Genetic variation of putative myokine signaling is dominated by biological sex and sex hormones
Figures
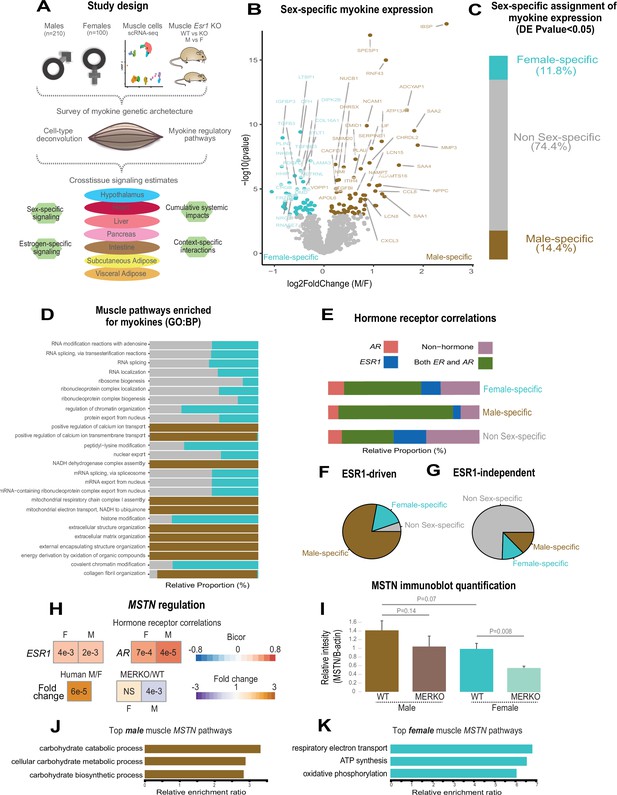
Sex and hormone effects on myokine regulation.
(A) Overall study design for integration of gene expression from muscle from 310 humans, single-cell RNA-sequencing (RNA-seq), muscle-specific deletion of Esr1 to infer interorgan coregulatory process across major metabolic tissues. (B–C) Differential expression analysis for sex was performed on all genes corresponding to secreted proteins in skeletal muscle (myokines). The specific genes which showed significant changes in each sex are shown as a volcano plot (B) and the relative proportions of myokines corresponding to each category at a least-stringent logistic regression p-value less than 0.05 (C). (D) For each differential expression category based on sex shown in C, myokines were correlated with all other muscle genes for pathway enrichment. Then the top 10 enriched pathways in males, females, or non-sex specific (by overall significance) were visualized together where number of genes corresponding to each category shown as a relative proportion. (E) The same analysis as in D, except instead of myokines being correlated with AR, ESR1, both hormone receptors, or neither, as compared to correlating with all genes. (F–G) Myokines were binned into two categories based on significant differential expression (logistic regression adjusted p-value < 0.05) between muscle-specific WT and MERKO mice (F) or those that showed no change (G), then visualized as relative proportions within each category shown in (C). (H) Midweight bicorrelation (bicor) coefficients (color scheme) and corresponding regression p-values (filled text) are shown for muscle MSTN ~ESR1 or AR in both sexes (top). Below, correlations are shown for differential expression log2FC (color scheme) and corresponding logistic regression p-values (text fill) for MSTN between sexes in humans or WT vs. MERKO mice. (I) Quantification of processed form of myostatin (Figure 1—figure supplement 2, bottom band) relative to β-actin in WT or MERKO muscle in male or female mice. p-Values calculated using a Student’s t-test. (J–K) The top three pathways of genes which significantly (p < 1e-4) correlated with muscle MSTN in males (J) or females (K). For human data, n = 210 males and n = 100 females. For mouse MERKO vs. WT comparisons, n = 3 mice per group per sex. p-Values from midweight bicorrelations were calculated using the Student’s p-value from WGCNA and logistic regression p-values were calculated using DESeq2.
-
Figure 1—source data 1
Skeletal muscle sex hormone receptor expression between sexes.
- https://cdn.elifesciences.org/articles/76887/elife-76887-fig1-data1-v2.zip
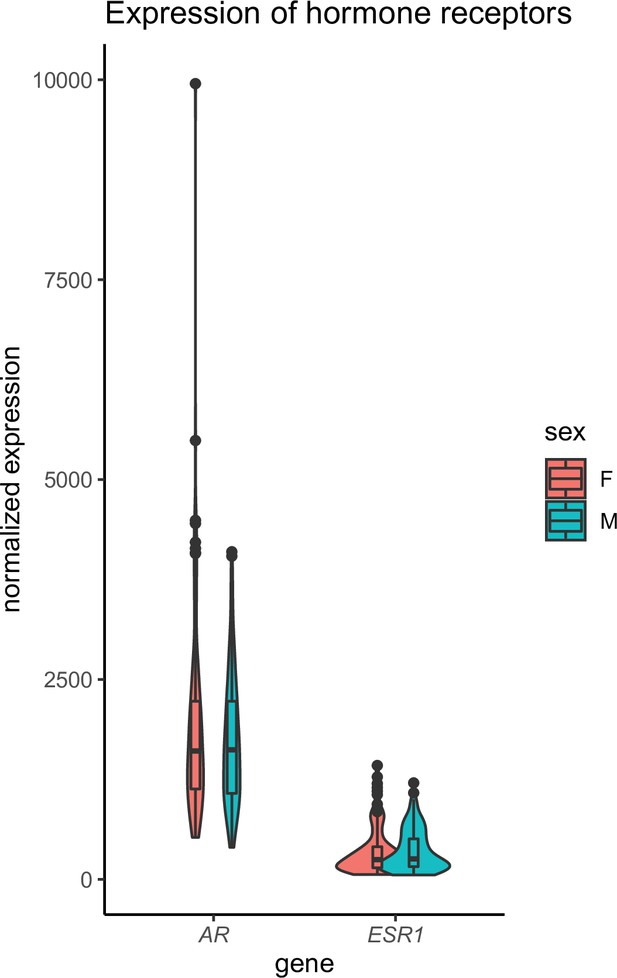
Skeletal muscle sex hormone receptor expression between sexes.
Normalized gene expression levels for androgen receptor (AR) or estrogen receptor (ESR1) (y-axis) in each sex (x-axis). None of the expression levels were significantly different between sexes (Student’s t-test).
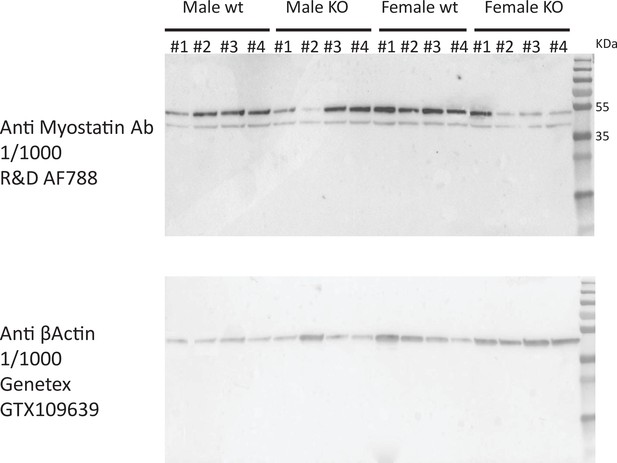
Immunoblot for myostatin in EDL muscle from WT and MERKO male and female mice.
Full immunoblots shown for skeletal muscle lysate blotted for myostatin (top) or β-actin (bottom) corresponding to different C57BL/6J male (left) or female (right) mice in either WT (floxed) or KO (floxed-cre) for skeletal muscle Esr1. Band sizes shown to indicate either precursor (top band) or processed/LAP form (bottom band) of myostatin.
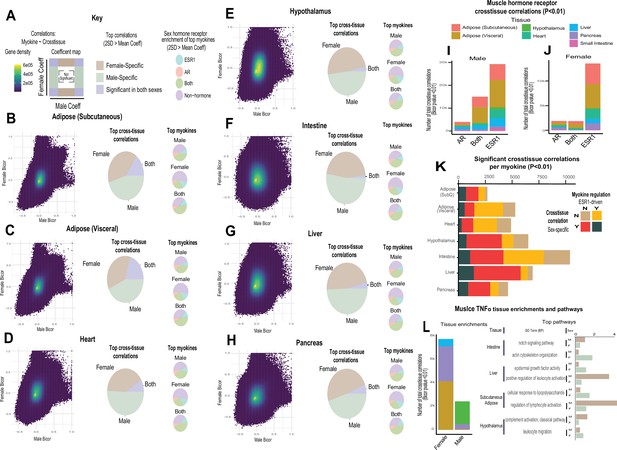
Sex and hormone effects on myokine regulation.
(A–H) Key illustrating analysis for distribution of midweight bicorrelation coefficients between all myokines in skeletal muscle and global transcriptome measures in each target tissue. Coefficients are plotted between sexes (left), where proportions for 2SD > mean are subdivided into occurrence uniquely in females, males, or shared (middle). The significant (2SD > mean) myokines identified in each category were then binned into hormone receptor correlations for ESR1, AR, both, or neither (right). This analysis was performed on all myokines across subcutaneous adipose tissue (B), visceral adipose (C), heart (D), hypothalamus (E), small intestine (F), liver (G), and pancreas (H). (I–J) Significant cross-tissue correlations between muscle ESR1, AR, or both hormone receptors are colored by tissue and shown for males (I) or females (J). (K) For each tissue (y-axis), the ratio of significant cross-tissue correlations per muscle myokine (x-axis) are shown and colored by categories of either the myokine regulated by ESR1 and/or a significant target tissue regression occurring specifically in one sex. (L) Number of significant cross-tissue correlations with muscle TNFα are shown for each sex and colored by tissue as in I–L (left). The −log10(p-value) of significance in an overrepresentation test (x-axis) are shown for top significant inter-tissue pathways for muscle TNFα in each sex (right).
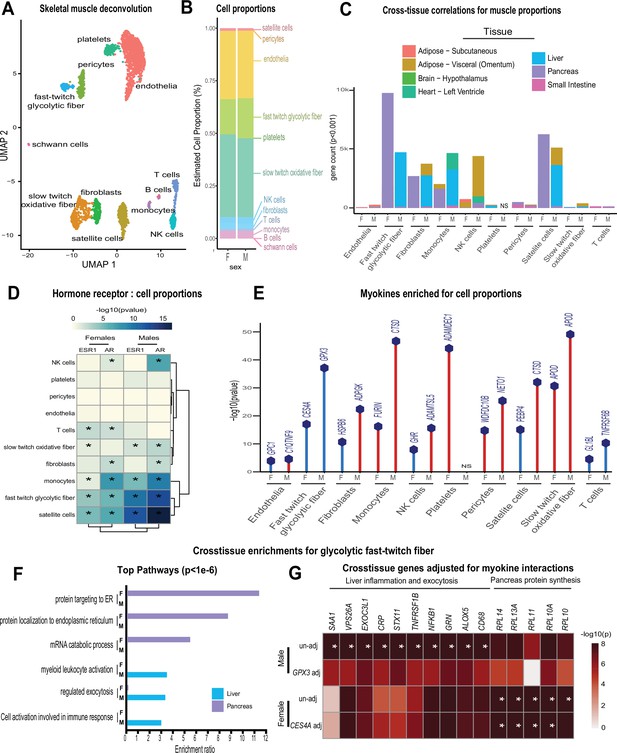
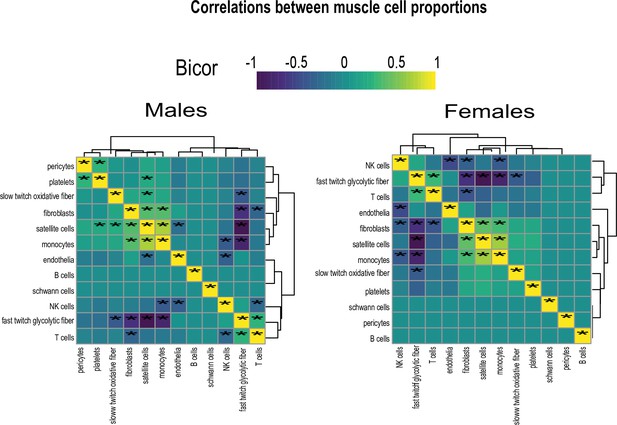
Genetic variation of muscle cell proportions and coregulated cross-tissue processes.
(A) Uniform Manifold Approximation and Projection (UMAP) for skeletal muscle single-cell sequencing used to deconvolute proportions. (B) Mean relative proportions of pseudo-single-cell muscle cell compositions (denoted by color) between sexes. (C) Number of significant cross-tissue correlations (y-axis) corresponding to each skeletal muscle type in each sex (x-axis). Target tissues are distinguished by color, where NS (male platelets) denotes that no significant cross-tissue correlations were observed. (D) Heatmap showing significance of correlations between skeletal muscle hormone receptors and cell proportions, * = p < 0.01. (E) The strongest enriched myokines are plotted for each myokine (y-axis, −log10 p-value of myokine ~ cell composition) are shown for each muscle proportion for each sex (x-axis). Gene symbols for myokines are shown above each line, where red lines indicate positive correlations between myokine and cell type and blue shows inverse relationships. (F) Significant cross-tissue correlated genes in liver (blue) and pancreas (purple) for muscle fast-twitch glycolytic fibers (p < 1e-6) were used for overrepresentation tests where enrichment ratio of significance (x-axis) is shown for each pathway and sex (y-axis). (G) Heatmap showing the regression significance of the top five genes corresponding to inflammation (liver), exocytosis (liver), and protein synthesis (pancreas) for proportions of fast-twitch fiber type (un-adj). Below each correlation between fast-twitch fiber and liver or pancreas gene, the same regressions were performed while adjusting for abundance of select myokines in each sex. * = p < 1e-6.
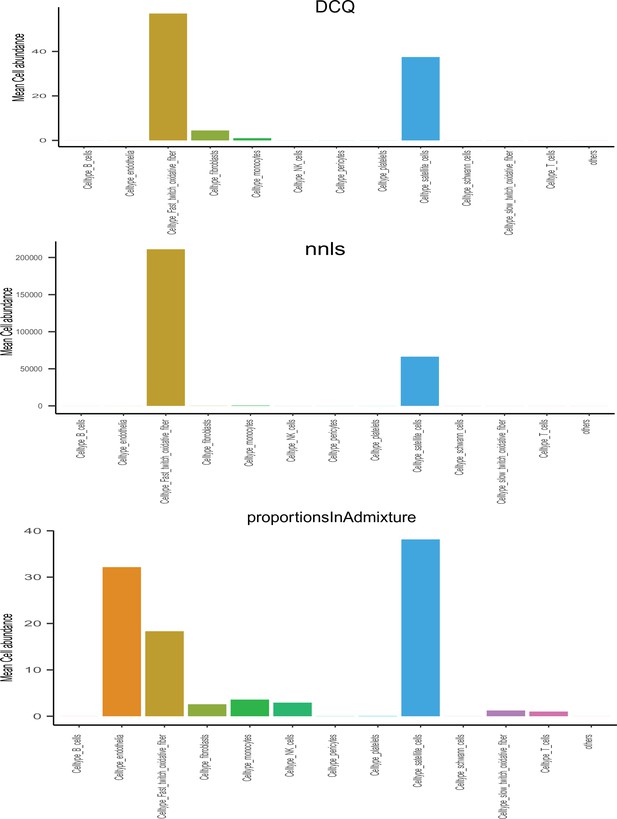
Comparisons of deconvolution methods.
Cell proportions were estimated from skeletal muscle sequencing across the 310 individuals in GTEx.
Here, comparisons of the three most common methods (DCQ, NNLS, and porportionsInAdmixture) were plotted for each pseudo-sc-proportion, where proportionsInAdmixture method captured the largest relative number of cell types.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-MSTN (Goat polyclonal) | R&D | AF788 | (1:1000) |
| Antibody | Rabbit anti-βactin (Rabbit polyclonal) | Genetex | GTX109639 | (1:1000) |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/76887/elife-76887-transrepform1-v2.docx
-
Supplementary file 1
DE statistics for myokines based on sex.
- https://cdn.elifesciences.org/articles/76887/elife-76887-supp1-v2.xlsx
-
Supplementary file 2
Cell type marker genes used.
- https://cdn.elifesciences.org/articles/76887/elife-76887-supp2-v2.xlsx