The scaffolding protein flot2 promotes cytoneme-based transport of wnt3 in gastric cancer
Figures
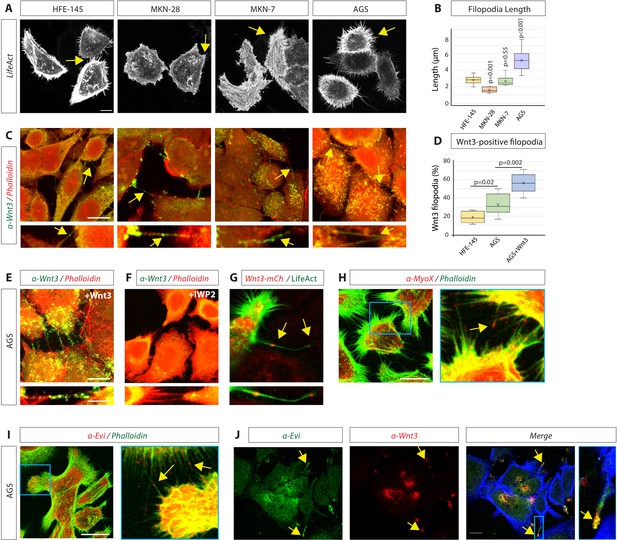
Gastric epithelial normal and cancer cell lines utilise cytonemes to transport Wnt3 intercellularly.
(A) Confocal images of normal gastric epithelial cell line (HFE-145) and gastric cancer (GC) cell lines (MKN-28, MKN-7, and AGS) expressing LifeAct-GFP to visualise actin-based structures. Yellow arrows indicate examples of filopodia. (B) Quantification of filopodia length in GC cell lines MKN-28, MKN-7, and AGS (n=7, 8, 25; n=number of cells). Significance is calculated by Student’s t-test. (C) Immunofluorescent images of HFE, MKN-28, MKN-7, and AGS, stained with antibodies against Wnt3 (green) and actin (Phalloidin-iFluor594, red). Scale bar 10 µm. High-magnification images indicate an example of a Wnt3-bearing cytonemes. Scale bar 2.5 µm. (D) Quantification of Wnt3-positive filopodia in gastric epithelial (HFE-145) and cancer (AGS) cells as a percentage of total filopodia (number of cells analysed = 6, 8, 6). Significance is calculated by Student’s t-test. (E) Immunohistochemistry (IHC) images of AGS cells overexpressing Wnt3 and stained with an antibody against Wnt3 (green) and actin (iFluor594, red). Scale bar 10µm. High-magnification images highlight cytonemes. Scale bar 2.5 µm. (F) IHC images of AGS cells treated with the Porcupine inhibitor IWP2 (100 µM, 48 hr) and stained with an antibody against Wnt3 (green) and actin (iFluor594, red). (G) Live confocal cell imaging of AGS cells expressing Wnt3-mCherry and LifeAct-GFP. Cytoneme-localised Wnt3-mCherry highlighted by yellow arrows. (H–J) IHC images of AGS cells stained with antibodies against (H) Myosin-X (MyoX) and (I) Evi/Wntless (red) and Wnt3 (red) and (J) Evi/Wntless (green Scale bars 10 µm). Phalloidin labels actin (FITC-Phalloidin, green; Phalloidin-iFluor350, blue).
-
Figure 1—source data 1
Western blot images of HFE, AGS, MKN7, and MKN28 cell lysates stained for Flot2.
Beta-actin was used as the loading control.
- https://cdn.elifesciences.org/articles/77376/elife-77376-fig1-data1-v2.xlsx
Wnt3 transport on cytonemes.
AGS cells were transfected with Wnt3-mCherry and LifeAct. After 24 hr, the AGS cells were imaged in a 35 mm dishes with a Leica TCS Leica SP8 confocal microscope, using 63×objective. This video lasts 70 s, containing 8 frames.
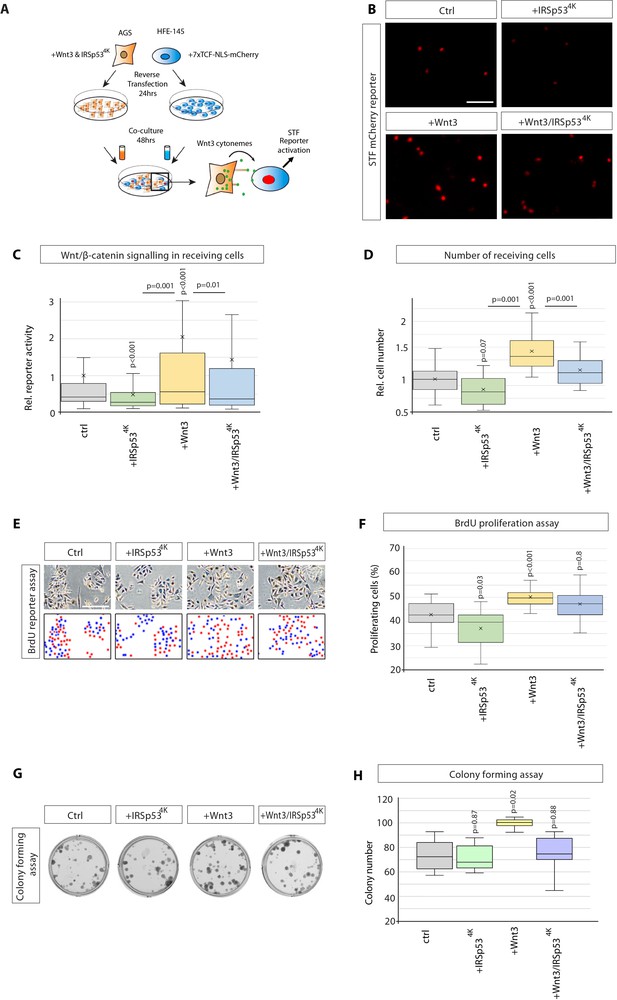
Wnt3 cytonemes regulate paracrine Wnt/β-catenin signalling and proliferation.
(A) Experimental protocol for measuring paracrine Wnt signalling activation. HFE cells expressing the SuperTOPFlash (STF) reporter, 7×TCF-NLS-mCherry, were cocultivated with AGS cells expressing indicated constructs. Fluorescence of STF mCherry reporter was measured after 48 hr and compared to untransfected control cells. (B) Representative images of STF reporter fluorescence for indicated conditions. Scale bar 100 µm. (C) Quantification of STF mCherry reporter fluorescence in HFE cells co-cultured with AGS (n per condition = 322, 394, 258, and 275). (D) Relative number of HFE cells per image after co-culture with AGS cells expressing indicated constructs. Significance calculated by Student’s t-test with Bonferroni correction for multiple comparisons. (n per condition = 28, 26, 27, and 17; n=number of images). (E) Representative images of proliferating, BrdU-stained (red); co-cultured AGS and HFE-145 cells, as described in (a). Cells were counterstained with haematoxylin (blue dots). Scale bar 100 µm. Complementary images show BrdU+ cells with red dots; blue dots mark BrdU- cells. (F) Quantification of BrdU-stained cells as a percentage of the population. Significance calculated by Student’s t-test with Bonferroni correction for multiple comparisons. (n per condition = 20, 20, 20, and 20; n=number of images). (G), Colony-forming assay of AGS cells. AGS cells were transfected with the indicated constructs and co-cultured with AGS-RFP cells for 2 days. After sorting, AGS-RFP expressing cells were plated at clonal density for 10–12 days; (H), Quantification of spherical colonies. Significance is calculated by Student’s t-test (n=9).
-
Figure 2—source data 1
Western blot images of HFE, AGS, MKN7, and MKN28 cell lysates stained for Flot2.
Beta-actin was used as the loading control.
- https://cdn.elifesciences.org/articles/77376/elife-77376-fig2-data1-v2.xlsx
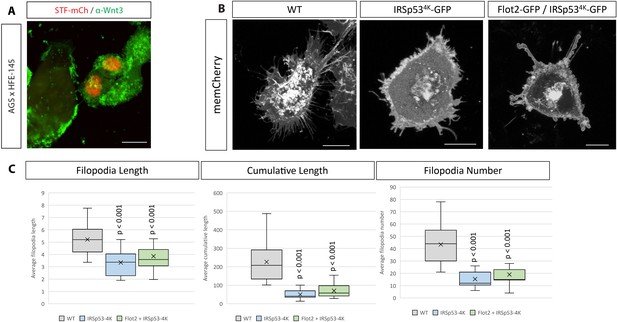
Wnt3 cytonemes depend on IRSp53 function.
(A) Antibody staining against Wnt3 (green) after co-cultivation of SuperTOPFlash (STF)-mCh-expressing HFE-145 cells with AGS cells, showing Wnt3 staining in receiving HFE-145 cells. Scale bar 20 µm. (B) Confocal images of AGS cells expressing the dominant-negative IRSp534K-GFP mutant and memCherry, showing inhibition of filopodia formation (even in the presence of Flot2-GFP). Scale bar 10 µm. (C) Quantification of filopodia of cells from (a). (n per condition = 25, 13, 13; n=number of cells). Significance is calculated by Student’s t-test.
-
Figure 2—figure supplement 1—source data 1
Western blot image of AGS cell lysates stained for Flot2.
Cells were treated with control or Flot2 siRNA (3 or 4 µl) for 48 hr prior to lysate collection. Beta-actin was used as the loading control.
- https://cdn.elifesciences.org/articles/77376/elife-77376-fig2-figsupp1-data1-v2.xlsx
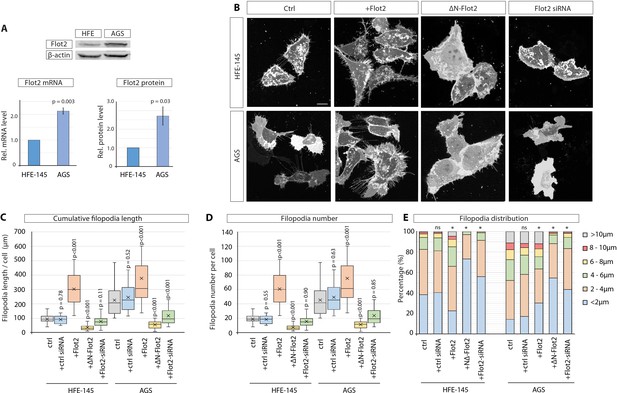
Flotillin-2 is over-expressed and promotes filopodia formation and elongation in gastric cancer cells.
(A) Flot2 protein levels in HFE-145 and AGS cells as quantified by Western blot after normalising to beta-actin levels (n=3) and by RT-qPCR after normalising to Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) as a housekeeping gene (n=4). Relative protein and mRNA levels are compared to HFE-145. Error bars represent SEM. Significance is calculated by Student’s t-test. (B) Representative images of HFE and AGS cells expressing membrane-mCherry and indicated Flotillin-2 (Flot2) constructs or siRNA after 48 hr. Scale bars 10 µm. (C–D) Filopodia quantifications of HFE and AGS cells transfected with indicated Flot2 plasmids or siRNA. Significance calculated by Student’s t-test with Bonferroni correction for multiple comparisons. Average cumulative filopodia length (C), average filopodia number per cell (D). (n per condition [HFE]=22, 19, 25, 23, 24). (n per condition [AGS]=25, 21, 25, 25, 25; n=number of cells measured). (E) Distribution of filopodia, categorised by length as a percentage of total filopodia per HFE or AGS cell 48 hr post-transfection with indicated Flot2 plasmids or siRNA. A Pearson’s χ2 test was performed to test for significance between control (ctrl) group (expected) and experimental groups (observed) with 5 degrees of freedom (df) and a p-value <0.05. The specific χ2 values are as follows, HFE: ctrl siRNA 0.86, Flot2 0.001, dnFlot2 <0.001, Flot2 siRNA <0.001, and for AGS: ctrl siRNA 0.65, Flot2 0.007, dnFlot2 <0.001, and Flot2 siRNA <0.001. Asterisks mark significant differences.
-
Figure 3—source data 1
Western blot images of HFE, AGS, MKN7, and MKN28 cell lysates stained for Flot2.
Beta-actin was used as the loading control.
- https://cdn.elifesciences.org/articles/77376/elife-77376-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Western blot images of HFE, AGS, MKN7, and MKN28 cell lysates stained for Flot2.
Beta-actin was used as the loading control.
- https://cdn.elifesciences.org/articles/77376/elife-77376-fig3-data2-v2.zip
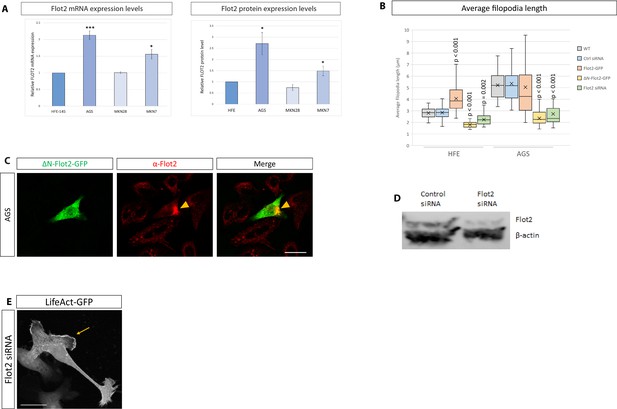
Effects of Flot2 on filopodia and Wnt3.
(A) Quantification of Flot2 expression levels (mRNA and protein) in HFE-145, AGS, MKN-28, and MKN-7 cells. Significance is calculated by Student’s t-test. (B) Quantification of filopodia length in HFE and AGS cells expressing indicated constructs. Significance is calculated by Student’s t-test with Bonferroni correction for multiple comparisons. (C) Antibody staining of AGS cells expressing ΔN-Flot2-GFP and stained for endogenous WT Flot2 (red), showing the effect of the dominant-negative mutant on Flot2 localisation. Scale bar 20 µm. (D) Analysis of efficiency of siRNA-mediated knock-down of Flot2 in AGS cells by Western blot. (E) Confocal image of an AGS cell with depleted Flot2 (siRNA-mediated) and expressing LifeAct-GFP to visualise actin. Yellow arrow highlights an example of lamellipodia frequently seen in Flot2-depleted cells. Scale bars 10 µm.
-
Figure 3—figure supplement 1—source data 1
Flotillin-2 promotes filopodia formation.
- https://cdn.elifesciences.org/articles/77376/elife-77376-fig3-figsupp1-data1-v2.xlsx
-
Figure 3—figure supplement 1—source data 2
Flotillin-2 promotes filopodia elongation.
- https://cdn.elifesciences.org/articles/77376/elife-77376-fig3-figsupp1-data2-v2.zip
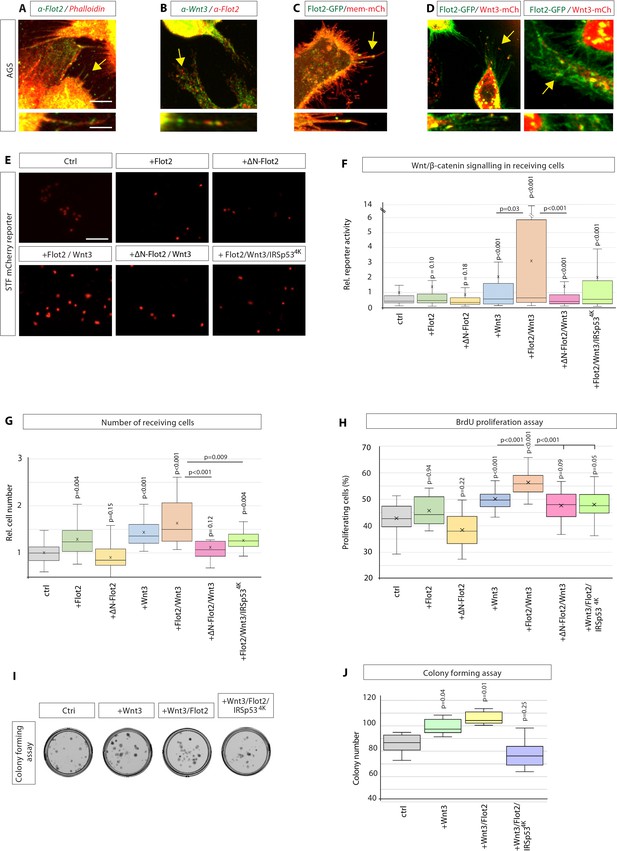
Flotillin-2 marks Wnt3 cytonemes and influences paracrine Wnt/β-catenin signalling and proliferation.
(A) Immunohistochemistry (IHC) analysis showing endogenous localisation of Flot2 (green) in AGS cells. TRITC phalloidin was used to visualise actin. Arrows indicate the localisation of Flot2 to filopodia. Scale bars 5 µm. High-magnification images indicate an example of a Flot2-bearing cytonemes. Scale bars 2.5 µm. (B), IHC analysis shows that Flot2 co-localises with Wnt3 on cytonemes. (C) Confocal images showing the subcellular localisation of Flot2-GFP in AGS cells. Arrows indicate the localisation of Flot2-GFP on cytonemes. (D) Confocal images highlighting co-localisation of Flot2-GFP and Wnt3-mCh on cytonemes in AGS cells (arrows). Flot2-GFP and Wnt3-mCherry also cluster and co-localising at a cytoneme contact point (arrow). (E) Representative images of SuperTOPFlash (STF) reporter fluorescence for indicated conditions. Scale bar 100 uM. (F) Relative quantification of STF mCherry reporter fluorescence in HFE cells co-cultivated with AGS cells expressing indicated constructs. Quantifications are relative to AGS control. (n per condition = 322, 443, 403, 258, 336, 306, and 297; n=number of nuclei measured). (G) Relative number of HFE cells per image after co-cultivation with AGS cells expressing indicated constructs. Significance calculated by Student’s t-test with Bonferroni correction for multiple comparisons. (n per condition = 28, 27, 26, 27, 22, 24, and 15; n=number of images). (H) Quantification of BrdU-stained cells as a percentage of the population, after co-cultivation of AGS and HFE cells, as described in Figure 2a. significance calculated by Student’s t-test with Bonferroni correction for multiple comparisons. (n per condition = 20; n=number of images). (I), Colony-forming assay of AGS cells. AGS cells were transfected with the indicated constructs and co-cultivated with AGS-RFP cells for 2 days. After sorting, AGS-RFP expressing cells were plated at clonal density; (J), Quantification of colonies after 10–12 days. Significance is calculated by Student’s t-test (n=9).
-
Figure 4—source data 1
Flotillin-2 regulates paracrine Wnt/β-catenin signalling and proliferation.
- https://cdn.elifesciences.org/articles/77376/elife-77376-fig4-data1-v2.xlsx
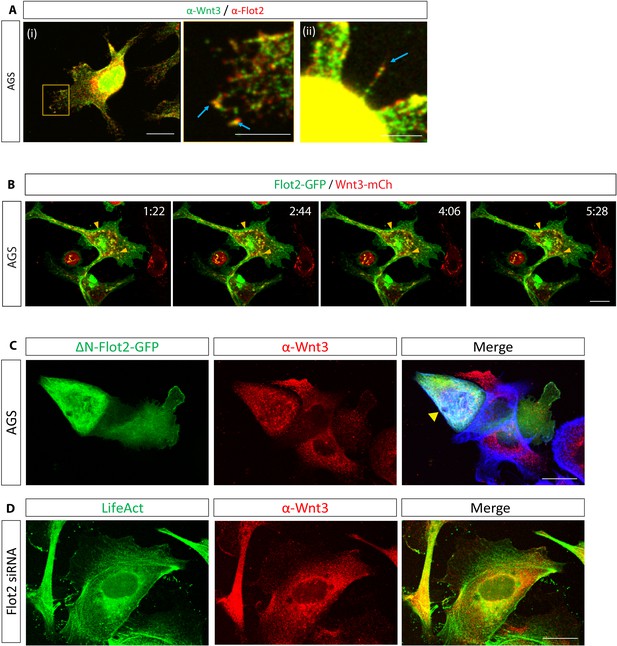
Co-localisation study of Flot2 and Wnt3.
(A) Immunofluorescent images of AGS cells stained for Wnt3 (green) and Flot2 (red). Co-localisation of Flot2 and Wnt3 highlighted by blue arrows. Scale bars represent (i) 10 µm (left) and 5 µm (right), (ii) 5 µm. (B) Confocal time-lapse images of AGS cells expressing Flot2-GFP and Wnt3-mCh. Yellow arrows highlight the co-localisation of Flot2 and Wnt3 intracellularly. Scale bar 10 µm. (C) AGS cells have been transfected with indicated constructs to compare Wnt3 localisation in AGS depleted for Flot2 function.
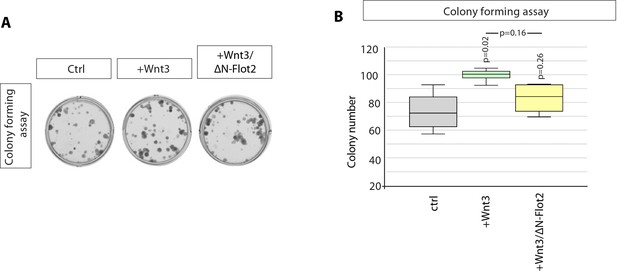
Colony formation assay.
(A) Quantification of spherical colonies after 8–10 days. Results for AGS-RFP cells co-cultured with Ctrl and Wnt3 expressing AGS cells are the same displayed in Figure 2G and (B), Significance calculated by Student’s t-test (n=9).
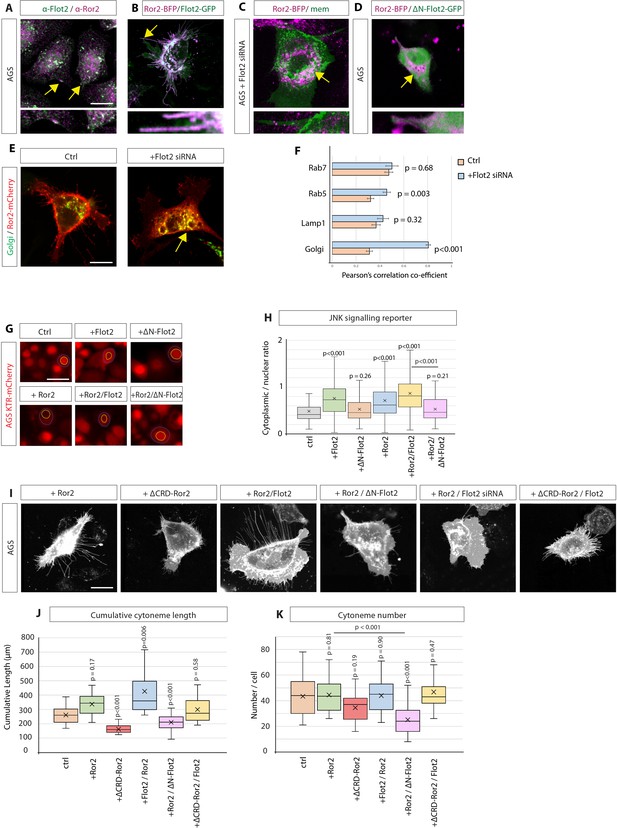
Flotillin-2 is required for Ror2 membrane localisation, Ror2/PCP signalling and Ror2-mediated cytoneme formation.
(A) Immunohistochemistry (IHC) analysis of AGS cells stained for Ror2 (red) and Flot2 (green). Flot2 and Ror2 show co-localisation with a Pearson’s correlation coefficient (PCC) of 0.65 (n=10), highlighted at the membrane by arrows. Scale bars represent 10 µm, and in high-magnification images, 2.5 µm (right). (B–D), Confocal live-cell imaging of AGS cells expressing Ror2-BFP with Flot2-GFP (B), Flot2 siRNA (c) or ∆N-Flot2-GFP (D) and memCherry. Arrows highlight subcellular regions of co-localisations. (E) Live confocal images of AGS cells expressing Ror2-mCherry and indicated organelle markers +/- Flot2 siRNA. Arrows highlight the co-localisation of Ror2-mCherry and mTurq2-Golgi. Scale bar 10 µm F, Quantification of co-localisation of Ror2-mCherry with indicated markers, assessed by PCC. Significance is calculated by Student’s t-test. (n per condition [WT]=7, 10, 8, and 10) (n per condition [Flot2 siRNA]=7, 6, 7, 8). (G) Representative images of AGS cells stably expressing the JNK-KTR-mCherry reporter and indicated constructs after 48 hr. Blue dotted line encircles the cytoplasm and yellow dotted line the nucleus of a representative cell. Scale bar 20 µm. (H) Quantification of the JNK-KTR-mCherry reporter. Nuclear and cytoplasmic fluorescence of cells were measured, and the cytoplasmic: nuclear ratio was calculated. Significance is calculated by one-way ANOVA with Bonferroni correction for multiple comparisons. (n per condition = 136, 109, 74, 109, 82, and 79). (I) Representative confocal images of AGS cells expressing memCherry and indicated constructs for 48 hr. Scale bars 10 µm. (J, K) Quantification of cytoneme length and number of AGS cells transfected with constructs indicated in (i). Significance calculated by Student’s t-test with Bonferroni correction for multiple comparisons. (n per condition = 25, 22, 21, 23, 25, and 21; n=number of cells).
-
Figure 5—source data 1
Flotillin-2 is required for Ror2-mediated cytoneme formation.
- https://cdn.elifesciences.org/articles/77376/elife-77376-fig5-data1-v2.xlsx
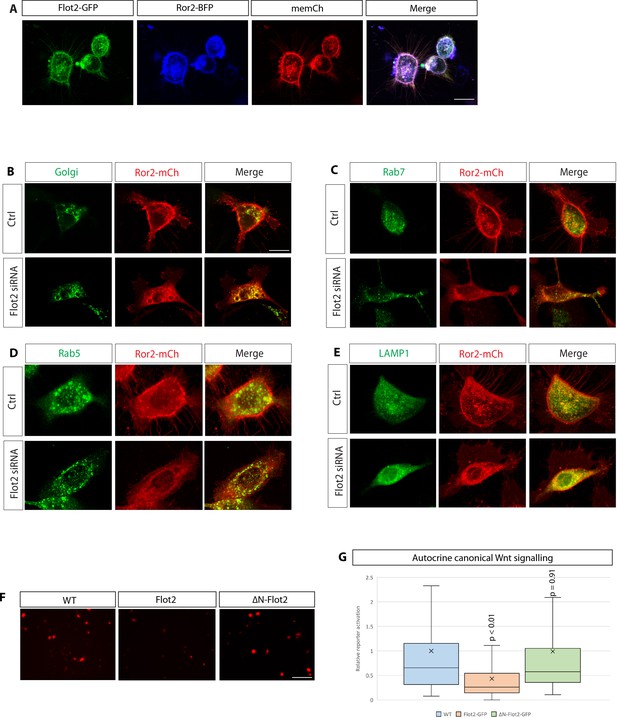
Flotillin-2 and Ror2 – localisation and signalling.
(A) Confocal images of AGS cells expressing Flot2-GFP, Ror2-BFP and membrane-mCherry to highlight membrane localisation of Flot2 and Ror2. Scale bar 20 µm. (B – E) Confocal images of AGS cells expressing Ror2-mCherry and indicated cell organelle markers. Scale bars represent 20 µm. (F) Representative images of AGS cells transfected with the 7×TCF-NLS-mCherry reporter and indicated constructs for 48 hr. Scale bar 20 µm G, Relative quantification of 7×TCF-NLS-mCherry fluorescence compared to untransfected control. Significance is calculated by Student’s t-test. (n per condition = 406, 212, and 469; n=number of nuclei measured).
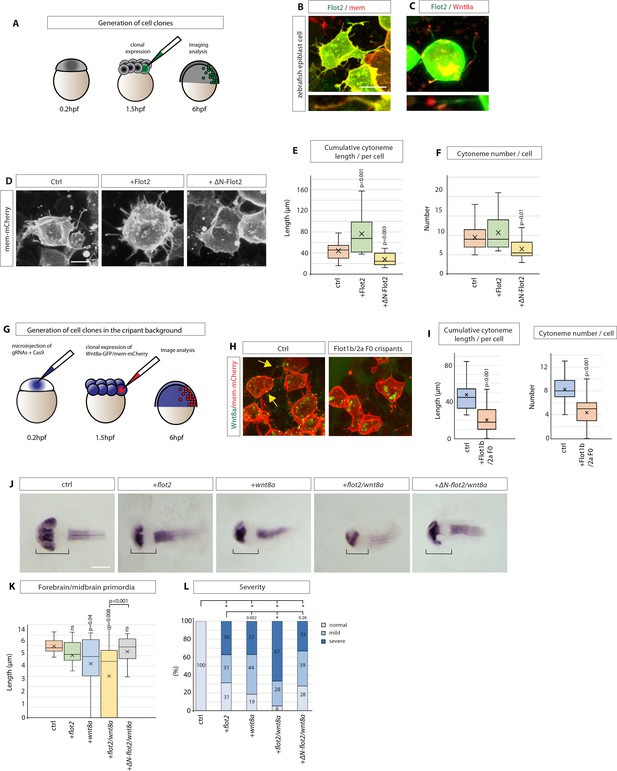
Flotillin-2 promotes cytoneme formation and Wnt8a signalling in zebrafish development.
(A) Experimental setting to generate small clones expressing indicated constructs in the zebrafish embryo. (B, C) Confocal images of zebrafish epiblast cells injected with Flot2-GFP / memCherry and Flot2-GFP / Wnt8a-mCherry and imaged at 8 hpf. Scale bars represent 10 µm (D) Representative images of zebrafish epiblast cells injected with memCherry indicated constructs. Scale bar 10 µm. (E), (F), Quantification of filopodia from epiblast cells injected. Significance is calculated by Student’s t-test. (n per condition = 17, 20, and 14). (G), Experimental strategy to generate Wnt8a-GFP/mCherry cell clones in F0 Flot1b/2 a Crispants background. (H), Confocal images of zebrafish epiblast cells expressing indicated constructs. (I) Quantification of cytoneme length and number in injected epiblast cells. Significance is calculated by Student’s t-test. (n per condition = 18, 15) (J) In situ hybridisation against pax6a in zebrafish embryos at 30hpf after microinjection of 100 ng/µl of indicated DNA constructs and imaged. Scale bar represents 100 µm. (K) Quantification of forebrain and midbrain primordia length in zebrafish embryos injected as in (A). Significance is calculated by Student’s t-test. (n per condition = 23, 16, 27, 18, and 18; n=number of embryos). (L) Qualitative analysis of phenotype severity in zebrafish embryos injected as indicated in (A). Phenotypes are classified into the categories normal, mild and severe. Numbers in bars represent percentages of total embryos. A Pearson’s χ2 test revealed a significant difference between the ctrl group (expected) and experimental groups (observed) with 2 degrees of freedom (df) and a p-value < 0.05 of χ2 <0.001. Distribution comparison of flot2 injected embryos revealed a significant difference to the wnt8a group (χ2=0.002) and the flot2 + wnt8 a group (χ2<0.001), but not to wnt8a+ΔN-flot2 (χ2=0.28). Asterisks mark significant differences.
-
Figure 6—source data 1
Flotillin-2 promotes cytoneme formation in zebrafish development.
- https://cdn.elifesciences.org/articles/77376/elife-77376-fig6-data1-v2.xlsx
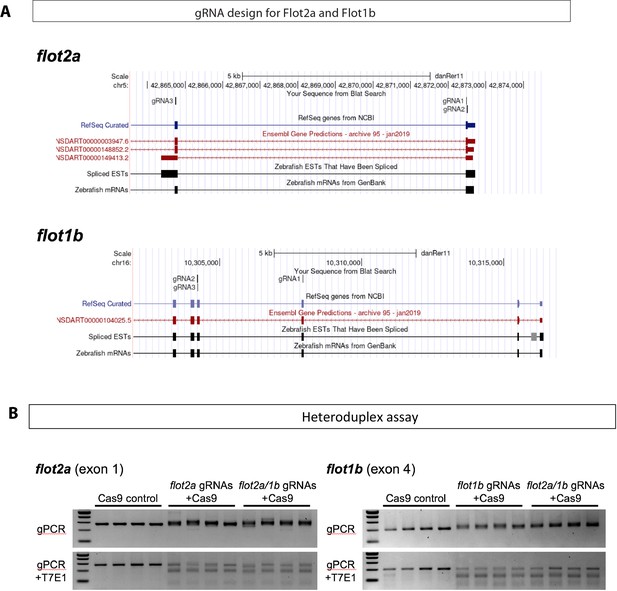
Site-specific mutagenesis of zebrafish flot2a and flot1b genes.
(A) Location of guide RNA (gRNA) target sites. Three individual gRNAs were designed to target coding exons of flot2a and flot1b genes (annotated with blue and red boxes). Two gRNAs and one gRNA target exon 1 and exon 2 of flot2a, respectively. For flot1b, one gRNA and two gRNAs were designed to target exon 3 and exon 4, respectively. (B) Site-specific mutagenesis was detected by T7 endonuclease I (T7E1) assay. Genomic sequences including gRNA target sites in flot2a exon 1 or flot1b exon 4 were amplified by PCR from genomic DNA extracted from single embryo injected with Cas9 (control) or gRNAs/Cas9 ribonucleoprotein complex (n=4 per condition). Heteroduplex DNA in the PCR products was cleaved by T7E1.





