Coagulation factors directly cleave SARS-CoV-2 spike and enhance viral entry
Figures
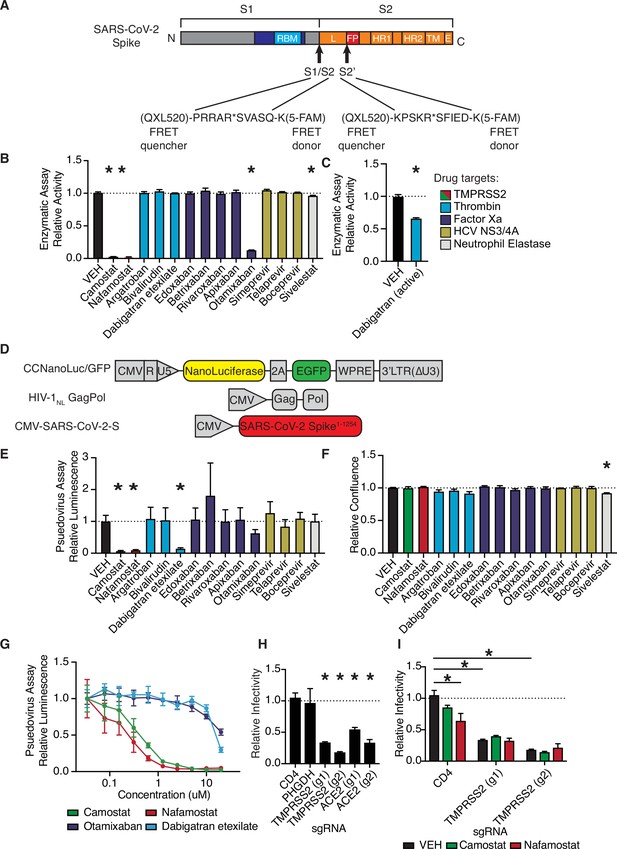
Anticoagulant serine protease inhibitors suppress SARS-CoV-2 entry via inhibition of TMPRSS2.
(A) Peptides derived from two known cleavage sites of SARS-CoV-2 spike were designed with C-terminal fluorophore 5-FAM and N-terminal fluorescence resonance energy transfer (FRET) quencher QXL-520. (B) FDA-approved and investigational serine protease inhibitors were screened by enzymatic assay to inhibit TMPRSS2 cleavage of SARS-CoV-2 S1/S2 peptide substrate. Relative change in fluorescence with respect to DMSO vehicle is shown. Colors indicate the described target of the drugs screened. All drugs screened at 10 µM final concentration. (C) Active form of dabigatran in enzymatic assay for TMPRSS2 inhibition. Relative fluorescence with respect to its corresponding 0.1 N HCl vehicle is shown. (D) Schematic of constructs used to generate SARS-CoV-2 spike-pseudotyped/HIV-1-based particles. (E) Calu3 cells were treated with 10 µM of the indicated drugs for 24 hr prior to infection with HIV-1NL/SARS-CoV-2 pseudovirus. Media was changed at 24 hr post infection and pseudoviral entry was measured by nanoluciferase luminescent signal at 40 hr. (F) Calu3 cells treated with 10 µM of the indicated drugs were monitored for confluence by Incucyte for 40 hr. (G) Pseudoviral entry was measured by nanoluciferase luminescent signal in Calu3 cells treated various concentrations of the indicated drugs for 4 hr prior to infection with SARS-CoV-2 pseudovirus. (H) Caco2 cells were infected with lenti-Cas9-blast and U6-sgRNA-EFS-puro-P2A-tRFP and selected. Neutral controls targeting CD4 (not endogenously expressed) or PHGDH intron 1, two sgRNAs each targeting different regions of ACE2 and TMPRSS2 were included. Cells were subsequently infected with HIV-1NL/SARS-CoV-2 pseudovirus. (I) Caco2 cells co-expressing Cas9 and sgRNAs targeting CD4 (not expressed) or TMPRSS2 were treated with 10 µM camostat, nafamostat, or DMSO vehicle. N = 3, *p < 0.05, two-tailed t-test. Data represented as mean ± SEM.
-
Figure 1—source data 1
Data and summary statistics for enzymatic and pseudovirus assays.
- https://cdn.elifesciences.org/articles/77444/elife-77444-fig1-data1-v2.xlsx

Optimization of fluorescence resonance energy transfer (FRET) enzymatic assay.
(A) TMPRSS2 enzymatic assay was performed in AB1 (20 mM Tris-HCl, pH 7.3, 100 mM NaCl, 1 mM EDTA, fresh 1 mM DTT) or AB2 (50 mM Tris-HCl, 150 mM NaCl, pH 8) using 10 µM of either S1/S2 or S2’ peptide substrate. (B) Titration of enzyme concentration was performed (0–1000 nM) with 10 µM S1/S2 substrate. Initial reaction velocity V0 (rate of change in fluorescent signal) each enzyme concentration with 10 µM S1/S2 peptide substrate.
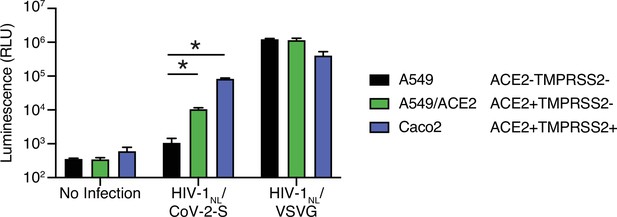
Further characterization of HIV-1/SARS-CoV-2 pseudovirus.
A549 cells (which do not express ACE2), A549/ACE2 cells (ectopic ACE2 expression from a lentiviral vector), and Caco2 cells (which express endogenous ACE2 and TMPRSS2) infected with HIV-1NL-based particles pseudotyped with SARS-CoV-2 S or VSV G. N = 3, *p < 0.05, two-tailed t-test. Data represented as mean ± SEM.
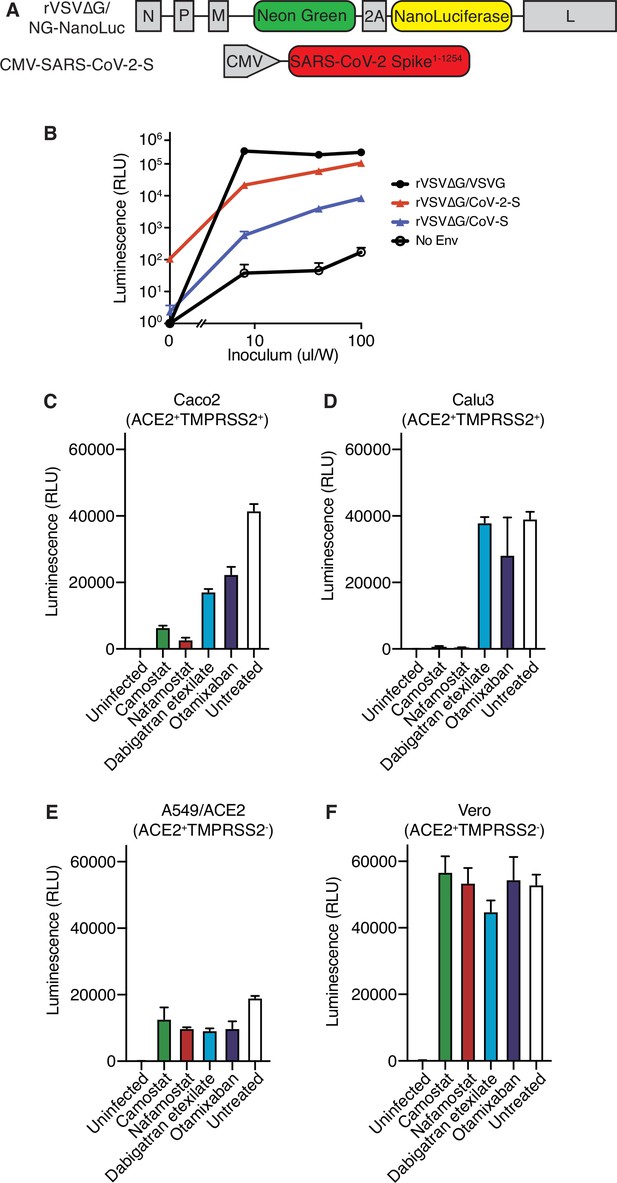
Further characterization of rVSV∆G/SARS-CoV-2 pseudovirus.
(A) Schematic of constructs used to generate SARS-CoV-2 spike-pseudotyped/VSV-based pseudovirus. (B) Nanoluciferase luminescent signal following addition of rVSV∆G pseudovirus complemented with VSV G, SARS-CoV-2 S, SARS-CoV S, or without complementation with any envelope protein to Calu3 cells. Each pseudovirus was titrated by adding the indicated volume of inoculum, supplemented with fresh media up to 200 µl/well in a 96-well plate. (C–F) Nanoluciferase luminescent signal following infection of (C) Caco2, (D) Calu3, (E) A549/ACE2, or (F) Vero cells with rVSV∆G/SARS-CoV-2 pseudovirus pretreated for 4 hr with 10 µM camostat, nafamostat, dabigatran, or otamixaban, compared with uninfected or infected/untreated cells. Expression status of ACE2 and TMPRSS2 for each cell line is indicated. N = 3. Data represented as mean ± SEM.
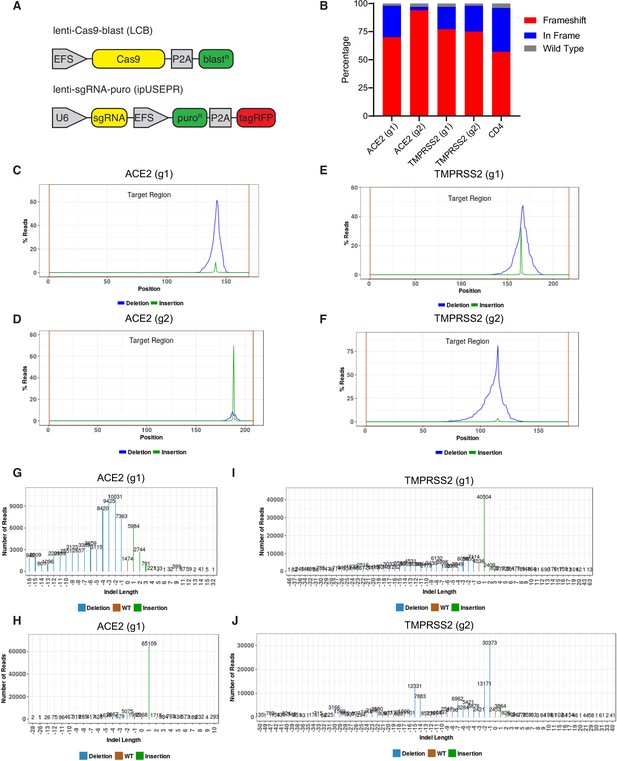
Evidence of CRISPR knockout efficiency.
(A) Constructs used for CRISPR experiments. (B) Percentage of reads exhibiting wild type, frameshift, or in-frame indels at each locus for the indicated sgRNAs. (C–F) Distribution of reads with deletion or insertion by position within amplicon. (G–J) Distribution of the size of insertions and deletions in each amplicon. Two sgRNAs targeting ACE2 (g1 and g2) and two sgRNAs targeting TMPRSS2 (g1 and g2) were analyzed.
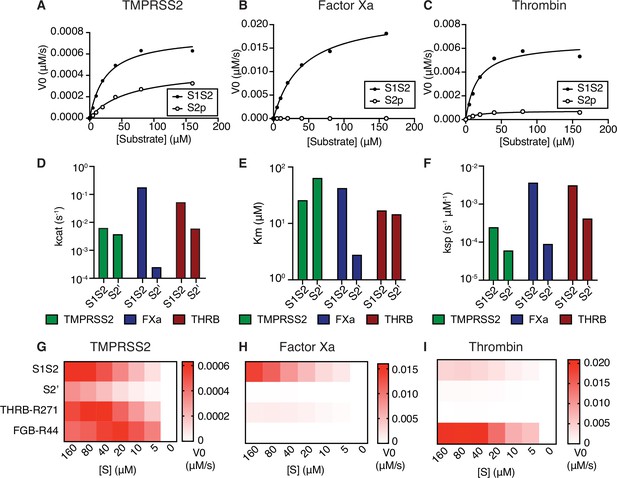
Coagulation factors directly cleave SARS-CoV-2 spike.
Initial velocities for the cleavage of SARS-CoV-2 spike S1/S2 and S2’ peptide substrates by (A) TMPRSS2, (B) factor Xa, and (C) thrombin were measured over a range of 0–160 µM substrate. From initial velocity values, enzyme kinetic constants (D) turnover rate Kcat (s–1), (E) affinity constant Km, and (F) specificity constant (Kcat/Km) were obtained for the indicated enzymes with S1/S2 and S2’ peptides. (G–I) Heatmaps depict the initial velocity V0 of cleavage of the indicated peptide substrates and concentrations by (G) TMPRSS2, (H) factor Xa, and (I) thrombin.
-
Figure 2—source data 1
Data and summary statistics for enzymatic assays.
- https://cdn.elifesciences.org/articles/77444/elife-77444-fig2-data1-v2.xlsx
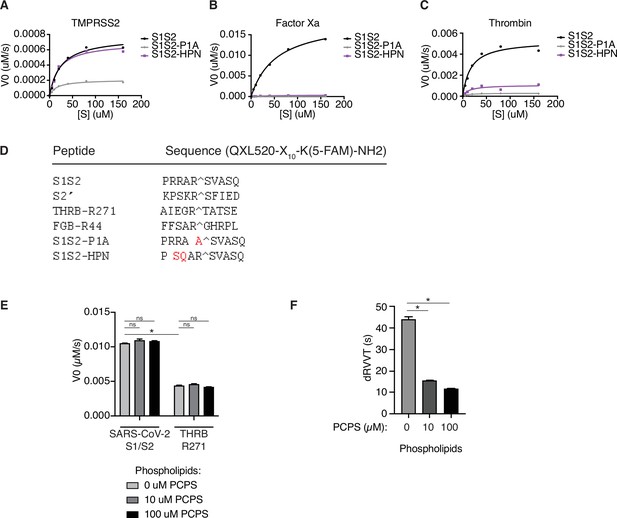
Fluorescence resonance energy transfer (FRET) enzymatic assay with modified peptide substrates.
(A–C) Initial reaction velocity with respect to enzyme concentration for peptide substrates of the SARS-CoV-2 spike S1/S2 site (S1S2), with P1 arginine substituted with alanine (S1S2-P1A), or with substitutions in the P3 and P4 position (RR > SQ) with (A) TMPRSS2, (B) factor Xa, or (C) thrombin. (D) List of peptide substrates used in this study. (E) Initial reaction velocity of factor Xa cleavage of SARS-CoV-2 S1/S2 or thrombin-R271 peptide substrates in the presence of 0–100 µM phosphatidylcholine/phosphatidylserine (PC/PS) phospholipid vesicles. (F) Dilute Russell’s viper venom clotting time (dRVVT) assay of pooled normal human plasma, supplemented with 0–100 µM PC/PS phospholipid vesicles. N = 3, *p < 0.05, two-tailed t-test. Data represented as mean ± SEM.
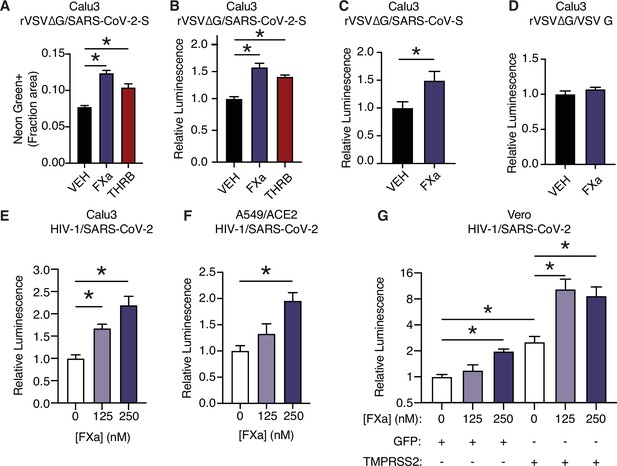
Factor Xa and thrombin facilitate SARS-CoV-2 spike-mediated entry.
(A) Calu3 cells were infected with rVSV∆G/SARS-CoV-2 pseudovirus with concomitant treatment with vehicle, 250 nM factor Xa, or 250 nM thrombin. Quantification of the ratio of green fluorescent area to total confluence (4 fields/replicate well, 4 wells/condition). (B) Nanoluciferase luminescent signal was measured following infection with rVSV∆G/SARS-CoV-2 pseudovirus and the addition of either vehicle, factor Xa, or thrombin. The effect of factor Xa on rVSV∆G complemented with either (C) SARS-CoV spike or (D) VSV-G was measured by luminescent signal. Luminescent signal was measured following HIV-1NL/SARS-CoV-2 pseudovirus infection and concomitant treatment with 125–250 nM factor Xa in (E) Calu3 cells, (F) A549/ACE2, and (G) Vero cells following transduction with lentiviral vectors to express GFP or TMPRSS2. Following selection, cells were infected with HIV-1NL/SARS-CoV-2 pseudovirus and concomitantly treated with 125–250 nM factor Xa. Subsequently, nanoluciferase luminescent signal was determined and plotted relative to vehicle-treated control. *p < 0.05, two-tailed t-test. Data represented as mean ± SEM.
-
Figure 3—source data 1
Data and summary statistics for pseudovirus assays with exogenous proteases.
- https://cdn.elifesciences.org/articles/77444/elife-77444-fig3-data1-v2.xlsx
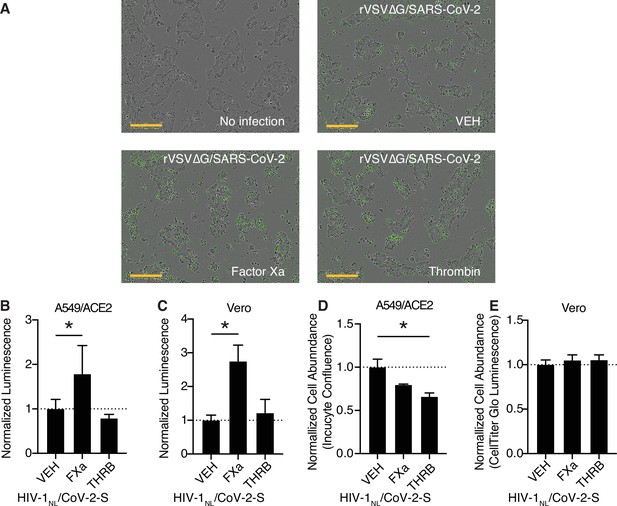
Further characterization of coagulation factor-induced SARS-CoV-2 pseudovirus infection.
(A) Representative merged brightfield and green fluorescence images of Calu3 cells without infection or following rVSV∆G/SARS-CoV-2 pseudovirus infection and concomitant treatment with vehicle, 250 nM factor Xa, or 250 nM thrombin (corresponding to Figure 3A). Scale bars represent 300 µm. (B–E) HIV-1NL/SARS-CoV-2 pseudovirus with addition of purified protease or vehicle. Nanoluciferase luminescent signal relative to vehicle-treated control was measured following infection in (B) A549/ACE2 and (C) Vero cells. Cell abundance following protease treatment, relative to vehicle control was determined for (D) A549/ACE2 and (E) Vero cells. N = 3, *p < 0.05, two-tailed t-test. Data represented as mean ± SEM.
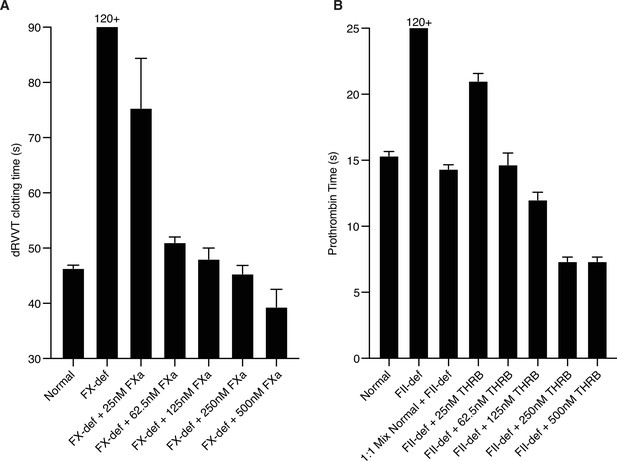
Assessing relevant protease levels with ex vivo clotting assays.
(A) Factor X activity was determined by dilute Russell’s viper venom clotting time (dRVVT). Pooled normal plasma or FX-deficient plasma with the addition of the indicated concentration of active purified factor Xa were assayed. (B) Prothrombin activity was determined by prothrombin time (PT) via activation with thromboplastin. Pooled normal plasma or prothrombin-deficient plasma with the addition of the indicated concentration of active purified thrombin were assayed. N = 3. Data represented as mean ± SEM.
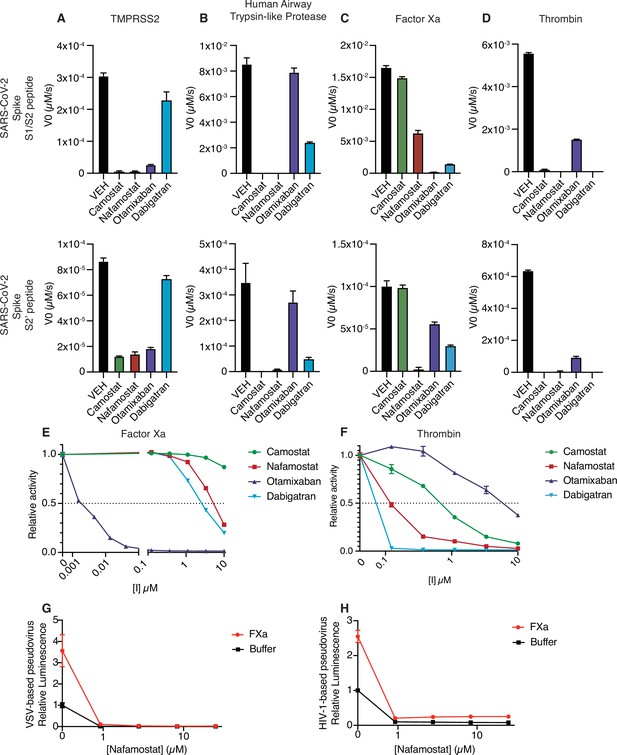
Nafamostat broadly inhibits cleavage of spike peptides by both coagulation factors and transmembrane serine proteases.
Initial velocities for the cleavage of 10 µM SARS-CoV-2 spike S1/S2 (top) and S2’ (bottom) peptide substrates by (A) TMPRSS2, (B) TMPRSS11D/human airway trypsin-like protease (C) factor Xa, and (D) thrombin were measured in the presence of DMSO vehicle, or 10 µM camostat, nafamostat, otamixaban, or dabigatran. The relative activity of (E) factor Xa and (F) thrombin were determined over a range of 0–10 µM of the indicated drugs. Calu3 cells were treated with a range of concentrations of nafamostat with or without addition of 250 nM exogenous factor Xa and infected with (G) rVSV∆G/SARS-CoV-2 pseudovirus or (H) HIV-1NL/SARS-CoV-2 pseudovirus and infectivity was measured by luminescence. N = 3, data represented as mean ± SEM.
-
Figure 4—source data 1
Data and summary statistics for enzymatic assays to determine the effects of protease inhibitors on host proteases.
- https://cdn.elifesciences.org/articles/77444/elife-77444-fig4-data1-v2.xlsx
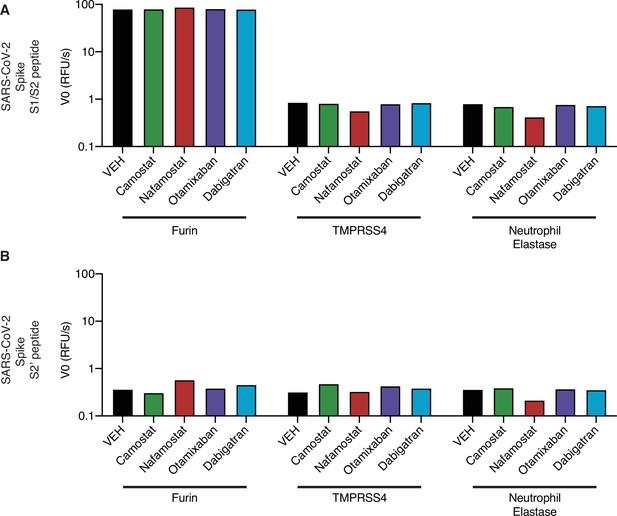
Activity of candidate inhibitors against other proteases.
Initial reaction velocity V0 of furin, TMPRSS4, or neutrophil elastase cleavage of (A) S1/S2 peptide substrate or (B) S2’ peptide substrate, treated with DMSO vehicle, camostat, nafamostat, otamixaban, or dabigatran. 10 µM substrate and 10 µM inhibitor were used.
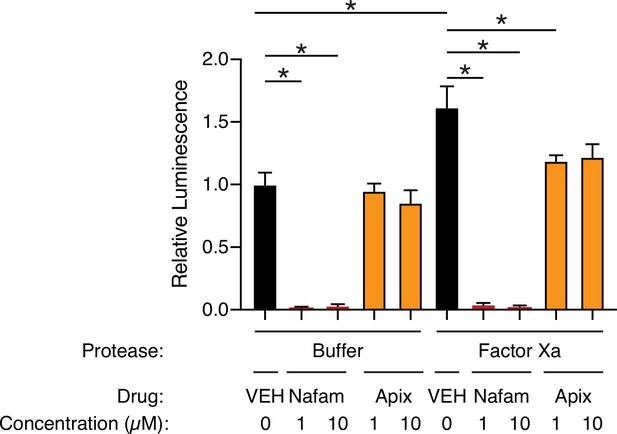
Apixaban rescues effect of factor Xa, related to Figure 4.
Calu3 cells were infected with rVSV∆G/SARS-CoV-2 pseudovirus with addition of protease buffer (left) or factor Xa (right). Cells were treated at the time of infection with DMSO vehicle (black), 1 or 10 µM nafamostat (red), or 1 or 10 µM apixaban (orange). N = 6, *p < 0.05, two-tailed t-test. Data represented as mean ± SEM.
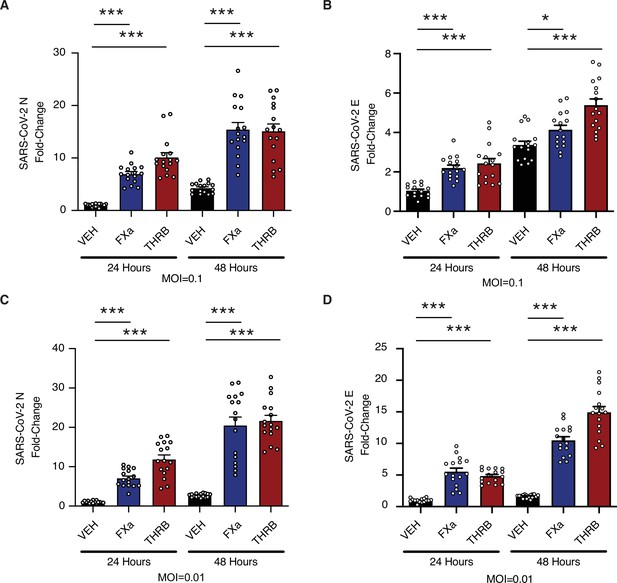
Factor Xa and thrombin increase SARS-CoV-2 infection in lung organoids.
Human pluripotent stem cell (hPSC)-derived lung organoids were infected with SARS-CoV-2. Upon infection, organoids were treated with 170 nM of purified factor Xa or thrombin. (A) Relative level of SARS-CoV-2-N RNA following infection at multiplicity of infection (MOI) = 0.1. (B) Relative level of SARS-CoV-2-E RNA following infection at MOI = 0.1. (C) Relative level of SARS-CoV-2-N RNA following infection at MOI = 0.01. (D) Relative level of SARS-CoV-2-E RNA following infection at MOI = 0.01. N = 16, *p = 0.0144, ***p < 0.0001, two-tailed t-test. Data represented as mean ± SEM.
-
Figure 5—source data 1
Data and summary statistics for infection assays with exogenous proteases.
- https://cdn.elifesciences.org/articles/77444/elife-77444-fig5-data1-v2.xlsx
Tables
Kinetics of SARS-CoV-2 spike peptide substrate cleavage.
Kinetic constants obtained from initial velocity studies with varying concentrations of SARS-CoV-2 spike S1/S2 and S2’ peptide substrates. Each estimate is based on seven different concentrations of substrate in 1:2 serial dilution (0–160 µM).
| Enzyme | Substrate | Vmax (µM/s) | Kcat (s–1) | Km (µM) | Ksp (s–1 µM–1) |
|---|---|---|---|---|---|
| TMPRSS2 | S1/S2 | 7.71E-04 | 6.17E-03 | 24.71 | 2.50E-04 |
| TMPRSS2 | S2' | 4.60E-04 | 3.68E-03 | 60.94 | 6.04E-05 |
| Factor Xa | S1/S2 | 2.24E-02 | 1.79E-01 | 40.35 | 4.43E-03 |
| Factor Xa | S2' | 3.04E-05 | 2.43E-04 | 2.711 | 8.97E-05 |
| Thrombin | S1/S2 | 6.50E-03 | 5.20E-02 | 16.34 | 3.18E-03 |
| Thrombin | S2' | 7.34E-04 | 5.87E-03 | 13.98 | 4.20E-04 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Chemical compound, drug | Camostat | Selleck | Cat# S2874 | |
| Chemical compound, drug | Nafamostat | Selleck | Cat# S1386 | |
| Chemical compound, drug | Apixaban | Medchem Express | Cat# HY-50667 | |
| Chemical compound, drug | Betrixaban | Medchem Express | Cat# HY-10268 | |
| Chemical compound, drug | Bivalirudin (TFA) | Medchem Express | Cat# HY-15664 | |
| Chemical compound, drug | Boceprevir | Medchem Express | Cat# HY-10237 | |
| Chemical compound, drug | Dabigatran etexilate | Medchem Express | Cat# HY-10274 | |
| Chemical compound, drug | Edoxaban | Medchem Express | Cat# HY-10264 | |
| Chemical compound, drug | Otamixaban | Medchem Express | Cat# HY-70035 | |
| Chemical compound, drug | Rivaroxaban | Medchem Express | Cat# HY-50903 | |
| Chemical compound, drug | Simeprevir | Medchem Express | Cat# HY-10241 | |
| Chemical compound, drug | Sivelestat | Medchem Express | Cat# HY-17443 | |
| Chemical compound, drug | Telaprevir | Medchem Express | Cat# HY-10235 | |
| Chemical compound, drug | Dabigatran | Medchem Express | Cat# HY-10163 | |
| Peptide, recombinant protein | Thrombin | Millipore Sigma | Cat# 605195 | |
| Peptide, recombinant protein | Factor Xa | Millipore Sigma | Cat# 69036 | |
| Peptide, recombinant protein | TMPRSS2 | LSBio | Cat# LS-G57269 | |
| Peptide, recombinant protein | TMPRSS4 | Aviva System Biology | Cat# OPCA0240 | |
| Peptide, recombinant protein | Furin | Thermo Fisher Scientific | Cat# 1503SE010 | |
| Peptide, recombinant protein | Neutrophil elastase | Thermo Fisher Scientific | Cat# 9167SE020 | |
| Peptide, recombinant protein | S1/S2 | Anaspec | QXL520-PRRARSVASQ-K(5-FAM)-NH2 | |
| Peptide, recombinant protein | S2’ | Anaspec | QXL520-KPSKRSFIED-K(5-FAM)-NH2 | |
| Peptide, recombinant protein | THRB-R271 | Anaspec | QXL520-AIEGRTATSE-K(5-FAM)-NH2 | |
| Peptide, recombinant protein | FGB-R44 | Anaspec | QXL520-FFSARGHRPL-K(5-FAM)-NH2 | |
| Peptide, recombinant protein | S1/S2-P1A | Anaspec | QXL520-PRRAASVASQ-K(5-FAM)-NH2 | |
| Peptide, recombinant protein | S1/S2-HPN | Anaspec | QXL520-PSQARSVASQ-K(5-FAM)-NH2 | |
| Chemical compound, drug | Phosphatidylcholine | Avanti Polar Lipids | Cat# 850375C | 1,2-Dioleoyl-sn-glycero-3-phosphocholine |
| Chemical compound, drug | phosphatidylserine | Avanti Polar Lipids | Cat# 840035C | 1,2-Dioleoyl-sn-glycero-3-phospho-L-serine |
| Cell line (Homo sapiens) | Calu3 | ATCC | Cat# HTB-55; RRID:CVCL_0609 | |
| Cell line (Homo sapiens) | A549 | ATCC | Cat# CCL-185; RRID:CVCL_0023 | |
| Cell line (Homo sapiens) | Caco2 | ATCC | Cat# HTB-37; RRID:CVCL_0025 | |
| Cell line (Chlorocebus sabaeus) | Vero | Laboratory of Benjamin tenOever | RRID:CVCL_0059 | |
| Cell line (Homo sapiens) | HEK293T | ATCC | Cat# CRL-3216; RRID:CVCL_0063 | |
| Recombinant DNA reagent | pEGPN | This paper | ||
| Recombinant DNA reagent | pEGPN-ACE2 | This paper | ||
| Recombinant DNA reagent | pEGPN-TMPRSS2 | This paper | ||
| Recombinant DNA reagent | Lenti-Cas9-blast | Addgene | Cat# 52962 | |
| Recombinant DNA reagent | ipUSEPR | Francisco Sanchez-Rivera & Scott Lowe | ||
| Recombinant DNA reagent | CMV-SARS-CoV-2-S | Schmidt et al., 2020 | ||
| Recombinant DNA reagent | CCNanoLuc/GFP | Schmidt et al., 2020 | ||
| Recombinant DNA reagent | HIV-1NL GagPol | Schmidt et al., 2020 | ||
| Commercial assay or kit | NEBuilder master mix | New England Biolabs | Cat# E2621 | |
| Chemical compound, drug | XtremeGene9 | Millipore Sigma | Cat# 6365787001 | |
| Chemical compound, drug | Polybrene | Santa Cruz Biotechnology | Cat# SC-134220 | |
| Chemical compound, drug | Lenti-X | Takara Bio | Cat# 631232 | |
| Chemical compound, drug | G418 | Sigma-Aldrich | Cat# # G8168 | |
| Chemical compound, drug | Blasticidin | Invivogen | Cat# ANT-BL-1 | |
| Chemical compound, drug | Puromycin | Thermo Fisher Scientific | Cat# A1113803 | |
| Commercial assay or kit | Cell Lysis Buffer | Promega | Cat# E1531 | |
| Commercial assay or kit | NanoGlo Luciferase Assay | Promega | Cat# N1130 | |
| Biological sample (Homo sapiens) | Normal human plasma | Pacific Hemostasis | Cat# 95059–698 | |
| Biological sample (Homo sapiens) | Factor X-deficient plasma | Haematologic Technologies | Cat# FX-ID | |
| Biological sample (Homo sapiens) | Prothrombin-deficient plasma | Haematologic Technologies | Cat# FII-ID | |
| Biological sample (Vipera russelli) | Russell’s Viper Venom | Sigma-Aldrich | Cat# V2501 | |
| Strain, strain background (Indiana vesiculovirus) | rVSVdG/NG-NanoLuc | Schmidt et al., 2020 | ||
| Strain, strain background (SARS-CoV-2) | SARS-CoV-2, isolate USA-WA1/2020 | BEI Resources, NIAID, NIH | Cat# NR-52281 | |
| Sequence-based reagent | sgRNA: CD4 | This study | sgRNA | GGTGCAATGTAGGAGTCCAA |
| Sequence-based reagent | sgRNA: PHGDH intron1 | This study | sgRNA | GGGCGAGAGAGAGAAAATTG |
| Sequence-based reagent | sgRNA: ACE2 g1 | This study | sgRNA | CACCGCAAAGGCGAGAGATAGTTG |
| Sequence-based reagent | sgRNA: ACE2 g2 | This study | sgRNA | CACCGACATCTTCATGCCTATGTG |
| Sequence-based reagent | sgRNA: TMPRSS2 g1 | This study | sgRNA | CACCGCTGGAACGAGAACTACGGG |
| Sequence-based reagent | sgRNA: TMPRSS2 g2 | This study | sgRNA | CACCGGGGACGGGTAGTACTGAGC |
| Sequence-based reagent | Primer: CD4-Forward | This study | PCR primers | GATAATGGAGAGATGTTGTTGGTTT |
| Sequence-based reagent | Primer: CD4- Reverse | This study | PCR primers | ATGTCCAGGTGCCACTATCCT |
| Sequence-based reagent | Primer: PHGDH intron 1 – Forward | This study | PCR primers | AAAGCAGAACCTTAGCAAAGAGG |
| Sequence-based reagent | Primer: PHGDH intron 1 – Reverse | This study | PCR primers | GAACTAATTGATACGGGGTGCAT |
| Sequence-based reagent | Primer: ACE2-g1- Forward | This study | PCR primers | TCCCTACTTTTTGTCGTTATTAGCA |
| Sequence-based reagent | Primer: ACE2-g1- Reverse | This study | PCR primers | GGTGATCCACAGCTAATGTATTGTT |
| Sequence-based reagent | Primer: ACE2-g2- Forward | This study | PCR primers | TCAAAATGCGATTTCTACAATGTTA |
| Sequence-based reagent | Primer: ACE2-g2- Reverse | This study | PCR primers | TGGGCTTTTCAGATTAAACCATTAT |
| Sequence-based reagent | Primer: TMPRSS2-g1-Forward | This study | PCR primers | ACAAATTCCACCTGCTGGTTATAG |
| Sequence-based reagent | Primer: TMPRSS2-g1- Reverse | This study | PCR primers | ACTTCATCCTTCAGGTGTACTCATC |
| Sequence-based reagent | Primer: TMPRSS2-g2- Forward | This study | PCR primers | CAGGAAATAAACACAAAGAGAATCC |
| Sequence-based reagent | Primer: TMPRSS2-g2-Reverse | This study | PCR primers | ACTATGAAAACCATGGATACCAACC |
| Sequence-based reagent | SARS-CoV-2-N-F | PCR primers | TAATCAGACAAGGAACTGATTA | |
| Sequence-based reagent | SARS-CoV-2-N-R | PCR primers | CGAAGGTGTGACTTCCATG | |
| Sequence-based reagent | SARS-CoV-2-E-F | PCR primers | ACAGGTACGTTAATAGTTAATAGCGT | |
| Sequence-based reagent | SARS-CoV-2-E-R | PCR primers | ATATTGCAGCAGTACGCACACA | |
| Sequence-based reagent | Human 18S-F | PCR primers | GGCCCTGTAATTGGAATGAGTC | |
| Sequence-based reagent | Human 18S-R | PCR primers | CCAAGATCCAACTACGAGCTT | |
| Software, algorithm | Prism 9 | GraphPad Software |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/77444/elife-77444-transrepform1-v2.docx
-
Supplementary file 1
Graphical abstract: Positive feedback in SARS-CoV-2 infection and coagulopathy.
In this study, we investigated the role of coagulation factors in SARS-CoV-2 infection. Hyperactivated coagulation is a feature of COVID-19 pathology. Coagulation factors, including factor Xa and thrombin, can cleave SARS-CoV-2 spike. This activity can exacerbate infection by enhancing viral entry. Lastly, we show that a subset of protease inhibitors with anticoagulant properties, such as nafamostat, also have the potential to block host-mediated spike activation by multiple human proteases.
- https://cdn.elifesciences.org/articles/77444/elife-77444-supp1-v2.zip





