Differences in pathways contributing to thyroid hormone effects on postnatal cartilage calcification versus secondary ossification center development
Figures
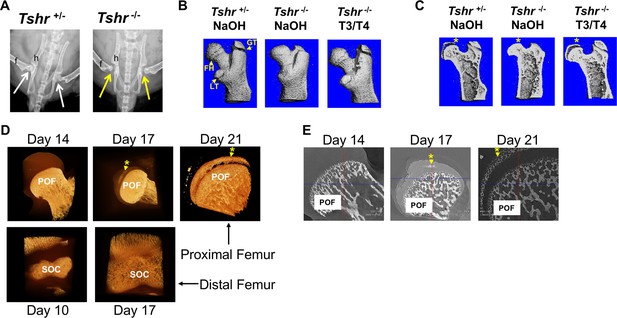
Tshr-/- proximal femur phenotype and onset of mineralization.
(Panel A) Ventral X-ray view of postnatal (P) 21-day-old mice. Arrows point to joint between hip (h) and femur (f). (Panel B) Lateral view of three-dimensional (3D) μCT scans from P21 proximal femurs. FH = femur head, GT = greater trochanter; LT = lesser trochanter. (Panel C) Lateral μCT hemi-section view of proximal femurs. Asterisk adjacent to region mineralized in FH. (Panel D) Lateral views of 3D nano-computed tomography (nano-CT) images of proximal FH (top row) and distal femur epiphysis (bottom row). Mineralized tissue is opaque ivory/gold colored. (Panel E) Two-dimensional sections of nano-CT. Mineralized tissue is white/bright gray. POF: primary ossification front, SOC: secondary ossification center. Asterisk and arrow point to mineralization at FH. Images shown are representative of three to five mice per group.
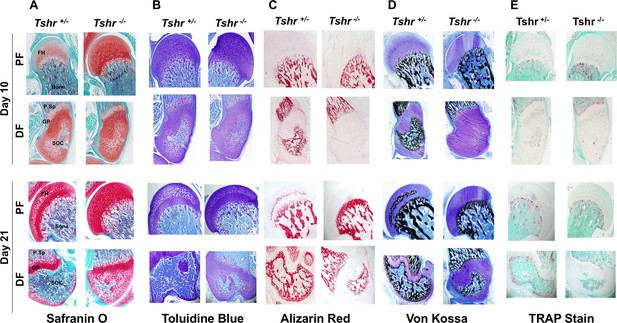
Histological analysis of femur head (FH) in euthyroid Tshr+/- and hypothyroid Tshr-/- mice.
Safranin O stained sections showing cartilage in red (Panel A), toluidine blue stained sections showing cartilage in violet (Panel B), Alizarin red stained sections showing mineral in red (Panel C), Von Kossa stained sections showing mineral in black (Panel D), and TRAP/ACP5 stained sections showing TRAP activity in red (Panel E) at the proximal femur, PF, and distal femur, DF, from Tshr+/- and Tshr-/- mice. P.Sp = primary spongiosa; GP = growth plate; SOC = secondary ossification center.
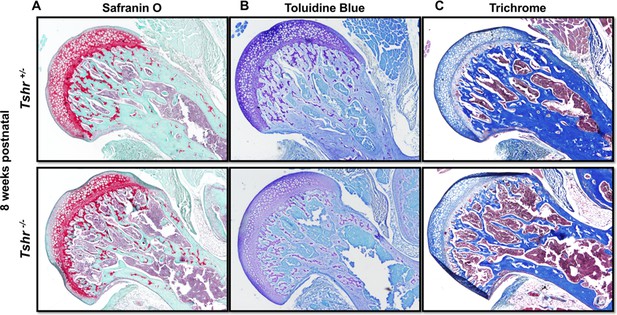
Delayed mineralization in Tshr-/- femur head (FH).
Postnatal week 8 proximal femur sections stained in red for Safranin O (Panel A), violet for toluidine blue (Panel B), dark blue-mineral, light blue-collagen, red-nuclei for trichrome (Panel C).
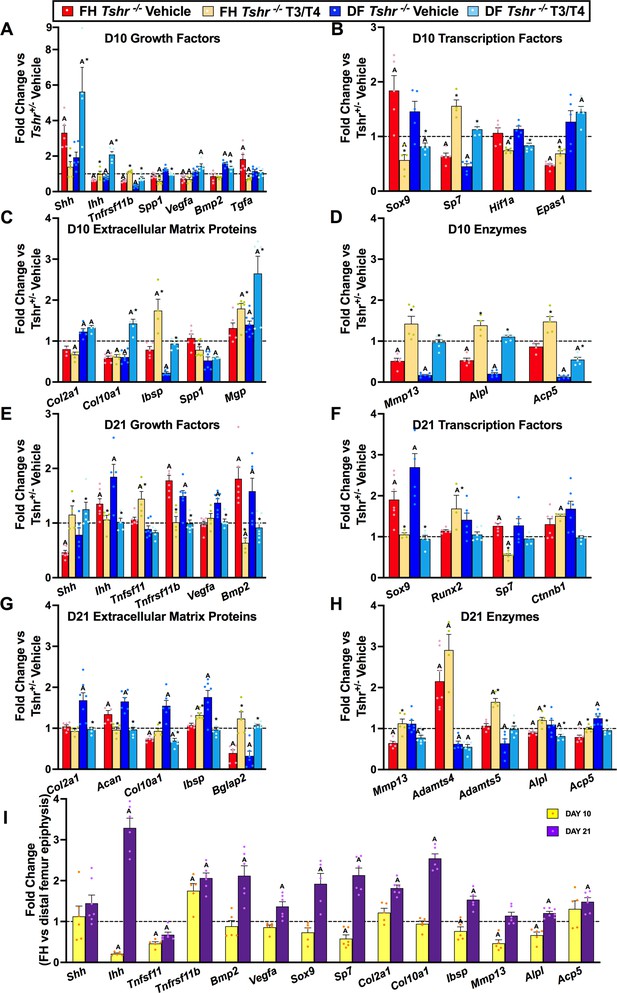
Thyroid hormone (TH)-dependent transcriptional differences at proximal and distal femur (DF).
(Panels A–G) Gene expression changes from femur head (FH) or DF between Tshr-/- injected with vehicle, or T3/T4, plotted as fold-change relative to anatomical and stage matched region in euthyroid Tshr+/- with vehicle (dashed line). Day 10 samples (Panels A–D) were treated on days 5–9, while day 21 samples (Panels E–G) were treated on days 5–14. (Panel I) Fold-change of mRNA expression at FH versus DF epiphysis in Tshr+/- days 10 and 21. Statistics analyzed by t-test where A = p < 0.05, * = p < 0.05 between T3/T4 treatment and vehicle at FH or DF. (n = 6) Sonic hedgehog (Shh), Indian hedgehog (Ihh), tumor necrosis factor receptor superfamily member 11b – synonym rank ligand (Tnfsf11b/Rankl), tumor necrosis factor receptor superfamily member 11b – synonym osteoprotegerin (Tnfrsf11b/opg), vascular endothelial growth factor a (Vegfa), bone morphogenetic protein 2 (Bmp2), transforming growth factor alpha (Tgfa), SRY-box transcription factor 9 (Sox9), runt-related transcription factor 2 (Runx2), Sp7 transcription factor – synonym osterix (Sp7/Osx), hypoxia inducible factor 1alpha (Hif1a), endothelial PAS domain protein 1 – synonym hypoxia inducible factor 2alpha (Epas1/Hif2a), collagen type 2 alpha 1 (Col2a1), collagen type 10 alpha 1 (Col10a1), integrin binding sialoprotein – synonym bone sialoprotein (Ibsp/bsp), secreted phosphoprotein 1 – synonym osteopontin (Spp1/Opn), bone gamma-carboxyglutamate protein 2 – synonym osteocalcin (Bglap2/Ocn), matrix gla protein (Mgp), matrix metallopeptidase 13 (Mmp13), alkaline phosphatase liver/bone/kidney (Alpl), acid phosphatase five tartrate resistant – synonym tartrate resistant acid phosphatase (Acp5/Trap), beta catenin (Ctnnb1), aggrecan (Acan), ADAM metallopeptidase with thrombospondin type 1 motif 4 (Adamts4), ADAM metallopeptidase with thrombospondin type 1 motif 5 (Adamts5).
-
Figure 3—source data 1
Source data for Figure 3A–D.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) data for day 10 Tshr femur head (FH) and distal femur (DF). Average fold changes plotted and t-test values are highlighted in yellow.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Source data for Figure 3E–H.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) data for day 21 Tshr femur head (FH) and distal femur (DF). Average fold changes plotted and t-test values are highlighted in yellow.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig3-data2-v2.xlsx
-
Figure 3—source data 3
Source data for Figure 3I.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) data for day 10 and day 21 comparing femur head (FH) versus distal femur (DF) from euthyroid mice. Average fold changes plotted and t-test values are highlighted in yellow.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig3-data3-v2.xlsx
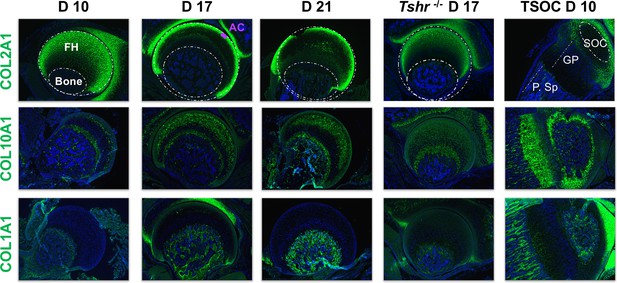
Immunohistochemical characterization of femur head (FH) development.
Longitudinal sections of Tshr+/- were probed for protein expression in FH on days 10, 17, 21, proximal tibia epiphysis on day 10, and FH of Tshr-/- on day 17 by immunofluorescence for collagens: COL2A1, COL10A1, COL1A1 (all green). Immunofluorescent images counterstained with DAPI (blue). Abbreviations are references for all immunohistology figures (Figures 4—9); FH, femur head; AC, articular cartilage; SOC, secondary ossification center; GP, growth plate; P.Sp, primary spongiosa.
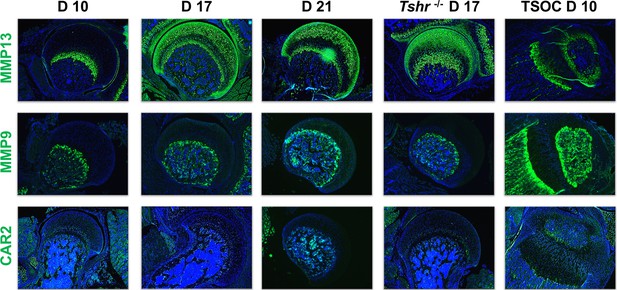
Immunohistochemical characterization of femur head (FH) development.
Longitudinal sections of Tshr+/- were probed for protein expression in FH on days 10, 17, 21, proximal tibia epiphysis on day 10, and FH of Tshr-/- on day 17 by immunofluorescence for enzymes: matrix metallopeptidase MMP13, MMP9, carbonic anhydrase 2 (CAR2) stained in green; counterstained with DAPI (blue).
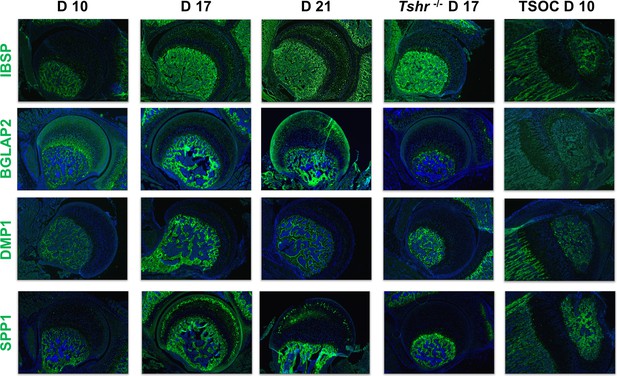
Immunohistochemical characterization of femur head (FH) development.
Longitudinal sections of Tshr+/- were probed for protein expression in FH on days 10, 17, 21, proximal tibia epiphysis on day 10, and FH of Tshr-/- on day 17 by immunofluorescence for non-collagenous extracellular matrix proteins: IBSP, BGLAP2, dentin matrix protein 1 (DMP1), and SPP1 stained in green; samples were counterstained with DAPI (blue).
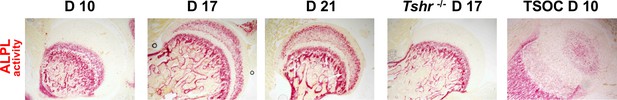
Alkaline phosphatase (ALPL) activity in femur head (FH) on days (D) 10, 17, 21, proximal tibia epiphysis on day 10 of Tshr+/-, and FH of Tshr-/- on day 17 FH (red stain).
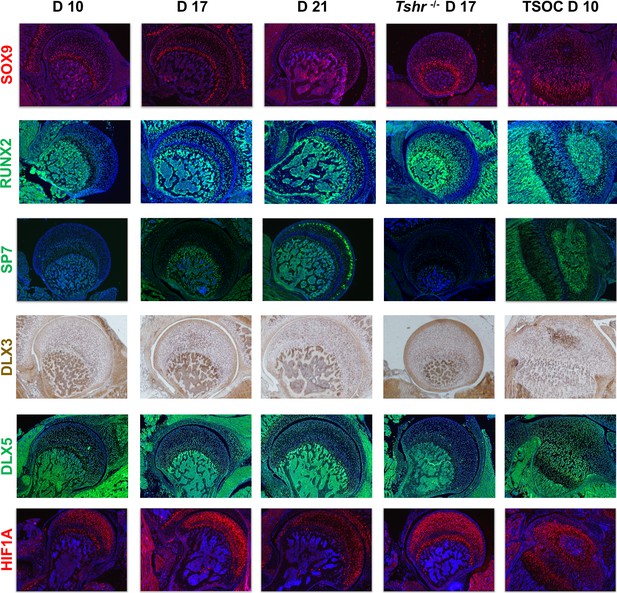
Immunohistochemical characterization of femur head (FH) development.
Longitudinal sections of Tshr+/- were probed for protein expression in FH on days 10, 17, 21, proximal tibia epiphysis on day 10, and FH of Tshr-/- on day 17 by immunofluorescence for transcription factors: SOX9, HIF1A are stained in red. RUNX2, SP7, and distal-less homeobox 5 (DLX5) are stained in green. Immunofluroescence images counterstained with DAPI (blue). DLX3 stained in brown by colorimetric immunohistochemistry.
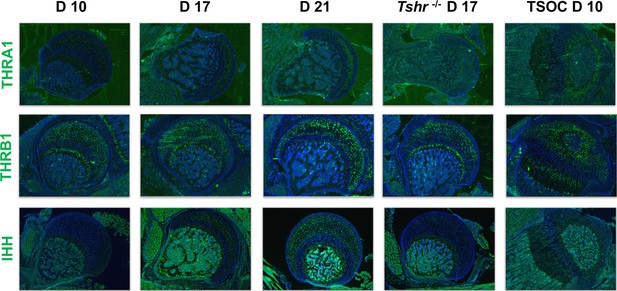
Immunohistochemical characterization of femur head (FH) development.
Longitudinal sections of Tshr+/- were probed for protein expression in FH on days 10, 17, 21, proximal tibia epiphysis on day 10, and FH of Tshr-/- on day 17 by immunofluorescence for thyroid hormone response factors: thyroid hormone receptor A1 (THRA1), THRB1, IHH stained green. Counterstained with DAPI (blue).
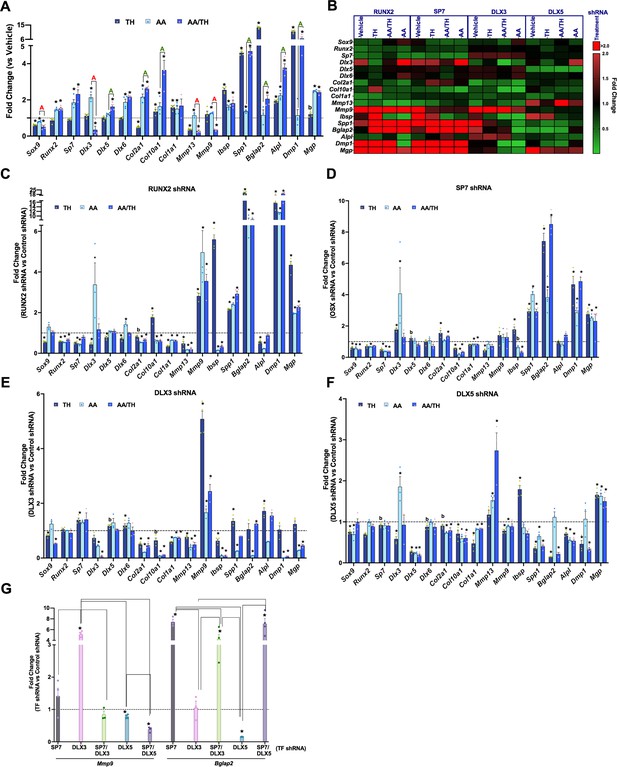
Thyroid hormone (TH) and transcription factor regulation of chondroprogenitor genes.
(Panels A–G) Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) on day 3 for ATDC5 cell line represented as fold-change for genes labeled on X-axis. (Panel A) ATDC5 with control shRNA following treatment with TH, ascorbic acid (AA), or AA and TH (AA/TH). (Panel B) Heat map of all 17 genes analyzed in ATDC5 with shRNA for RUNX2, SP7, DLX3, DLX5. Graphed are the response to treatment with vehicle, TH, AA, or AA/TH versus control shRNA. Reduction in expression signifies positive regulation (green), while increased expression signifies negative regulation (red). (Panels C–F) Individual transcription factor knockdown versus control shRNA following treatments versus vehicle. (Panel G) Comparison between groups knocked down for a given transcription factor combination on X-axis versus control shRNA in TH treatment at either Mmp9 or Bglap2. Statistics analyzed by t-test where * = p < 0.05 or less; b = p < 0.05–0.10, and in Panel G one-way ANOVA where comparison bars indicate p < 0.05 or less (n = 4).
-
Figure 9—source data 1
Source data for Figure 9A.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) data for control shRNA. Average fold changes plotted and t-test values are highlighted in yellow.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig9-data1-v2.xlsx
-
Figure 9—source data 2
Source data for Figure 9C.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) data for RUNX2 shRNA. Average fold changes plotted and t-test values are highlighted in yellow.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig9-data2-v2.xlsx
-
Figure 9—source data 3
Source data for Figure 9D.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) data for SP7 shRNA. Average fold changes plotted and t-test values are highlighted in yellow.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig9-data3-v2.xlsx
-
Figure 9—source data 4
Soure data for Figure 9E.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) data for DLX3 shRNA. Average fold changes plotted and t-test values are highlighted in yellow.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig9-data4-v2.xlsx
-
Figure 9—source data 5
Source data for Figure 9F.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) data for DLX5 shRNA. Average fold changes plotted and t-test values are highlighted in yellow.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig9-data5-v2.xlsx
-
Figure 9—source data 6
Source data for Figure 9G.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) data for SP7 shRNA without or with either DLX3 shRNA and/or DLX5 shRNA. Average fold changes plotted and t-test values are highlighted in yellow.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig9-data6-v2.xlsx
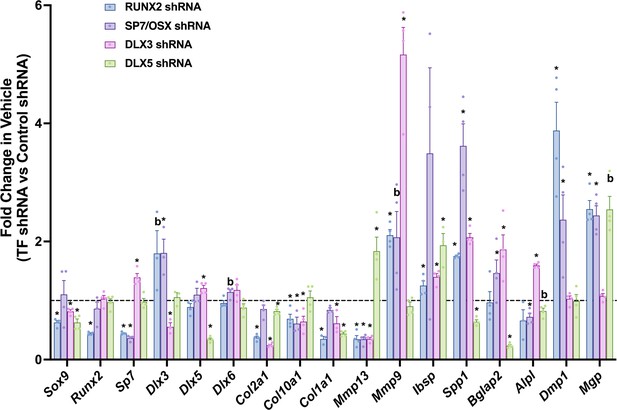
Effect of individual transcription factor knockdown in vehicle treatment.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) on day 3 for ATDC5 transduced with shRNA for individual transcription factors RUNX2, SP7, DLX3, or DLX5 comparing fold changes at genes labeled on x-axis versus control shRNA in vehicle treatment condition. Statistics analyzed by t-test where * = p < 0.05 or less; b = p < 0.05–0.10 (n = 4).
-
Figure 9—figure supplement 1—source data 1
RUNX2 shRNA.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig9-figsupp1-data1-v2.xlsx
-
Figure 9—figure supplement 1—source data 2
SP7 shRNA.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig9-figsupp1-data2-v2.xlsx
-
Figure 9—figure supplement 1—source data 3
DLX3 shRNA.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig9-figsupp1-data3-v2.xlsx
-
Figure 9—figure supplement 1—source data 4
DLX5 shRNA.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig9-figsupp1-data4-v2.xlsx
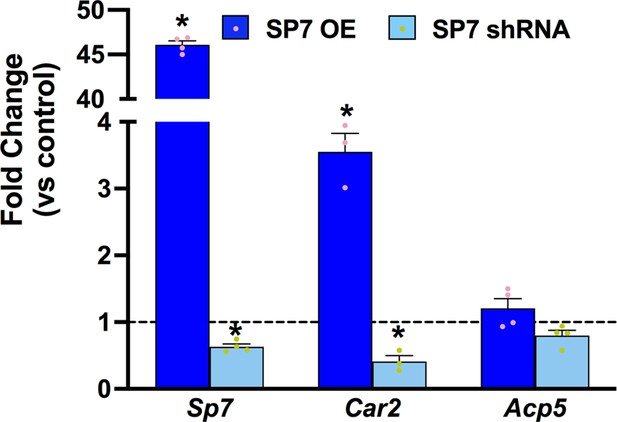
Regulation of mineralization enzymes by SP7.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) on day 3 for ATDC5 with SP7 overexpression (OE) or knockdown with SP7 shRNA. Fold changes are relative to empty vector GFP control for OE, or control shRNA in vehicle treated cells. Statistics analyzed by t-test where * = p < 0.05 or less (n = 3).
-
Figure 9—figure supplement 2—source data 1
SP7 OE and shRNA.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig9-figsupp2-data1-v2.xlsx
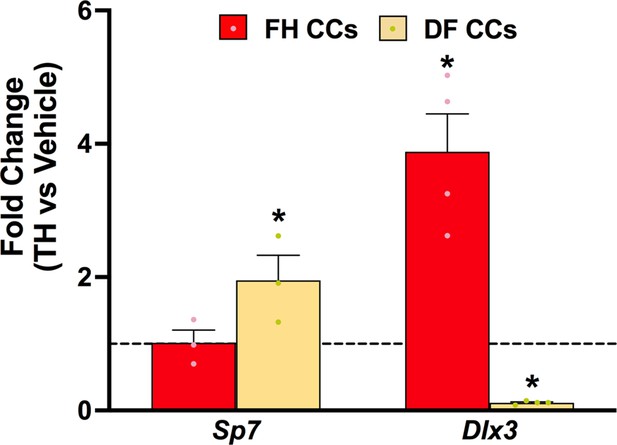
Thyroid hormone (TH) regulation of Sp7 and Dlx3 expression in chondrocytes ex vivo.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) for Sp7 or Dlx3 from cultured chondrocytes (CCs) isolated from femur head (FH) or distal femur (DF) treated with TH for 22 days. Statistics analyzed by t-test where * = p < 0.05 or less (n = 4).
-
Figure 9—figure supplement 3—source data 1
FH vs DF chondrocyte response to TH.
- https://cdn.elifesciences.org/articles/76730/elife-76730-fig9-figsupp3-data1-v2.xlsx
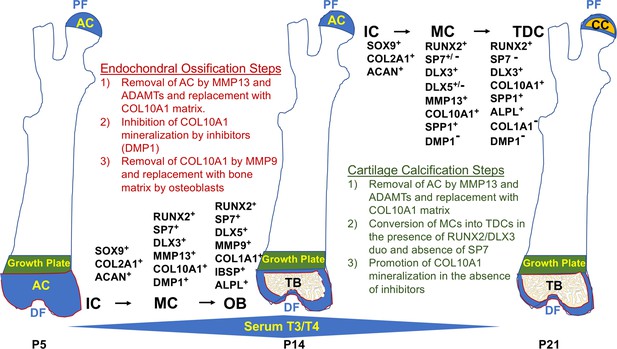
Spatiotemporal model of proximal-distal femoral chondrocyte fate acquisition.
Please see text in the discussion for explanation. Abbreviations: PF, proximal femur; AC, articular cartilage; CC, calcified cartilage; DF, distal femur; IC, immature chondrocyte; MC, mature chondrocyte; TDC, terminally differentiated chondrocyte; OB, osteoblast; TB, trabecular bone; P5–21, postnatal day; T3/T4, thyroid hormone.
Tables
Comparison of genes regulated by different transcription factors with a p-value < 0.05 from Figure 9 panel B after treatment with vehicle, TH, AA + TH, or AA.
‘Negative’ indicates knockdown of a given transcription factor results in a significant increase of gene in question, while ‘positive’ indicates the opposite. 2 = RUNX2; 3 = DLX3; 5 = DLX5; 7 = SP7/OSX shRNAs.
| Vehicle treatment | TH treatment | AA + TH treatment | AA treatment | |||||
|---|---|---|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | |
| Sox9 | 2,3,5 | 2,3,5,7 | 7 | 5,7 | ||||
| Runx2 | 7 | |||||||
| Sp7 | 3 | 2 | 3 | 2 | 2 | |||
| Dlx3 | 7 | 7 | 2,5 | 2,5,7 | ||||
| Dlx5 | 3 | 2 | ||||||
| Dlx6 | 2 | 3 | 2 | |||||
| Col2a1 | 2,3 | 7 | 3 | 7 | 2,3,5 | 2,3,5 | ||
| Col10a1 | 2,3,7 | 2 | 5,7 | 2,3,5,7 | 2,3,5,7 | |||
| Col1a1 | 2,3,5 | 2,3,5 | 2,3,5,7 | 2,3,5,7 | ||||
| Mmp13 | 5 | 2,3,7 | 2,3,7 | 5 | 2,3 | 5 | 2,3 | |
| Mmp9 | 2,3 | 2,3 | 5 | 2,3 | 2,3 | 5 | ||
| Ibsp | 2,3 | 2,5,7 | 3 | 2,3,7 | 2,3 | |||
| Spp1 | 2,3,7 | 5 | 2,3,7 | 5 | 2,7 | 5 | 2,7 | 3,5 |
| Bglap2 | 3 | 5 | 2,7 | 5 | 2,3,7 | 5 | 2,7 | 3 |
| Alpl | 3 | 7 | 3 | 2 | 5 | 2,5 | ||
| Dmp1 | 2,7 | 2,7 | 5 | 2,7 | 3,5 | 2,7 | 3 | |
| Mgp | 2,7 | 2,5,7 | 2,5,7 | 3 | 2,5,7 | 3 | ||
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/76730/elife-76730-transrepform1-v2.pdf
-
Supplementary file 1
Antibodies.used.
Antigen retrieval with: HD = 2 mg/mL hyaluronidase (Sigma-Aldrich) in PBS at 37°C for 45 min. S = 10 mM sodium citrate with 2 mM EDTA and 0.05% Tween-20 (pH 6.0) at 95°C for 6 min. L = 10 mM sodium citrate with 2 mM EDTA and 0.05% Tween-20 (pH 6.0) at 60°C for 72 hr.
- https://cdn.elifesciences.org/articles/76730/elife-76730-supp1-v2.xlsx
-
Supplementary file 2
Real Time quantitative PCR primers used.
- https://cdn.elifesciences.org/articles/76730/elife-76730-supp2-v2.xlsx





