HA-Ric at attP-86Fb landing site | |
| Genetic reagent (M. musculus) | Lztr1-/- | EUCOMM | Lztr1tm1a(EUCOMM)Wtsi; RRID: IMSR_EM:06794 | Lztr1 knockout |
| Genetic reagent (M. musculus) | Rit1-/- | This paper | Rit1 knockout | |
| Genetic reagent (M. musculus) | Lztr1-/-;Rit1-/- | This paper | Lztr1 and Rit1 double knockout | |
| Genetic reagent (M. musculus) | 129S1/Svlmj | Jackson Laboratories | 002448; RRID:IMSR_JAX:002448 | |
| Recombinant DNA reagent | pcDNA3-DEST-Flag-SOScat (H. sapiens) | This paper | Vector to express Flag-SOS1 (residues 564–1049) in mammalian cells. | |
| Recombinant DNA reagent | pcDNA3-DEST-HA-LZTR1 (H. sapiens) | This paper | Vector to express HA-LZTR1 in mammalian cells. | |
| Recombinant DNA reagent | pcDNA3-DEST-HA-LZTR1 (M. musculus) | This paper | Vector to express HA-LZTR1 in mammalian cells. | |
| Recombinant DNA reagent | pcDNA3-DEST-HA-LZTR1 (D. rerio) | This paper | Vector to express HA-LZTR1 in mammalian cells. | |
| Recombinant DNA reagent | pcDNA3-DEST-HA-LZTR1 (D. melanogaster) | This paper | Vector to express HA-LZTR1 in mammalian cells. | |
| Recombinant DNA reagent | pGEX-6P-DEST-KRAS (H. sapiens) | This paper | Vector to express GST-KRAS in E. coli cells. | |
| Recombinant DNA reagent | pGEX-6P-DEST-RIT1 (H. sapiens) | This paper | Vector to express GST-RIT1 in E. coli cells. | |
| Recombinant DNA reagent | pGEX-6P-DEST-KRAS (M. musculus) | This paper | Vector to express GST-KRAS in E. coli cells. | |
| Recombinant DNA reagent | pGEX-6P-DEST-RIT1 (M. musculus) | This paper | Vector to express GST-RIT1 in E. coli cells. | |
| Recombinant DNA reagent | pGEX-6P-DEST-KRAS (D. rerio) | This paper | Vector to express GST-KRAS in E. coli cells. | |
| Recombinant DNA reagent | pGEX-6P-DEST-RIT1 (D. rerio) | This paper | Vector to express GST-RIT1 in E. coli cells. | |
| Recombinant DNA reagent | pGEX-6P-DEST-RAS (D. melanogaster) | This paper | Vector to express GST-KRAS in E. coli cells. | |
| Recombinant DNA reagent | pGEX-6P-DEST-RIC (D. melanogaster) | This paper | Vector to express GST-RIT1 in E. coli cells. | |
| Antibody | Anti-HA (Rabbit monoclonal) | Cell Signalling Technology | Cat#: 3724; RRID: AB_1549585 | WB (1:3,000) |
| Antibody | Anti-Flag (Rabbit monoclonal) | Cell Signalling Technology | Cat#: 14793; RRID: AB_2572291 | WB (1:3,000) |
| Antibody | Anti-p-ERK1/2 (Rabbit monoclonal) | Cell Signalling Technology | Cat#: 4370; RRID: AB_2315112 | WB (1:1,000) |
| Antibody | Anti-ERK1/2 (Rabbit monoclonal) | Cell Signalling Technology | Cat#: 4696; RRID: AB_390780 | WB (1:2,000) |
| Antibody | Anti-p-MEK1/2 (Rabbit monoclonal) | Cell Signalling Technology | Cat#: 9154; RRID: AB_2138017 | WB (1:1,000) |
| Antibody | Anti-MEK1/2 (Mouse monoclonal) | Cell Signalling Technology | Cat#: 4694; RRID: AB_10695868 | WB (1:1,000) |
| Antibody | Anti-RIT1 (Rabbit polyclonal) | Abcam | Cat#: ab53720; RRID: AB_882379 | WB (1:1,000) |
| Antibody | Anti-b-Actin (Mouse monoclonal) | Sigma-Aldrich | Cat#: A2228; RRID: AB_476697 | WB (1:10,000) |
| Antibody | Anti-a-Tubulin (Mouse monoclonal) | Sigma-Aldrich | Cat#: T6199; RRID: AB_477583 | WB (1:10,000) |
| Antibody | Anti-KRAS (Mouse monoclonal) | Sigma-Aldrich | Cat#: WH0003845M1; RRID: AB_1842235 | WB (1:500) |
| Antibody | Anti-Ras (Rabbit monoclonal) | Cell Signalling Technology | Cat#: 4370; RRID: AB_2910195 | WB (1:1,000) |
| Antibody | Anti-NRAS (Mouse monoclonal) | Santa Cruz Biotechnology | Cat#: sc-31; RRID: AB_628041 | WB (1:1,000) |
| Antibody | Anti-HRAS (Rabbit polyclonal) | Santa Cruz Biotechnology | Cat#: sc-520; RRID: AB_631670 | WB (1:500) |
| Antibody | Anti-LZTR1 (Mouse monoclonal) | Santa Cruz Biotechnology | Cat#: sc-390166X; RRID: AB_2910196 | WB (1:1,000) |
| Sequence-based reagent | Dusp6_F | This paper | PCR primers | TCCTATCTCGGATCACTGGAG |
| Sequence-based reagent | Dusp6_R | This paper | PCR primers | GCTGATACCTGCCAAGCAAT |
| Sequence-based reagent | Spry2_F | This paper | PCR primers | CATCGCTGGAAGAAGAGGAT |
| Sequence-based reagent | Spry2_R | This paper | PCR primers | CATCAGGTCTTGGCAGTGT |
| Sequence-based reagent | Tbp_F | This paper | PCR primers | CCTTGTACCCTTCACCAATGAC |
| Sequence-based reagent | Tbp_R | This paper | PCR primers | ACAGCCAACATTCACGGTAGA |





