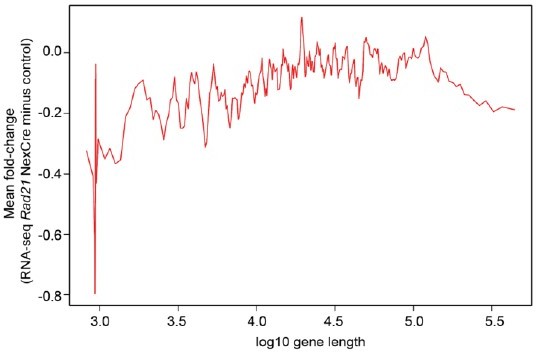
Cohesin-dependence of neuronal gene expression relates to chromatin loop length
Figures
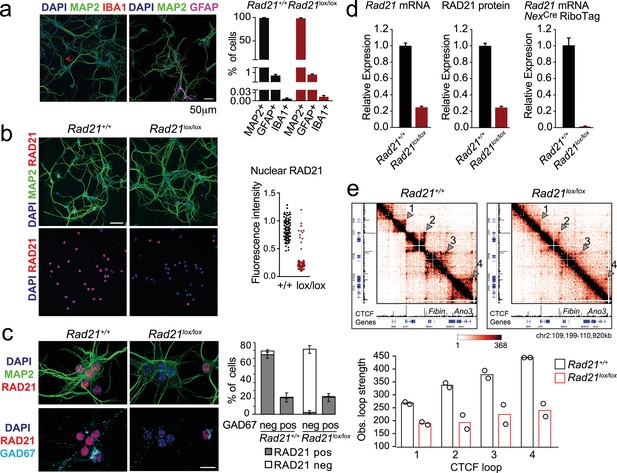
Conditional cohesin deletion in post-mitotic neurons.
(a) E17.5–E18.5 cortices were dissociated and plated on poly-D-lysine. After 10 days, cultures were stained for pan neuronal (MAP2), astrocyte (GFAP), and microglia (IBA1) markers, and cell type composition was determined by quantitative analysis of immunofluorescence images. Based on 6 Rad21+/+ NexCre and 8 Rad21lox/lox NexCre different samples analyzed in four independent experiments. (b) Immunofluorescence staining of Rad21+/+ NexCre and Rad21lox/lox NexCre neuronal explant cultures for RAD21 and MAP2 (left) and distribution of RAD21 expression by MAP+ neurons (right). Note the discontinuous distribution of RAD21 expression in Rad21lox/lox NexCre neurons. Three independent experiments per genotype. DAPI marks nuclei. Scale bar = 60 μm. (c) Immunofluorescence staining for RAD21, MAP2, and the marker of GABAergic inhibitory neurons, GAD67 (left). Distribution of RAD21 expression in GAD67+ and GAD67- neurons (right). Note that the discontinuous distribution of RAD21 expression in Rad21lox/lox NexCre neuronal explant cultures is due to GAD67+ GABAergic inhibitory neurons. Three independent experiments for Rad21+/+ NexCre and six independent experiments for Rad21lox/lox NexCre. DAPI marks nuclei. Scale bar = 20 μm. (d) Quantitative RT-PCR analysis of Rad21 mRNA expression in Rad21+/+ NexCre and Rad21lox/lox NexCre cortical explant cultures (mean ± SEM, n=18). Hprt and Ubc were used for normalization (left). RAD21 protein expression in Rad21+/+ NexCre and Rad21lox/lox NexCre cortical explant cultures was quantified by fluorescent immunoblots (mean ± SEM, n=6, a representative blot is shown in Figure 1—figure supplement 1) and normalized to LaminB (center). NexCre RiboTag RNA-seq of analysis of Rad21 mRNA expression in Rad21+/+ NexCre and Rad21lox/lox NexCre cortical explant cultures (right, three independent biological replicates). (e) 5C heat maps of Rad21+/+ NexCre and Rad21lox/lox NexCre cortical explant cultures. Shown is a 1.72 Mb region covered by 5C analysis of chr2 107601077–110913077 (Beagan et al., 2020). One of two independent biological replicates with similar results. CTCF ChIP-seq in cortical neurons (Bonev et al., 2017) and mm9 coordinates are shown for reference. Arrowheads mark the position of CTCF-based loops. Results were consistent across two replicates and three chromosomal regions. Histograms below show the quantification of representative CTCF-based loops (arrowheads) in two independent biological replicates for control and Rad21lox/lox NexCre neurons.
-
Figure 1—source data 1
Figure 1: Conditional cohesin deletion in post-mitotic neurons.
- https://cdn.elifesciences.org/articles/76539/elife-76539-fig1-data1-v2.xlsx
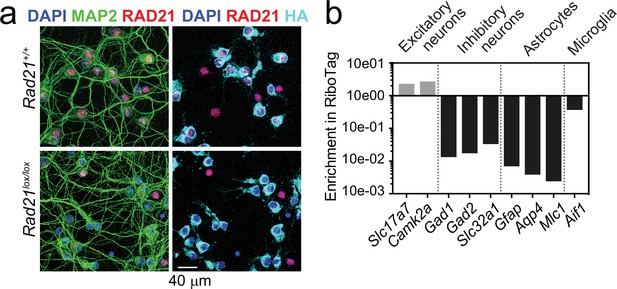
Rad21 NexCre RiboTag validation.
(a) RAD21 and LAMIN B protein expression in Rad21+/+ NexCre and Rad21lox/lox NexCre cortical explant cultures was analyzed by fluorescent immunoblots. One representative blot of six is shown here, the quantification of all six blots is shown in Figure 1d. (b) NexCre-dependent Rpl22-HA (RiboTag) expression is restricted to RAD21-negative cells in Rad21lox/lox NexCre neurons. Immunofluorescence staining for RAD21, the pan-neuronal marker MAP2 and HA (RiboTag) in explant culture. DAPI marks nuclei. Scale bar = 40 μm. (c) NexCre RiboTag captures excitatory neuron-specific transcripts such as Slc17a7 and Camk2a and depletes cell type-specific transcripts expressed in inhibitory neurons (Gad1, Gad2, Slc32a1), astrocytes (Gfap, Aqp4, Mlc1), and microglia (Aif). Transcript enrichment (or depletion) was calculated using the normalized counts from NexCre RiboTag versus standard RNA-seq in Rad21+/+ NexCre neurons.
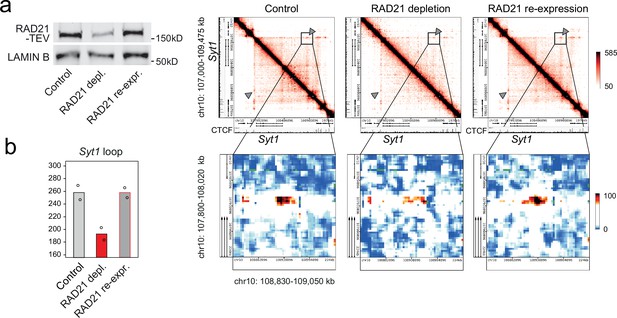
Restoration of cohesin rescues chromatin loops.
(a) Transient RAD21 depletion by Dox-inducible TEV expression and recovery after Dox washout (Weiss et al., 2021). RAD21-TEV western blots (left) and heat maps of chromatin contacts at the cohesin-dependent neuronal genes Syt1 in RAD21-TEV neurons (right). (b) Quantification of chromatin loops at the Syt1 locus obtained by 5C analysis of chr10 107002896–109474896 (Beagan et al., 2020) in control neurons, cohesin-depleted neurons 24 hr after Dox-dependent TEV induction, and after re-expression of RAD21. One of two independent biological replicates with similar results.
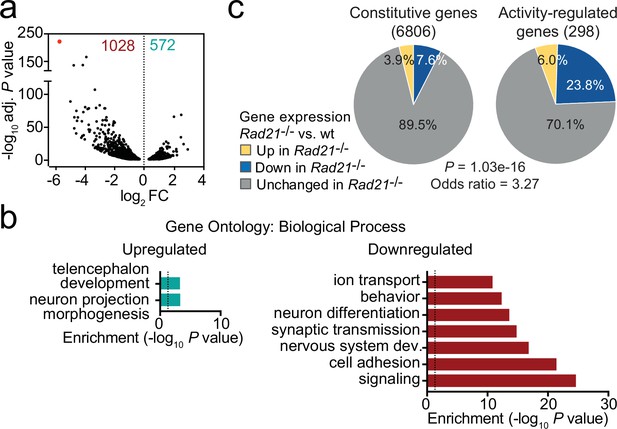
Loss of cohesin from immature post-mitotic neurons perturbs neuronal gene expression.
(a) Volcano plot representing log2 fold-change (FC) versus significance (-log10 of adjusted p values) of downregulated genes (1028) and upregulated genes (572) in RiboTag RNA-seq of Rad21lox/lox NexCre versus Rad21+/+ NexCre neurons (Supplementary file 1). Red marks Rad21. (b) Analysis of gene ontology of biological functions of deregulated genes in Rad21lox/lox NexCre neurons. Enrichment is calculated relative to expressed genes (Supplementary file 3). (c) The percentage of constitutive (adj. p>0.05 in KCl 1 hr versus TTX and KCl 6 hr versus TTX, see methods) and activity-regulated genes Kim et al., 2010 found deregulated in Rad21lox/lox NexCre neurons in explant culture at baseline as determined by RiboTag RNA-seq. The p-value (Fisher Exact Test) and Odds ratio indicate that ARGs are more frequently deregulated than constitutive genes.
-
Figure 2—source data 1
Figure 2: Loss of cohesin from immature post-mitotic neurons perturbs neuronal gene expression.
- https://cdn.elifesciences.org/articles/76539/elife-76539-fig2-data1-v2.xlsx
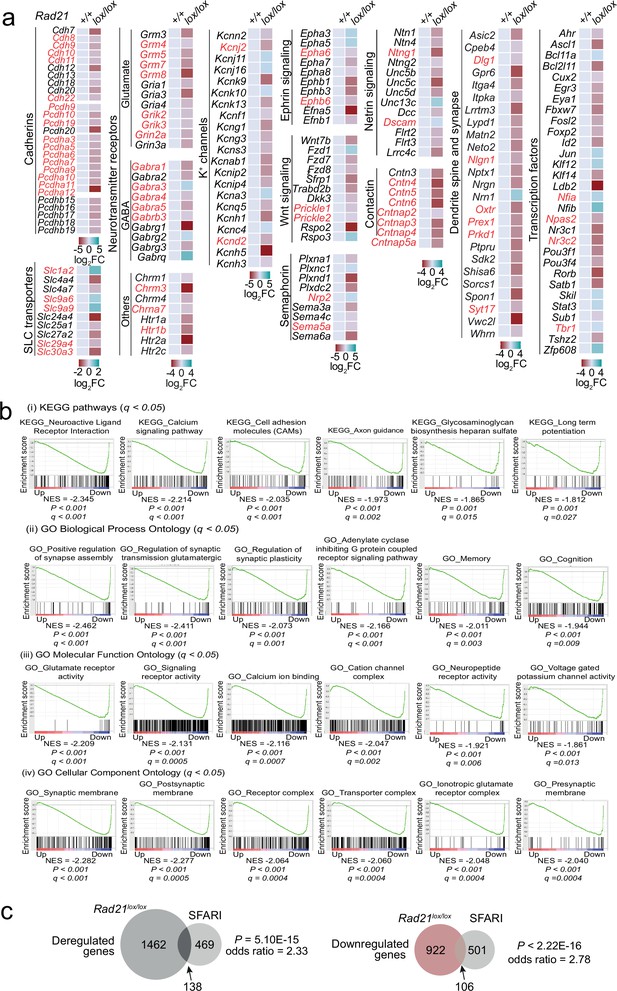
Gene expression in Rad21lox/lox NexCre neurons.
(a) Examples of deregulated genes in Rad21lox/lox NexCre neurons. Genes associated with autism spectrum disorders are highlighted in red. (c) Overlap between human genes associated with autism spectrum disorders from the SFARI database and differentially expressed genes (left), downregulated genes (middle) and upregulated genes (right) in Rad21lox/lox NexCre cortical neurons. (b) GSEA for downregulated genes in NexCre/+ Rad21lox/lox neurons using gene sets derived from (i) KEGG pathway database, (ii) GO Biological Process Ontology, (iii) GO Molecular Function Ontology, (iv) GO Cellular Component Ontology in the Molecular Signatures Database (MSigDB).
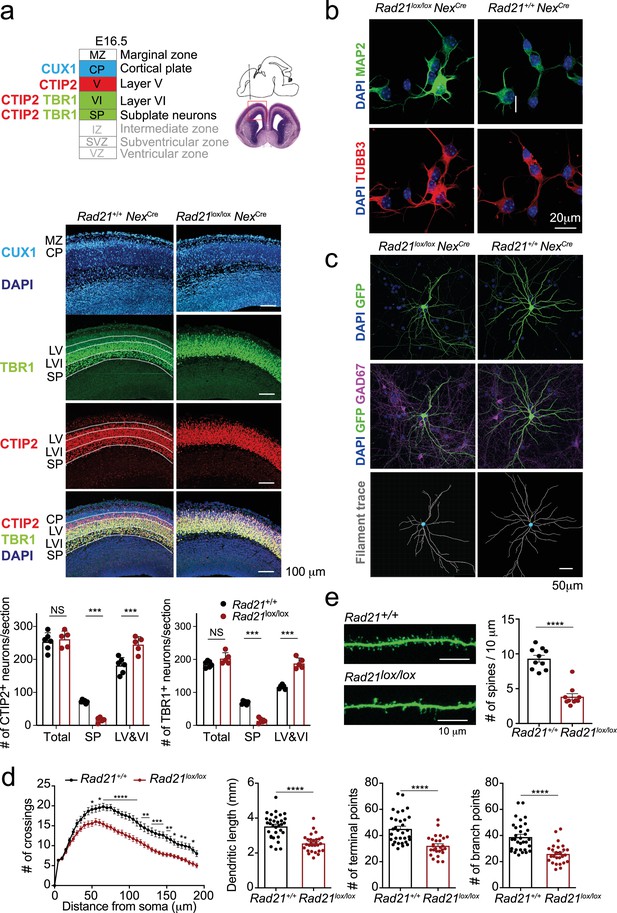
Cohesin contributes to the maturation of post-mitotic neurons.
(a) Top: Schema of cortical layers (Greig et al., 2013) showing subplate (SP), layer 6 (VI), layer 5 (V), the cortical plate (CP), and the marginal zone (MZ). Middle: Immunofluorescence analysis of the neuronal transcription factors CUX1, TBR1, and CTIP2 at E16.5. Scale bar = 100 μm. Representative of three biological replicates. Bottom: Quantification of TBR1+ and CTIP2+ neurons in the subplate (SP) and in layers 5 and 6 (LV and VI). Neuron counts per 150 × 300 μm field are shown for five comparable sections from two embryos per genotype. Mean ± SE, *** adj. p<0.0001, two-way ANOVA with Sidak’s multiple comparisons test. (b) Morphology of E18.5 neurons after 1d in explant culture. Immunofluorescence staining for the pan-neuronal marker MAP2, tubulin beta 3 (TUBB3), and DAPI. Scale bar = 20 μm. (c) Morphology of Rad21+/+ NexCre and Rad21lox/lox NexCre cortical neurons in explant culture on rat glia (Kaech and Banker, 2006). Cultures were sparsely labeled with GFP to visualize individual cells and their processes, and stained for GAD67 to exclude GABAergic neurons. Dendritic traces of GFP+ neurons. Scale bar = 50 μm. (d) Sholl analysis of Rad21+/+ NexCre and Rad21lox/lox NexCre cortical neurons in explant cultures shown in (c). Shown is the number of crossings, dendritic length, terminal points, and branch points per 10 μm. Three independent experiments, 32 Rad21lox/lox NexCre and 28 Rad21+/+ NexCre neurons. * adj. p<0.05, ** adj. p<0.01, *** adj. p<0.001, **** adj. p<0.0001. Scale bar = 10 μm. (e) Quantification of spines per 10 μm for Rad21+/+ NexCre and Rad21lox/lox NexCre cortical neurons. Two independent experiments, 10 Rad21lox/lox NexCre and 10 Rad21+/+ NexCre neuron. **** adj. p<0.0001. Scale bar = 10 μm.
-
Figure 3—source data 1
Figure 3: Cohesin contributes to the maturation of post-mitotic neurons.
- https://cdn.elifesciences.org/articles/76539/elife-76539-fig3-data1-v2.xlsx
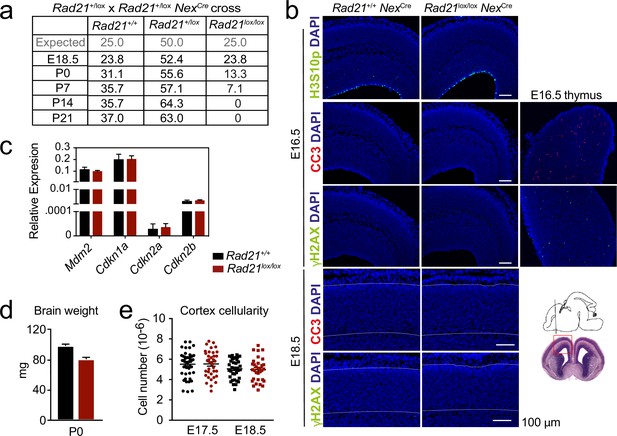
Impact of cohesin loss in immature post-mitotic neurons in vivo.
(a) Expected Mendelian ratios and observed percentages of live Rad21+/+ NexCre, Rad21lox/+ NexCre, Rad21lox/lox NexCre mice at the indicated developmental stages, n=217. (b) Immunofluorescence analysis shows that neither the mitotic marker phosphorylated Serine 10 on histone H3 (H3S10p) nor the apoptosis marker activated caspase 3 (CC3) or the DNA damage marker γH2AX in E16.5 (top) and E18.5 Rad21lox/lox NexCre (bottom, white lines demarcate the cortex). Wild type E16.5 thymi are shown as positive controls for CC3 and γH2AX. Two biological replicates. Scale bar = 100 μm. Photomicrographs of coronal brain sections at gestational age E16 modified from the Atlas of the prenatal mouse brain (Paxinos et al., 2020) are shown for orientation. (c) Quantitative RT-PCR analysis of gene expression in Rad21+/+ NexCre and Rad21lox/lox NexCre E17.5/18.5 cortical explant cultures 10 days after plating. Hprt and Ubc were used for normalization. Mean ± SEM of three cultures per genotype. (d) Brain weights of Rad21+/+ NexCre and Rad21lox/+ NexCre, Rad21lox/lox NexCre mice at birth (P0). Mean ± SEM of between three and six mice per genotype. (e) Embryonic cortices from wild type and Rad21lox/lox NexCre mice were dissected at E17.5 and E18.5 and dissociated. Cortical cell numbers were determined by counting in Neubauer chambers. Each symbol denotes an independent experiment. Mean ± SEM are also shown.
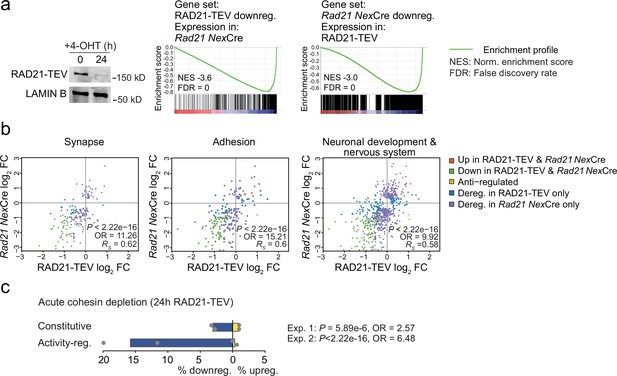
Acute cohesin depletion deregulates ARG expression.
(a) Western blot documenting acute RAD21 depletion by 4-OHT-inducible RAD-TEV cleavage (left). GSEA of the gene set downregulated (DEseq2, adj. p<0.05) in RAD21-TEV neurons in Rad21lox/lox NexCre neurons (center). GSEA of genes downregulated in Rad21lox/lox NexCre neurons (DEseq2, adj. p<0.05) in RAD21-TEV neurons (right). NES: normalized enrichment score. FDR: false discovery rate. (b) Scatter plots of gene expression within aggregate GO terms, comparing RAD21-TEV with Rad21lox/lox NexCre neurons. Genes that were found deregulated in at least one of the genotypes are shown. p-values and odds ratios refer to the probability of finding the observed patterns of co-regulation by chance. RS: Spearman’s rank coefficient. (c) Deregulation of constitutive and activity-regulated genes 24 hr after acute cohesin depletion by inducible proteolytic cleavage of RAD21-TEV; adj. p<0.05 based on DEseq2 analysis of three RNA-seq replicates per experiment. Blue indicates downregulation and yellow indicates upregulation in RAD21-TEV versus wild type. Two independent experiments are shown (Weiss et al., 2021 and Supplementary file 4).
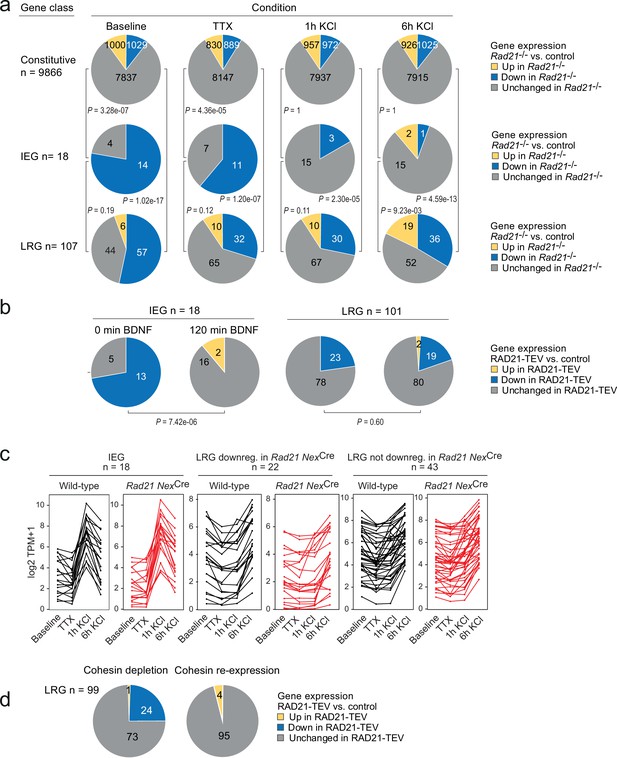
Activity-regulated neuronal gene (ARG) classes differ in their reliance on cohesin.
(a) Pie charts show the expression of constitutive genes (top), immediate early genes (IEGs) (center), and late response genes (LRGs) (bottom) in Rad21 NexCre neurons under four different conditions: baseline, TTX and D-AP5 (TTX), and in response to KCl stimulation for 1 hr or 6 hr. Numbers of expressed constitutive genes, IEGs, and LRGs are given on the left. Note that KCl stimulation normalizes the expression of most (10 out of 11) IEGs downregulated in TTX, but a fraction of LRGs remain downregulated. p-values test the prevalence of deregulated genes in each class under each condition, two-sided Fisher exact test. (b) Pie charts show the expression of IEGs (top) and LRGs (bottom) in RAD21-TEV neurons under baseline conditions 24 hr after ERt2-TEV induction and in response to BDNF (120 min). RAD21-TEV cleavage led to the downregulation (adj. p<0.05) of 13 out of 18 expressed IEGs and of 23 out of 101 expressed LRGs. p-values test the prevalence of downregulated IEGs and LRGs with and without BDNF stimulation, two-tailed Fisher exact test. Note that BDNF stimulation reversed the downregulation of IEGs but not LRGs in cohesin-depleted neurons. (c) Strip plots depict the expression of IEGs, LRGs that are downregulated in Rad21 NexCre neurons compared to control across conditions (TTX and 6 hr KCl), and LRGs that are not downregulated in Rad21 NexCre neurons compared to control across conditions. (d) Transient cohesin depletion and re-expression as in Figure 1—figure supplement 2. Pie charts show the expression of LRGs in RAD21-TEV relative to control neurons. 24 out of 97 LRGs expressed in RAD21-TEV neurons were downregulated 24 hr after Dox-dependent TEV induction (adj. p<0.05). The downregulation of LRGs was reversible upon Dox washout and restoration of RAD21 expression.
-
Figure 5—source data 1
Figure 5: Activity-regulated neuronal gene (ARG) classes differ in their reliance on cohesin.
- https://cdn.elifesciences.org/articles/76539/elife-76539-fig5-data1-v2.xlsx
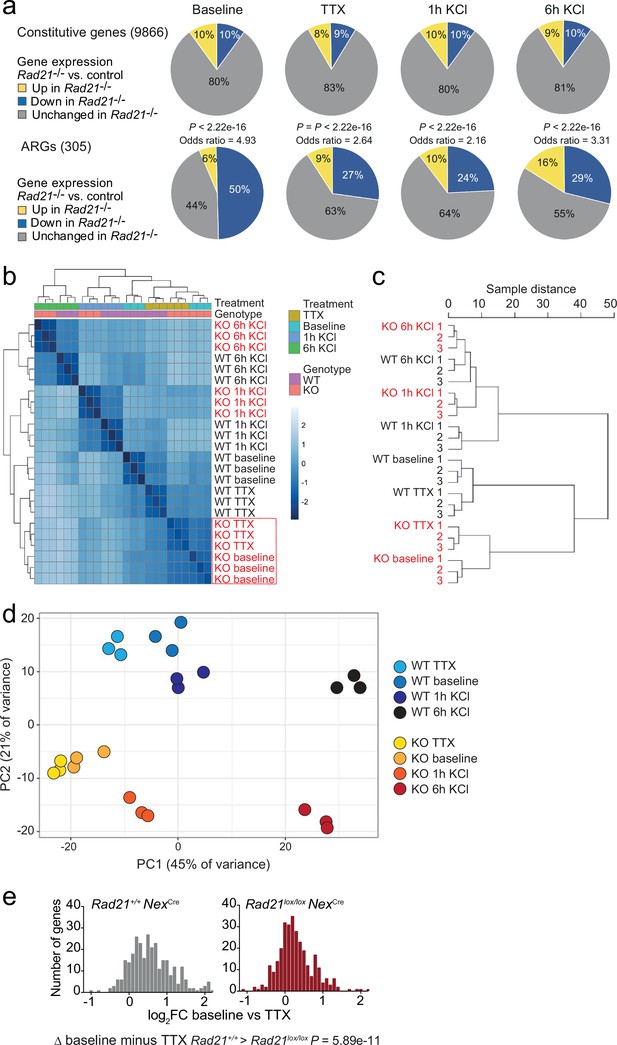
Gene expression and genotype interaction analysis.
(a) The expression of constitutive and previously defined activity-regulated genes (Kim et al., 2010) was determined by RNA-seq in explant cultures of Rad21lox/lox NexCre neurons at baseline, in the presence of TTX and D-AP5 (TTX), and after stimulation with KCl for 1 hr and 6 hr. Odds ratios show the enrichment of Activity-regulated neuronal genes (ARGs). p-values for deviation of odds ratios from one were determined by two-tailed Fisher’s exact test. (b) Heatmap of sample distances for genotype interaction analysis of Rad21+/+ and Rad21lox/lox NexCre neurons across conditions. Note the clustering of baseline and TTX conditions for Rad21lox/lox NexCre neurons (red box). (d) Principal component analysis of Rad21+/+ and Rad21lox/lox NexCre neurons across conditions (all genes). PC1 (45% of variance) separates activation conditions and PC2 separates genotype (21% of variance). (c) Sample distance dendrograms indicate that ARG expression in Rad21lox/lox NexCre neurons shows little difference between spontaneous activity at baseline and TTX. (e) ARG expression in explant cultures of Rad21+/+ and Rad21lox/lox NexCre neurons under baseline conditions that allow for cell-cell communication versus TTX/D-AP5 (TTX). The difference in ARG expression between baseline and TTX was greater for wild-type than for Rad21lox/lox NexCre neurons (p=5.89e-11, Kolmogorov-Smirnov test).
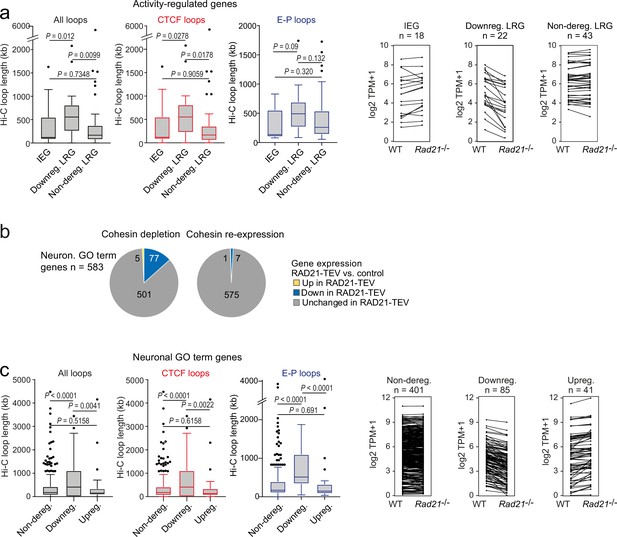
The genomic distance traversed by chromatin contacts formed by neuronal genes predicts whether or not cohesin is required for their full expression.
(a) The span of Hi-C loops (left), Hi-C loops with CTCF bound to at least one of the loop anchors (middle) and Hi-C loops between promoters and inducible enhancers (right) for Immediate early genes (IEGs) (n=18) and late response genes (LRGs) downregulated in Rad21 NexCre versus control neurons in both resting (TTX) and activation conditions (6 hr KCl, adj p<0.05 in both TTX and 6 hr KCl conditions, n=22, ‘Downreg. LRG’), and LRGs not deregulated in either resting (TTX) or activation conditions (6 hr KCl, adj p>0.05 in both TTX and 6 hr KCl conditions) in Rad21 NexCre relative to control neurons (adj. p>0.05, n=43, 'Non-dereg. LRG’). Box plots show the longest loop for each gene rather than average loop length, as Hi-C loop calling at 10 kb resolution precludes detection of loops <40 kb (Beagan et al., 2020). However, analysis of average loop length confirmed that downregulated genes form longer loops than non-deregulated genes among both ARGs and neuronal GO term genes (p=0.0056 and p<0.0001, respectively). Genes without loops are included except for analysis of enhancer loops. Box plots show the longest loop recorded for each gene. Boxes show upper and lower quartiles and whiskers show 1.5 of the interquartile range. p-values were determined by non-parametric Kolmogorov-Smirnov test. Strip plots depict the expression of IEGs, downregulated LRGs, and non-deregulated LRGs in wild-type and Rad21 NexCre neurons. (b) Transient cohesin depletion and re-expression as in Figure 1—figure supplement 2. Pie charts show the expression of neuronal genes related to synaptic transmission (GO:0007268) and glutamate receptor signaling pathway (GO:0007215). 77 out of 583 expressed genes in these GO terms (Neur. GO term genes) were downregulated 24 hr after Dox-dependent TEV induction (adj. p<0.05). The downregulation of 76 of 77 neuronal GO term genes was reversible upon Dox washout and restoration of RAD21 expression, an additional 6 genes were downregulated after Dox washout but not at 24 hr of TEV induction. (c) The span of Hi-C loops (left), Hi-C loops with CTCF bound to at least one of the loop anchors (middle) and Hi-C loops between promoters and constitutive or inducible enhancers (right) for genes in the neuronal GO terms synaptic transmission and glutamate receptor signaling. Gene expression in Rad21 NexCre versus control neurons was assessed in both resting and activation conditions: not deregulated in TTX or 6 hr KCl, n=401, Downregulated in both TTX and 6 hr KCl, n=85, Upregulated in both TTX and 6 hr KCl, n=41. Genes do not form loops are included except for analysis of enhancer loops. Boxes show upper and lower quartiles and whiskers show 1.5 of the interquartile range. p-values were determined by non-parametric Kolmogorov-Smirnov test. Strip plots show the expression of the depicted GO term genes in control and Rad21 NexCre neurons.
-
Figure 6—source data 1
Figure 6: The genomic distance traversed by chromatin contacts formed by neuronal genes predicts whether or not cohesin is required for their full expression.
- https://cdn.elifesciences.org/articles/76539/elife-76539-fig6-data1-v2.xlsx
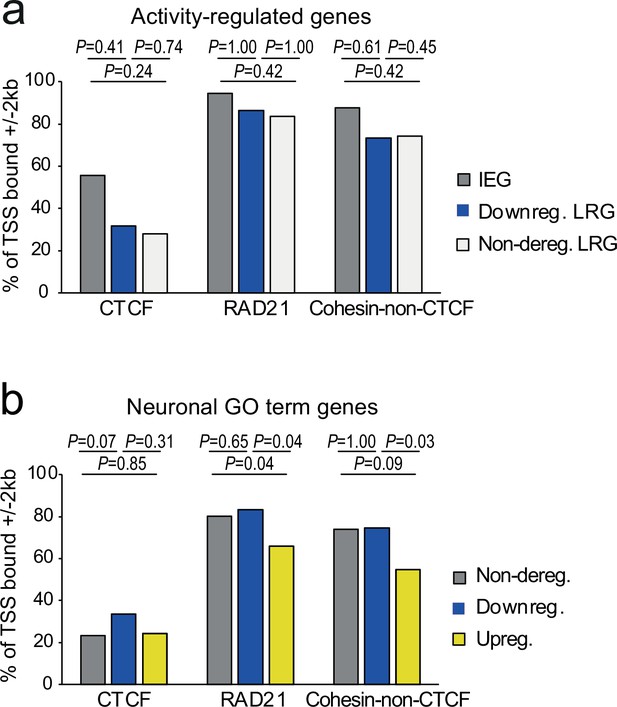
Promoter binding of CTCF or cohesin does not distinguish cohesin-dependent from cohesin-independent genes.
(a) Presence of CTCF (Bonev et al., 2017), RAD21 (Fujita et al., 2017), or cohesin-non-CTCF binding at gene promoters (TSS ± 2 kb) at immediate early genes (IEGs), late response genes (LRGs) that are downregulated across conditions (TTX, 6 h KCl), and LRGs that are not deregulated across conditions in Rad21 NexCre versus control neurons (left). p-values: two-tailed Fisher exact test. (b) Presence of CTCF (Bonev et al., 2017), RAD21 (Fujita et al., 2017), or cohesin-non-CTCF binding at gene promoters (TSS ± 2 kb) at neuronal genes related to ‘synaptic transmission’ (GO:0007268) and ‘glutamate receptor signaling pathway’ (GO:0007215) that are not deregulated across conditions, downregulated across conditions, or upregulated across conditions (TTX, 6 hr KCl) in Rad21 NexCre versus control neurons (right). p-values: two-tailed Fisher exact test.
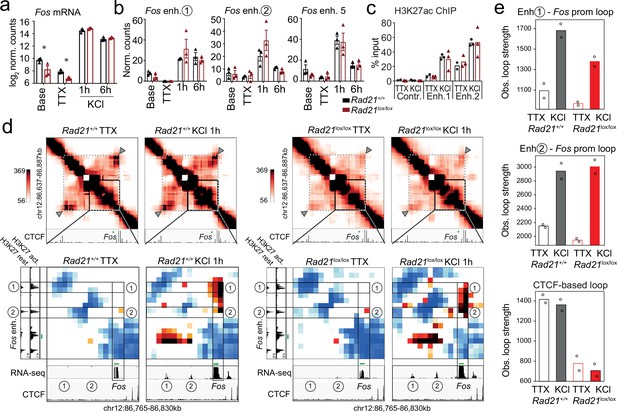
Fos enhancer-promoter contacts are robustly induced in cohesin-deficient neurons.
(a) Expression of the immediate early genes (IEG) Fos at baseline, after TTX/D-AP5 (TTX), and KCl-stimulation (left, mean log2-transformed counts from 3 biological replicates, * adj. p<0.05). (b) Enhancer transcripts in control and Rad21lox/lox NexCre neurons were quantified based on normalized RNA-seq reads within 1 kb of the enhancer. An intergenic region on chr11 was used as a negative control (71.177.622–71.177.792) . (c) H3K27ac ChIP normalized to H3 in control and Rad21lox/lox NexCre neurons at a control site, Fos enhancer 1 and Fos enhancer 2 after TTX/D-AP5 (TTX) or 1 hr KCl (KCl). (d) Interaction frequency (top) and interaction score (bottom) heatmaps of the region immediately surrounding Fos obtained by 5C analysis of chr12 86201802–87697802 (Beagan et al., 2020). Black frames highlight interactions between the Fos gene and upstream enhancers 1 and 2. CTCF ChIP-seq in cortical neurons (Bonev et al., 2017) is shown for orientation and H3K27ac ChIP-seq in inactive (TTX-treated) and activated neurons is shown to annotate enhancer regions (Beagan and Phillips-Cremins, 2020; Beagan et al., 2020). RNA-seq in TTX-treated and 1 hr KCl-activated control and Rad21lox/lox NexCre neurons shows KCl-inducible transcription of Fos enhancers in wild -type and cohesin-deficient neurons. Two independent biological replicates are shown in Figure 7—figure supplement 3a. (e) Quantification of the interaction frequencies between the Fos promoter and Fos enhancer 1 (top), the Fos promoter and Fos enhancer 2 (middle), and CTCF-marked boundaries of the sub-TAD containing Fos (bottom, grey arrowhead). Two replicates per genotype and condition.
-
Figure 7—source data 1
Figure 7: Fos enhancer-promoter contacts are robustly induced in cohesin-deficient neurons.
- https://cdn.elifesciences.org/articles/76539/elife-76539-fig7-data1-v2.xlsx
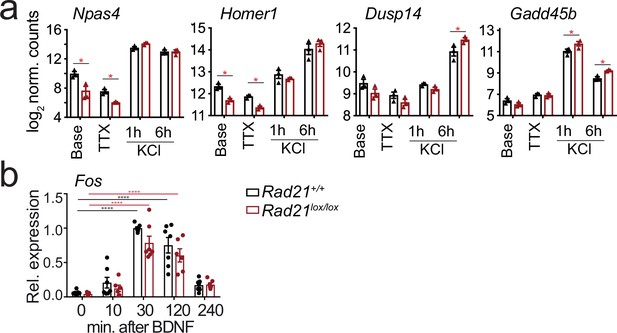
Inducible gene expression in cohesin-deficient neurons.
(a) Examples of Activity-regulated neuronal gene (ARG) expression at baseline, after TTX/D-AP5 (TTX), and KCl-stimulation. Mean log2-transformed counts from three biological replicates (* adj. p<0.05). (b) Expression of Fos mRNA at baseline and at the indicated times after BDNF stimulation. Data points represent biological RT-PCR replicates. p-values refer to induction relative to 0 min. *** p<0.001.
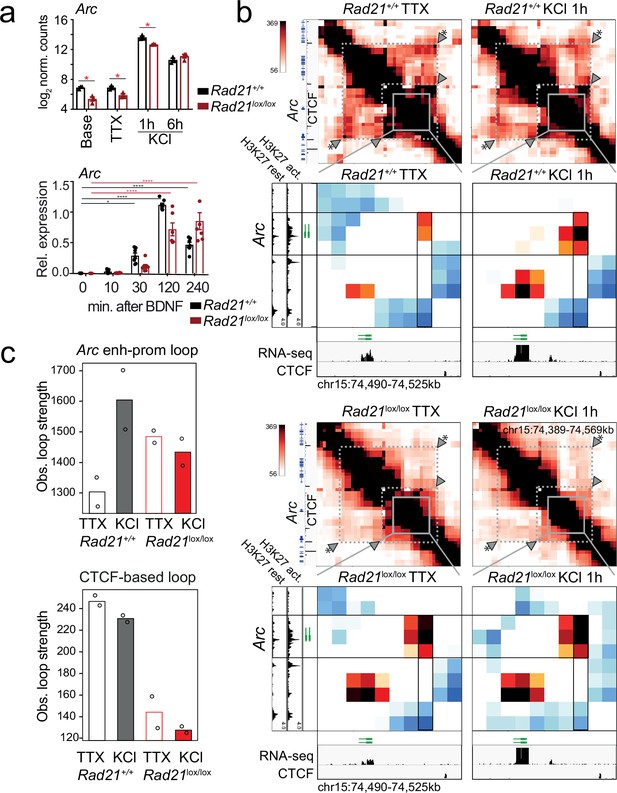
Contacts between the Arc promoter and an inducible enhancer in wild -type and cohesin-deficient neurons.
(a) Expression of Arc mRNA at baseline, after TTX/D-AP5 (TTX), and KCl-stimulation (top, mean log2-transformed counts from three biological RNA-seq replicates, * adj. p<0.05) and at the indicated time after BDNF stimulation (bottom, data points represent biological RT-PCR replicates). p-values refer to induction relative to 0 min. * p<0.05, *** p<0.001. (b) Interaction frequency (top) and interaction score (bottom) heatmaps of the region immediately surrounding Arc obtained by 5C analysis of chr15 73376037–74580037 (Beagan et al., 2020) in resting (TTX) and 1 hr KCl-activated wild -type (top) and Rad21lox/lox NexCre neurons (bottom). Black frames highlight interaction between the Arc gene (y-axis) and a nearby downstream enhancer (x-axis). CTCF-ChIP-seq for cortical neurons is shown (Bonev et al., 2017). H3K27ac ChIP-seq in inactive (TTX-treated) and activated neurons is shown to annotate enhancer regions (Beagan et al., 2020). Two independent biological replicates are shown in Figure 7—figure supplement 3. (c) Quantification of the interaction frequencies of the Arc enhancer-promoter loop (top). and a CTCF-based loop that braces the Arc locus (arrowhead marked with * in panel b) for comparison (bottom).
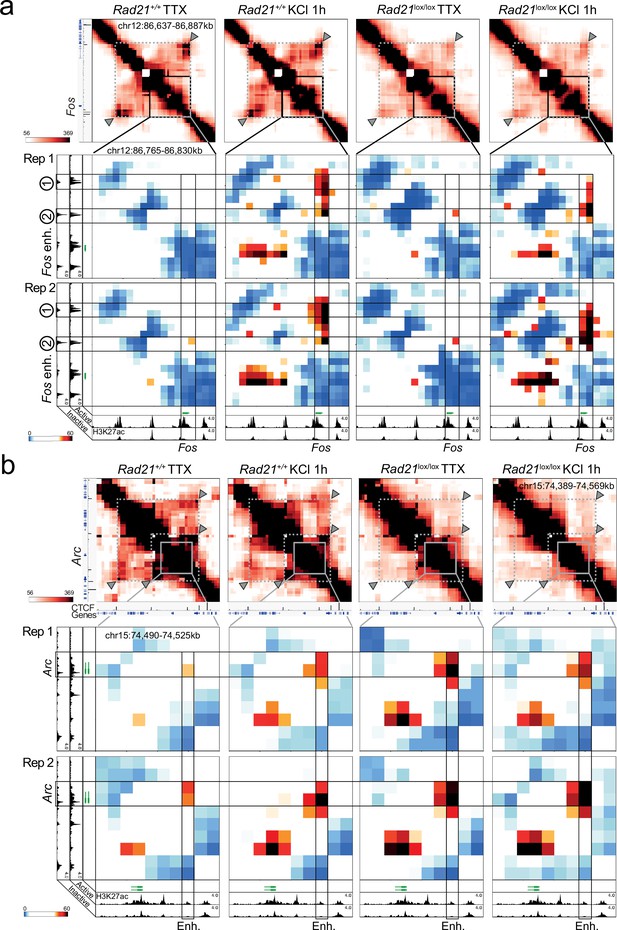
Replicate 5 C experiments.
(a) Top: Interaction frequency zoom-in heatmaps of 250 kb region surrounding the Fos gene obtained by 5C analysis of chr12 86201802–87697802. Dashed lines and arrow heads mark major CTCF binding sites at the boundaries of the domain that contains Fos. Note the weakening of these contacts in Rad21lox/lox NexCre neurons. Bottom: Interaction score heatmaps of the 65 kb region immediately surrounding the Fos gene. Black frames highlight interactions between the Fos gene and upstream enhancers 1 and 2. Two independent biological replicates are shown. H3K27ac ChIP-seq data (Beagan et al., 2020) from Bicuculline-treated (active) and TTX-treated (inactive) neurons annotate enhancer regions. (b) Top: Interaction frequency zoom-in heatmaps of ~200 kb region surrounding the Arc gene obtained by 5C analysis of chr15 73376037–74580037. Dashed lines and arrow heads mark major CTCF binding sites at the boundaries of domains that contain the Arc locus. Note the weakening of these contacts in Rad21lox/lox NexCre neurons. Bottom: Interaction score heatmaps of the ~40 kb region immediately surrounding the Arc gene. Black frames highlight interaction between the Arc gene (y-axis) and a nearby downstream enhancer (x-axis). H3K27ac ChIP-seq data25 from Bicuculline-treated (active) and TTX-treated (inactive) neurons annotate enhancer regions. Two independent biological replicates are shown.
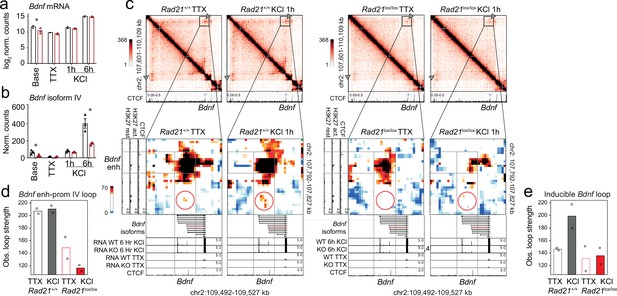
Bdnf enhancer-promoter contacts are weakened in the absence of cohesin.
(a) Total Bdnf transcripts at baseline, after TTX/D-AP5 (TTX), and KCl-stimulation (left, mean log2-transformed counts from three biological replicates, * adj. p<0.05). (b) Bdnf promoter IV transcripts at baseline, after TTX/D-AP5 (TTX), and KCl-stimulation (left, mean log2-transformed counts from three biological replicates, * adj. p<0.05). (c) Interaction frequency (top) and interaction score (bottom) heatmaps of the Bdnf region obtained by 5C analysis of chr2 107601077-110913077 (Beagan et al., 2020). CTCF ChIP-seq in cortical neurons (Bonev et al., 2017) and the position of Bdnf are displayed (top). Below: Zoom-in of constitutive Bdnf enhancer-promoter loop (gray frame). Shown on the side is H3K27ac ChIP-seq in resting and activated neurons, marking an activity-dependent enhancer, and CTCF ChIP-seq. RNA-seq in resting and activated wild-type and cohesin deficient neurons and CTCF ChIP-seq are shown underneath. A circle marks an inducible 1.68Mb 5C loop between Bdnf and an activity-induced enhancer (Bdnf enhancer 1 in Beagan et al., 2020). (d) Quantification of 5C interaction frequencies between Bdnf promoter IV and the activity-dependent enhancer. (e) Quantification of inducible 5C loop between Bdnf and Bdnf enhancer 1 (Beagan et al., 2020).
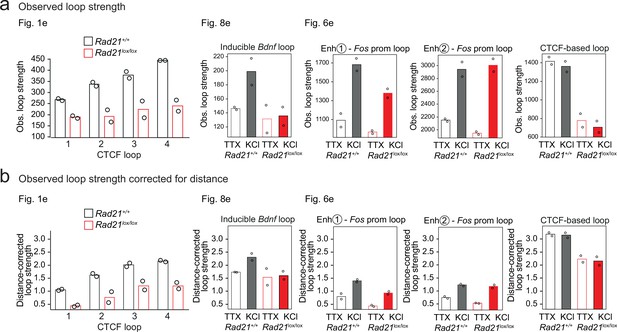
Quantification of observed and distance-corrected loop strength.
(a) Observed values loop strength, as presented throughout the paper. (b) Observed values loop strength corrected for the distance-dependent background signal and shown on the same y-axis scale for all loops shown.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | Rad21lox Rad21tm1.1Mmk | DOI: 10.1038/nature10312 | MGI:5293824 | |
| Genetic reagent (Mus musculus) | NexCre Neurod6tm1(cre)Kan | DOI: 10.1002/dvg.20256 | MGI:2668659 | |
| Genetic reagent (Mus musculus) | Rpl22(HA)lox (RiboTag) Rpl22tm1.1Psam | DOI: 10.1073/pnas.0907143106 | MGI:4355967 | |
| Genetic reagent (Mus musculus) | Rad21tev Rad21tm1.1Kktk | DOI: 10.1101/gad.605910 | MGI:4840469 | |
| antibody | anti-RAD21 (rabbit polyclonal) | Abcam | Cat #. ab154769 | WB: (dilution 1:1000) IF: (dilution 1:500) |
| antibody | anti-LAMIN B (goat polyclonal) | Santa Cruz Biotechnology | Cat #. sc-6216 | WB: (dilution 1:10,000) |
| antibody | anti-rabbit IgG (H + L) Alexa Fluor 680 (goat polyclonal) | ThermoFisher Scientific | Cat #. A-21109 | WB: (dilution 1:10,000) |
| antibody | anti-goat IgG (H + L) Alexa Fluor 680 (donkey polyclonal) | ThermoFisher Scientific | Cat #. A-21084 | WB: (dilution 1:10,000) |
| antibody | anti-HA (rabbit polyclonal) | Sigma | Cat #. H6908 | polysome immunoprecipitation |
| antibody | anti-GFAP (rabbit polyclonal) | Wako | Cat #. Z0334 | IF: (dilution 1:500) |
| antibody | anti-MAP2 (chicken polyclonal) | Abcam | Cat #. ab611203 | IF: (dilution 1:5000) |
| antibody | anti-GAD67 (mouse monoclonal) | Millipore | Cat #. MAB5406 | IF: (dilution 1:500) |
| antibody | anti-HA (mouse monoclonal) | Covance | Cat #. MMS-101R | IF: (dilution 1:1000) |
| antibody | IBA1 (rabbit polyclonal) | Wako | Cat #. 019–19741 | IF: (dilution 1:250) |
| antibody | anti-TUBB3 (Tuj1, mouse monoclonal) | Biolegend | Cat #. 801,202 | IF: (dilution 1:500) |
| antibody | anti-gamma-H2AX (rabbit polyclonal) | Bethyl Laboratories | Cat #. A300-081A | IF: (dilution 1:3000) |
| antibody | anti-Cleaved Caspase-3 (Asp175) (rabbit polyclonal) | Cell signalling | Cat #. 9,661 | IF: (dilution 1:400) |
| antibody | anti-TBR1 (rabbit polyclonal) | Abcam | Cat #. ab31940 | IF: (dilution 1:1000) |
| antibody | anti-CTIP2 (25B6, rat monoclonal) | Abcam | Cat #. ab18465 | IF: (dilution 1:500) |
| antibody | anti-CUX-1 (rabbit polyclonal) | Santa Cruz Biotechnology | Cat #. sc-13024 | IF: (dilution 1:400) |
| antibody | anti–Phospho-Histone H3 S10 Alexa Fluor 647 conjugate (rabbit polyclonal) | Cell signalling | Cat #. 9,716 | IF: (dilution 1:50) |
| antibody | anti-rabbit IgG (H + L) Alexa Fluor 647 (goat polyclonal) | ThermoFisher Scientific | Cat #. A-21244 | IF: (dilution 1:500) |
| antibody | anti-Rabbit IgG (H + L) Alexa Fluor 568 (goat polyclonal) | ThermoFisher Scientific | Cat #. A-11011 | IF: (dilution 1:500) |
| antibody | goat anti-mouse IgG (H + L) Alexa Fluor 488 (goat polyclonal) | ThermoFisher Scientific | A-11001 | IF: (dilution 1:500) |
| antibody | anti-chicken IgY (H + L) Alexa Fluor 568 (goat polyclonal) | Abcam | ab175711 | IF: (dilution 1:500) |
| Software, algorithm | ImageJ software | (http://imagej.nih.gov/ij/) | ||
| Software, algorithm | GraphPad Prism software | (https://graphpad.com) | ||
| Software, algorithm | FilamentTracer, Imaris software, Bitplane AG | https://imaris.oxinst.com | ||
| Software, algorithm | GSEA Desktop v3.0 | https://www.gsea-msigdb.org/gsea/index.jsp | ||
| Software, algorithm | Leica Application Suite X (LAS X, v2.7) software | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ | ||
| Software, algorithm | CellProfiler v2.2 | https://cellprofiler.org | ||
| Software, algorithm | Image Studio Software (v5.2) | Li-cor Image Studio https://www.licor.com/bio/image-studio/ |
Additional files
-
Supplementary file 1
Gene expression analysis NexCre Ribotag RNA-seq Rad21 lox/lox minus wild-type.
- https://cdn.elifesciences.org/articles/76539/elife-76539-supp1-v2.xlsx
-
Supplementary file 2
Gene expression analysis total RNA-seq NexCre Rad21 lox/lox minus wild-type baseline ('at rest').
- https://cdn.elifesciences.org/articles/76539/elife-76539-supp2-v2.xlsx
-
Supplementary file 3
Gene ontology analysis of NexCre Ribotag RNA-seq Rad21 lox/lox minus wild-type.
- https://cdn.elifesciences.org/articles/76539/elife-76539-supp3-v2.xlsx
-
Supplementary file 4
Gene expression analysis RAD21-TEV 24 h cleaved vs control at baseline.
- https://cdn.elifesciences.org/articles/76539/elife-76539-supp4-v2.xlsx
-
Supplementary file 5
Gene expression analysis total RNA-seq NexCre Rad21 lox/lox minus wild-type TTX, KCl 1 h, KCl 6 h.
- https://cdn.elifesciences.org/articles/76539/elife-76539-supp5-v2.xlsx
-
Supplementary file 6
Gene expression analysis RAD21-TEV 24 h cleaved vs control BDNF 30 min and BDNF 120 min.
- https://cdn.elifesciences.org/articles/76539/elife-76539-supp6-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/76539/elife-76539-transrepform1-v2.docx