Inner membrane complex proteomics reveals a palmitoylation regulation critical for intraerythrocytic development of malaria parasite
Figures
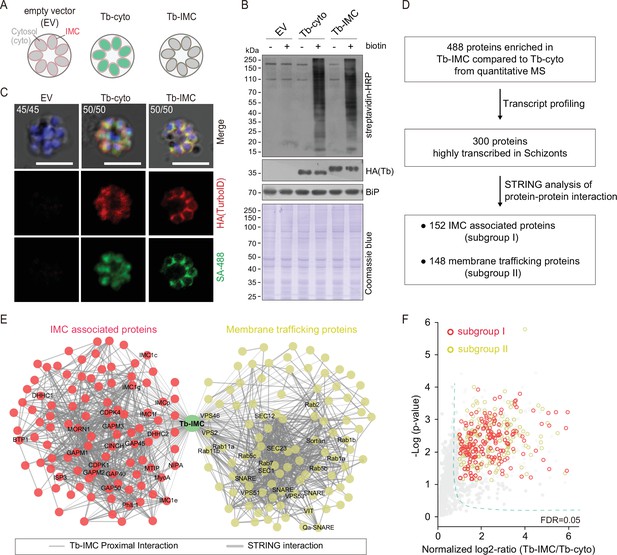
Proteomic of P. yoelii schizont IMC by TurboID and quantitative mass spectrometry.
(A) Schematic of schizonts with TurboID ligase localizing in the cytoplasm (Tb-cyto) and IMC (Tb-IMC). EV indicates the schizonts expressing empty vector (EV). See the detailed information of Tb-IMC and Tb-cyto in Figure 1—figure supplement 2. (B) Immunoblot and streptavidin blot of total lysate from the schizonts expressing the EV, Tb-cyto and Tb-IMC. Tagged ligase was detected by anti-HA antibody while biotinylated proteins were detected by streptavidin-conjugated horseradish peroxidase (streptavidin-HRP). Comparable loaded lysate was indicated by BiP control and Coomassie blue stain. (C) Costaining of TurboID ligase and biotinylated proteins in the schizonts expressing the ligase of the EV, Tb-cyto, and Tb-IMC. The schizonts incubated with or without 100 µM biotin were costained with the SA-488 and anti-HA antibody. x/y in the figure is the number of cells displaying signal/the number of cells analyzed. Scale bar = 5 μm. (D) Workflow for filtering the Tb-IMC interacting proteins (proximal interactors). 488 biotinylated proteins that were at least two times more abundant in Tb-IMC than that in Tb-cyto control among three replicates and with an adjusted p-value<0.05, were significantly enriched. Detailed information in Figure 1—figure supplement 2. (E) Interaction network of 300 Tb-IMC interacting proteins (STRING, p-value <1.0e-16, bold lines). Two subgroups (I: left, II: right) were functionally clustered. Many known inner membrane complex (IMC) or IMC-associated proteins were clustered into the subgroup I while many annotated ER/Golgi secretory or vesicle trafficking proteins were clustered into the subgroup II. (F) Volcano plots showing the 300 Tb-IMC interacting proteins. Relative biotinylation of each protein was calculated by quantifying protein intensity in Tb-IMC relative to Tb-cyto schizonts (n=3). Proteins in the subgroup I (red circle) and subgroup II (yellow circle) are indicated. MS: mass spectrometry; HA: hemagglutinin.
-
Figure 1—source data 1
(related to Figure 1B) Immunoblot and streptavidin blot of total lysate from the schizonts expressing the EV, Tb-cyto and Tb-IMC.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig1-data1-v3.xls
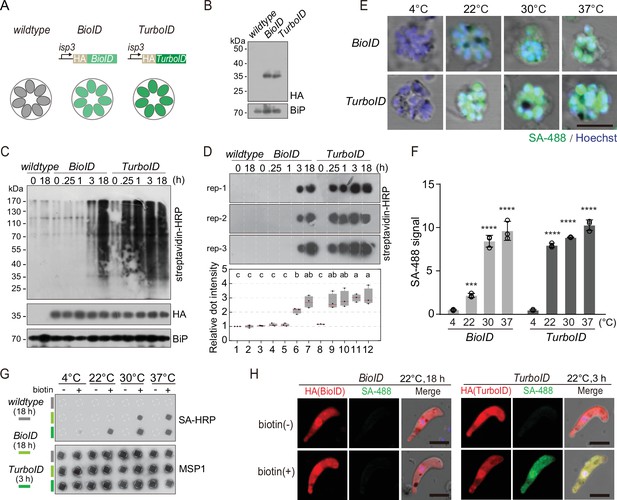
Protein proximity labeling by biotin ligases BioID and TurboID in the P. yoelii parasites.
(A) Schematic of the expressing cassettes for transient expression of BioID and TurboID. The ligase was fused with hemagglutinin (HA) and driven by the promoter of the isp3 gene. (B) Immunoblot of the HA-tagged ligases episomally expressed in the asexual blood stages of the P. yoelii. BiP as the loading control. (C) Streptavidin blot detecting the biotinylated proteins from the wildtype (WT), BioID or TurboID-expressing parasites incubated with exogenous biotin for different times (0, 0.25, 1, 3, and 18 hr) at 37°C. ER protein BiP is a loading control. (D) Streptavidin dot blot of the biotinylated proteins from total lysate of parasites in (C). Low panel shows the quantification of dot signals. Signals of WT parasite at 0 hr are set as 1.0, red bars indicate mean value. Different letters above the boxes indicate significant difference with a p<0.05. ANOVA analysis, followed by Tukey’s multiple comparison tests (n=3 biological replicates). (E) IFA of the biotinylated proteins in the schizonts from the BioID or TurboID-expressing parasites incubated at different temperatures (4, 22, 30, and 37°C). WT and BioID parasites were incubated with biotin for 18 hr while TurboID parasites were incubated with biotin for 3 hr. Parasites were stained with the Alexa Fluor 488 conjugated streptavidin (SA-488). Scale bar = 5 μm. (F) Quantitation of IFA signal intensity in (E). Values are means ± SD (n=3 biological replicates), two-tailed t-test, ***p<0.001, ****p<0.0001. (G) Streptavidin dot blot of the biotinylated proteins from the parasite lysates in (E). Merozoite surface protein MSP1 is used as the loading control. (H) IFA detecting the biotinylated proteins in the ligase-expressing ookinetes with or without biotin inoculation at 22°C. Cultured ookinetes expressing BioID and TurboID ligase were incubated for 18 and 3 hr, respectively, with or without 100 µM biotin at 22°C. The ookinetes were co-stained with the SA-488 and anti-HA antibody. Scale bar = 5 μm.
-
Figure 1—figure supplement 1—source data 1
(related to Figure 1—figure supplement 1B) Immunoblot of the HA-tagged ligases episomally expressed in the asexual blood stages of the P. yoelii.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig1-figsupp1-data1-v3.xls
-
Figure 1—figure supplement 1—source data 2
(related to Figure 1—figure supplement 1C) Streptavidin blot detecting the biotinylated proteins from the wildtype, BioID or TurboID-expressing parasites incubated with exogenous biotin for different times (0, 0.25, 1, 3, and 18 hr) at 37°C.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig1-figsupp1-data2-v3.xls
-
Figure 1—figure supplement 1—source data 3
(related to Figure 1—figure supplement 1D) Streptavidin dot blot of the biotinylated proteins from total lysate of parasites in C.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig1-figsupp1-data3-v3.xls
-
Figure 1—figure supplement 1—source data 4
(related to Figure 1—figure supplement 1D) Quantification of streptavidin dot blot of the biotinylated proteins.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig1-figsupp1-data4-v3.xls
-
Figure 1—figure supplement 1—source data 5
(related to Figure 1—figure supplement 1F) Quantification of IFA signal intensity.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig1-figsupp1-data5-v3.xls
-
Figure 1—figure supplement 1—source data 6
(related to Figure 1—figure supplement 1G) Streptavidin dot blot of the biotinylated proteins from the parasite lysates.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig1-figsupp1-data6-v3.xls
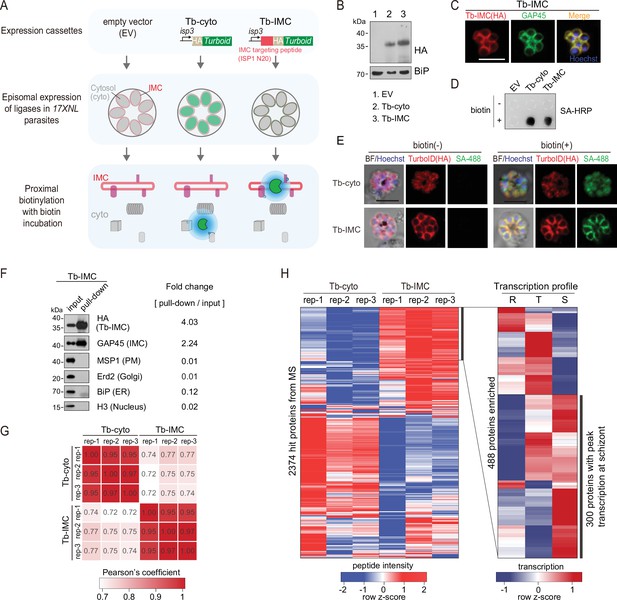
TurboID-mediated labeling of IMC proteins in the schizonts.
(A) Experimental workflow for TurboID proximity labeling of the inner membrane complex (IMC) proteins in the schizont. The hemagglutinin (HA)-tagged TurboID is fused with an IMC targeting peptide, the N-terminal 20 residues of ISP1 (Tb-IMC). ISP1 is highly expressed in ookinete, but not expressed or in a quite low expression level in the asexual blood stage. The HA-tagged TurboID alone (Tb-cyto) serves as a control for non-specific biotinylation. Both ligases (Tb-IMC and Tb-cyto) were driven by the promoter of gene isp3 and episomally expressed in the asexual blood stages. The schizonts expressing Tb-IMC, Tb-cyto, or empty vector (EV: construct without ligase gene) were purified and cultivated with 100 µM biotin at 37°C for 3 hr. (B) Immunoblot of the HA-tagged ligase from the total lysate of schizonts expressing the Tb-cyto or Tb-IMC shown in (A). ER protein BiP used as a loading control. (C) IFA of the Tb-IMC expressing schizonts with anti-HA antibody, anti-GAP45 antibody, and DNA stain Hoechst 33342. Scale bar = 5 μm. (D) Streptavidin dot blot of biotinylated proteins in the EV, Tb-cyto, Tb-IMC expressing schizonts incubated with or without 100 µM biotin. (E) Costaining of TurboID ligase and biotinylated proteins in the schizonts expressing the Tb-cyto or Tb-IMC. The schizonts incubated with or without 100 µM biotin were costained with anti-HA antibody and streptavidin-488 (SA-488). Scale bar = 5 μm. (F) Immunoblot of streptavidin-affinity purified biotinylated proteins in Tb-IMC schizonts. Tb-IMC ligase and several organelle marker proteins were probed with the indicated antibodies. Relative band intensity of each protein in the pull-down compared to the input indicates the enrichment ratio. (G) Correlation analysis of change in protein abundance among biological replicates between Tb-cyto and Tb-IMC. Three biological replicates were prepared from Tb-IMC and Tb-cyto schizonts, and the streptavidin-affinity purified proteins were for proteomic analysis by mass spectrometry. (H) Identification of 300 Tb-IMC interacting protein by quantitative MS and comparative transcription profiling. Clustered heatmap (left panel) of MS peptide intensity revealed 488 enriched proteins with high confidence (an adjusted p-value<0.05) in Tb-IMC compared to Tb-cyto. Comparative analysis of transcription pattern (right panel) based on the P. berghei transcriptome further narrow the candidates to 300 proteins, in which many known IMC or IMC-associated proteins were indicated. R: ring, T: trophozoite, and S: schizont.
-
Figure 1—figure supplement 2—source data 1
(related to Figure 1—figure supplement 2B) Immunoblot of the HA-tagged ligase from the total lysate of schizonts expressing the Tb-cyto or Tb-IMC.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig1-figsupp2-data1-v3.xls
-
Figure 1—figure supplement 2—source data 2
(related to Figure 1—figure supplement 2D) Streptavidin dot blot of biotinylated proteins in the EV, Tb-cyto, Tb-IMC expressing schizonts incubated with or without 100 µM biotin.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig1-figsupp2-data2-v3.xls
-
Figure 1—figure supplement 2—source data 3
(related to Figure 1—figure supplement 2F) Immunoblot of streptavidin-affinity purified biotinylated proteins in Tb-IMC schizonts.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig1-figsupp2-data3-v3.xls
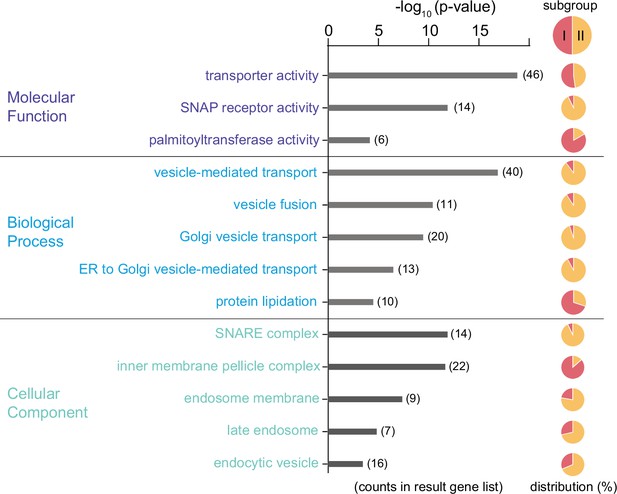
Predicated functional profile of the Tb-IMC interacting proteins Gene ontology analysis of the 300 Tb-IMC interacting proteins.
Bar plot showing the significantly enriched gene ontology (GO) terms from ‘Molecular Function’ (top panel), ‘Biological Process’ (middle panel) and ‘Cellular Component’ (bottom panel). For each of GO terms, the distribution of proteins from the subgroup I and II was indicated in the pie chart.
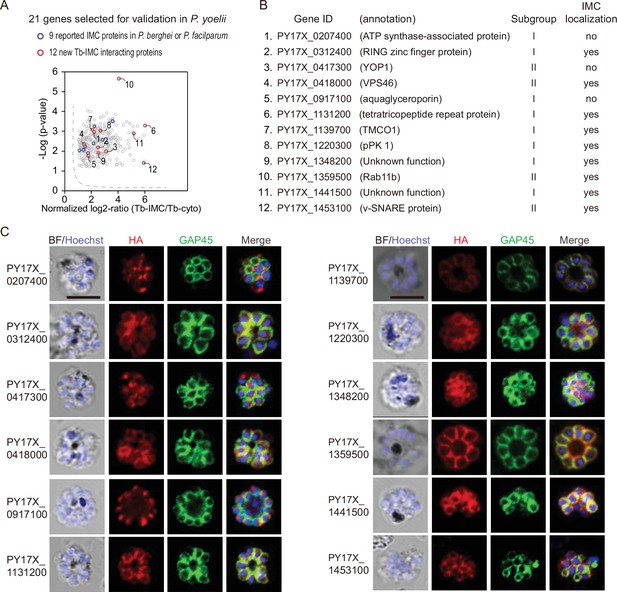
Validation of 9 new IMC proteins by localization analysis.
(A) 21 candidates selected from the Tb-IMC interacting proteins for subcellular localization analysis in the P. yoelii. The orthologues of 9 proteins (blue dot) have been experimentally validated to be IMC-residing or -associated in the schizonts of P. berghei or P. falciparum, while subcellular localization of other 12 proteins (red dot) have not been well-characterized in Plasmodium species.(B) Information and subcellular localization summary of the 12 newly tested Tb-IMC interacting proteins shown in A. (C) Immunofluorescence assays (IFA) analysis of 12 Tb-IMC interacting proteins in the P. yoelii schizonts. Each protein was tagged with a 6 HA at the N- or C-terminus and episomally expressed in the schizonts. The schizonts were costained with antibodies against GAP45 and HA. Nuclei were stained with Hoechst 33342. Scale bar = 5 μm.
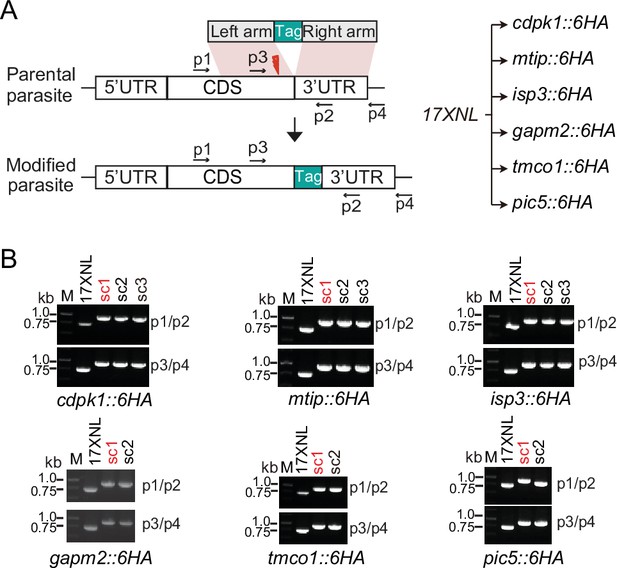
Genotyping of genetically modified parasites.
(A) Schematic of gene tagging with a 6 HA at the C-terminus using the CRISPR-Cas9 method (left panel). The parasite lines generated were listed at the right panel. (B) Diagnostic PCR of parasite single clone (sc). Parasites shown in red text were used in the experiments.
-
Figure 2—figure supplement 1—source data 1
(related to Figure 2—figure supplement 1B) Diagnostic PCR of Tb-IMC interacting protein candidates.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig2-figsupp1-data1-v3.xls
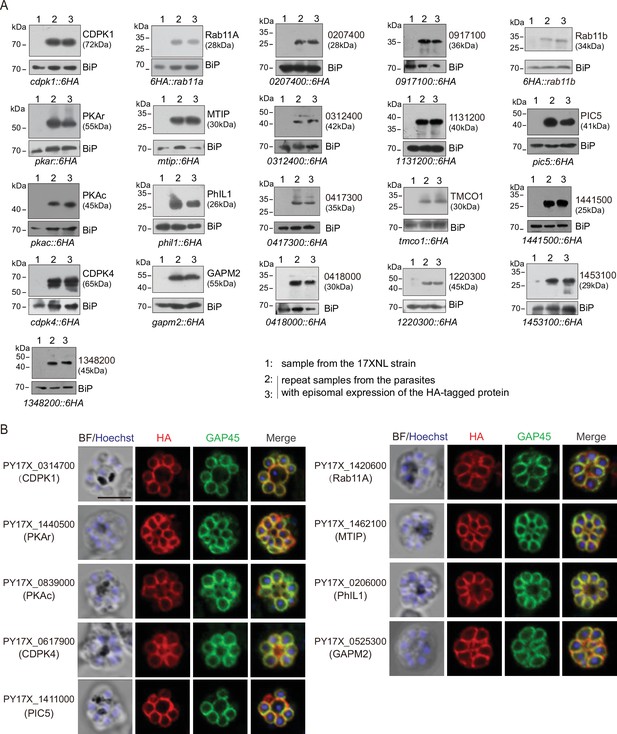
Expression and localization analysis of IMC protein candidates selected in this study.
(A) Immunoblot of 21 IMC protein candidates in the schizonts of P. yoelii. Among the 21 candidates, 6 out of them were endogenously tagged with a 6 HA at the C-terminus (see the Figure 2—figure supplement 1A,B), while others were tagged with a 6 HA at the N- or C-terminus and driven by the promoter of gene isp3 for episomal expression in the schizonts. Except the Rab11a and Rab11b (PY17X_1359500) were tagged at the N-terminus, other genes were tagged at the C-terminus. BiP as the loading control. (B) Immunofluorescence assays (IFA) of 9 known IMC proteins in the P. yoelii schizonts. The orthologues of 9 proteins, including CDPK1, CDPK4, PKAr, PKAc, Rab11A, MTIP, PhIL1, GAPM2, and PIC5 have been experimentally validated to be IMC-residing in the schizonts of P. berghei or P. falciparum. Each protein was tagged with a 6 HA at the N- or C-terminus and episomally expressed in the schizonts. The schizonts were co-stained with antibodies against GAP45 and HA. Nuclei were stained with Hoechst 33342. Scale bar = 5 μm.
-
Figure 2—figure supplement 2—source data 1
(related to Figure 2—figure supplement 2A) Immunoblot of 21 IMC protein candidates in the schizonts of P. yoelii.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig2-figsupp2-data1-v3.xls
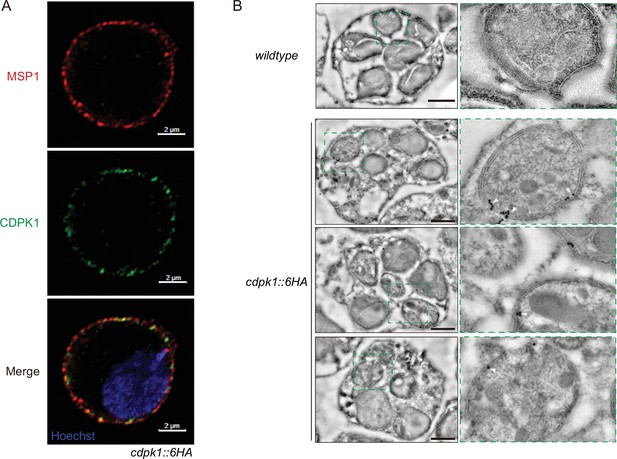
CDPK1 localizes at IMC in the schizonts of P.yoelii.
(A) Ultrastructural expansion microscopy (U-ExM) analysis of CDPK1 in schizonts of the cdpk1::6 HA parasite. Expanded schizonts were stained with antibodies against HA and MSP1 (parasite plasma membrane protein), and observed under the Airyscan model of confocal microscopy. The results showed that most of CDPK1 signals were localized in the inner side of MSP1 signals. Scale bar = 2 μm. (B) Immunoelectron microscopy images of cdpk1::6 HA parasite. White triangles denote the 15 nm gold particles coupled with the anti-HA antibody. Wildtype (WT) parasite was used as a control. Scale bar = 1 μm. The results showed that CDPK1 was primarily localized in the inner layer, but not the out layer of parasite periphery of the cdpk1::6 HA schizonts.
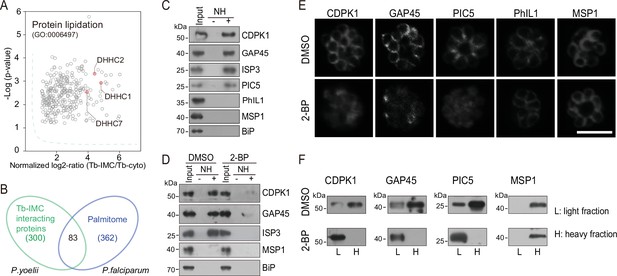
Palmitoylation regulates IMC protein localization.
(A) Enrichment of enzymes for protein lipidation in the Tb-IMC interacting proteins shown in Figure 1F, including three palmitoyl-S-acyl-transferases DHHC2, DHHC1, and DHHC7. (B) Venn diagram showing overlap between Tb-IMC interactors (green) identified in this study and the orthologs within the P. falciparum palmitome (blue). 83 Tb-IMC interactors (overlap, 28%) were considered to be potentially palmitoylated. Numbers indicate the number of proteins identified. (C) Acyl-RAC method detecting palmitoylation of CDPK1, GAP45, ISP3, and PIC5, but not PhIL1 in schizonts. NH: NH2OH. MSP1 and BiP served as loading controls. Total proteins were treated with MMTs to block the thiol side chain in free cysteine. Proteins with palmitoylated cysteine were re-exposed the thiol side chain with removal of palmitic acid by NH2OH, purified via Thiopropyl Sepharose, and eluted by DTT for immunoblot. Representative of two independent replicates. (D) Palmitoylation analysis of CDPK1, GAP45, and ISP3 in schizonts treated with 2-BP. NH: NH2OH. MSP1 and BiP served as loading controls. Representative of two independent replicates. (E) Immunofluorescence assays (IFA) analysis of CDPK1, GAP45, PIC5, PhIL1, and MSP1 in schizonts treated with 2-BP and DMSO, respectively. Purified mature schizonts were treated with 2-BP for 12 hr. Scale bar = 5 μm. (F) Fractionation analysis of CDPK1, GAP45, PIC5, and MSP1 in schizonts treated with 2-BP and DMSO, respectively. Light fraction includes cytosolic proteins while heavy fraction includes membrane proteins and cytoskeleton proteins. Representative of two independent replicates.
-
Figure 3—source data 1
(related to Figure 3C) Acyl-RAC method detecting palmitoylation of CDPK1, GAP45, ISP3, and PIC5, but not PhIL1 in schizonts.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig3-data1-v3.xls
-
Figure 3—source data 2
(related to Figure 3D) Palmitoylation analysis of CDPK1, GAP45, and ISP3 in schizonts treated with 2-BP.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig3-data2-v3.xls
-
Figure 3—source data 3
(related to Figure 3F) Fractionation analysis of CDPK1, GAP45, PIC5, and MSP1 in schizonts treated with 2-BP and DMSO, respectively.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig3-data3-v3.xls
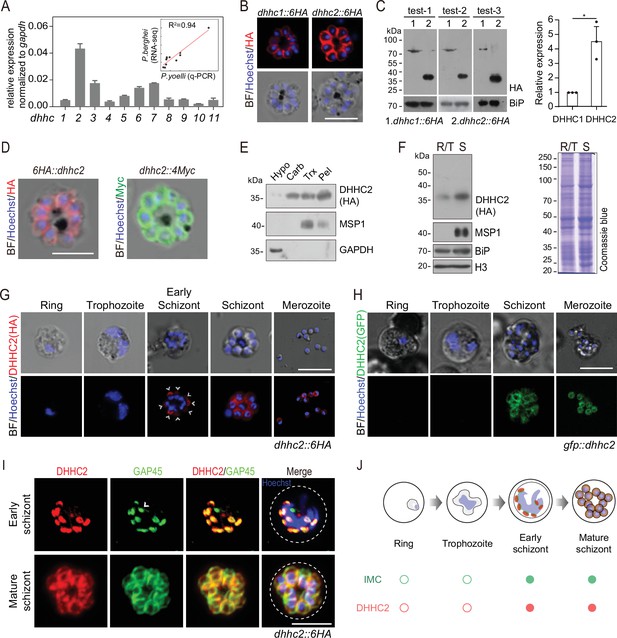
DHHC2 is an IMC-residing palmitoyl-S-acyl-transferase in schizonts.
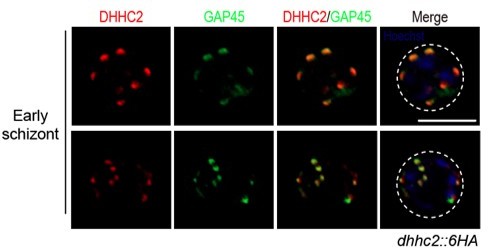
(A) RT-qPCR of transcripts for 11 dhhc (dhhc1- dhhc11) in schizonts. Gene expression was normalized to the gapdh transcript. The inlet indicates positive correlation between mRNA levels of these P. yoelii dhhc genes and mRNA levels of their dhhc orthologs in P. berghei determined via RNA-seq. Values are means ± SD (n=3). (B) Immunofluorescence assays (IFA) analysis of DHHC1 and DHHC2 in schizonts from two tagged parasite strains dhhc1::6 HA and dhhc2::6 HA. Scale bar = 5 μm. (C) Immunoblot of DHHC1 and DHHC2 from the cell lysate of similar number of schizonts from the dhhc1::6 HA and dhhc2::6 HA parasite, respectively. Right panel: the quantification of band intensity. Values are means ± SEM (n=3 biological replicates), two-tailed t-test, *p<0.05. (D) IFA analysis of DHHC2 in schizonts of another two tagged parasite strains 6 HA::dhhc2 and dhhc2::4Myc. Scale bar = 5 μm. (E) Solubility assay detected membrane association of DHHC2 in schizonts using different detergents. Cytosolic soluble proteins are in hypotonic buffer (Hypo), peripheral membrane proteins in carbonate buffer (Carb), integral membrane proteins in Triton X-100 buffer (Trx), and insoluble proteins in pellet (Pel). DHHC2 is in the membrane-associated fractions as IMC protein GAP45 and PM protein MSP1, while cytoplasm protein GAPDH is in the soluble fraction. Representative of two independent replicates. (F) Immunoblot of DHHC2 from early stages containing ring and trophozoite (R/T) and late stages containing schizont (S) of the dhhc2::6 HA parasites. Merozoite surface protein MSP1 was mainly expressed in the schizonts. BiP, histone H3, and Coomassie blue staining of total lysate were used as loading control. Representative of two independent replicates. (G) DHHC2 expression dynamics in different asexual blood stages (ring, trophozoite, schizont, and merozoite) of the parasite dhhc2::6 HA by IFA. Scale bar = 5 μm. (H) Fluorescent microscopy of GFP::DHHC2 in different asexual blood stages of the parasites gfp::dhhc2. Scale bar = 5 μm. (I) Maximum intensity projections of super-resolution immunofluorescence microscopy (Airyscan) of early and mature dhhc2::6 HA schizonts stained with anti-HA and anti-GAP45 antibodies. Arrow in the graph indicated a ‘GAP45-only spot’, which likely resulted from the difference in protein stain efficiency for DHHC2 and GAP45. Scale bar = 5 μm. (J) Model showing the IMC-associated localization of DHHC2 in the schizonts.
-
Figure 4—source data 1
(related to Figure 4A) RT-qPCR of transcripts for 11 dhhc (dhhc1- dhhc11) in schizonts.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig4-data1-v3.xls
-
Figure 4—source data 2
(related to Figure 4C) Immunoblot of DHHC1 and DHHC2 from the cell lysate of similar number of schizonts from the dhhc1::6 HA and dhhc2::6 HA parasite, respectively.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig4-data2-v3.xls
-
Figure 4—source data 3
(related to Figure 4C) Quantification of DHHC1 and DHHC2 intensity in immunoblot.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig4-data3-v3.xls
-
Figure 4—source data 4
(related to Figure 4E) Solubility assay detected membrane association of DHHC2 in schizonts using different detergents.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig4-data4-v3.xls
-
Figure 4—source data 5
(related to Figure 4F) Immunoblot of DHHC2 from early stages containing ring and trophozoite (R/T) and late stages containing schizont (S) of the dhhc2::6 HA parasites.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig4-data5-v3.xls
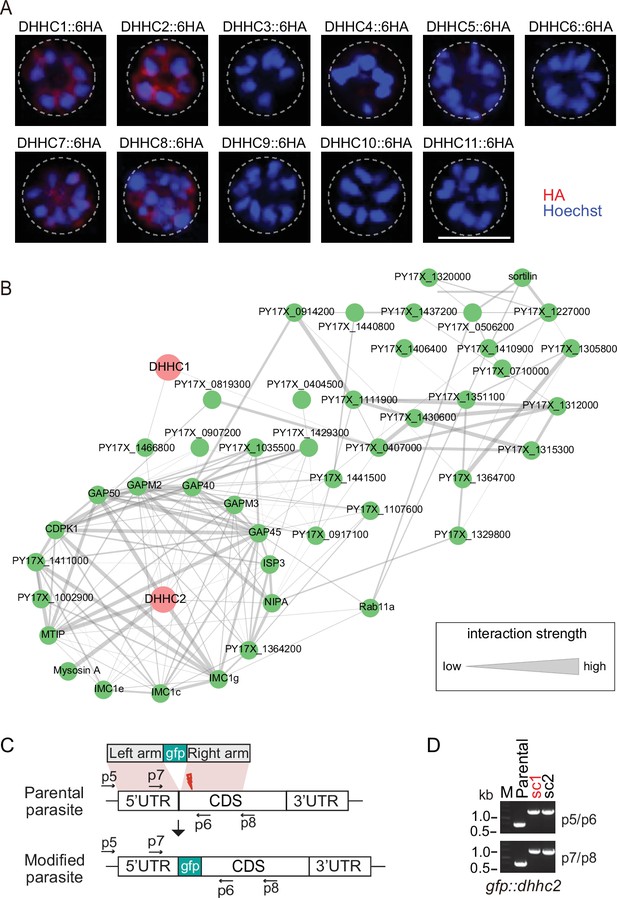
Localization analysis of 11 PATs (DHHC1-11) in schizonts.
(A) Immunofluorescence assays (IFA) analysis of 11 PATs (DHHC1-11) expression in the schizonts. Each of individual P. yoelii PATs was endogenously tagged with a 6 HA in the transgenic strains generated previously. The schizonts were costained with the anti-HA antibody and Hoechst 33342. Scale bar = 5 μm. (B) Predicted protein interaction network between DHHC1/DHHC2 and the putatively palmitoylated Tb-IMC interacting proteins (STRING; p<1.0e-16). (C) Schematic of gene-engineered gfp::dhhc2 via CRISPR-Cas9 method. (D) Diagnostic PCR of gfp::dhhc2 parasite line. Single clone (sc) shown in red was used for analysis.
-
Figure 4—figure supplement 1—source data 1
(related to Figure 4—figure supplement 1D) Diagnostic PCR of gfp::dhhc2 parasite line.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig4-figsupp1-data1-v3.xls
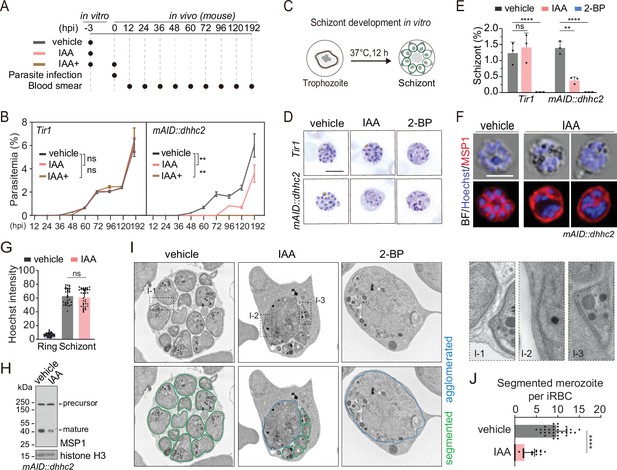
DHHC2 is essential for parasite proliferation in mice and regulates schizont segmentation and merozoite invasion.
(A) Experimental design of in vivo test of DHHC2 essentiality using AID. Parasites of Tir1 and mAID::dhhc2 were pretreated with vehicle or indole-3-acetic acid (IAA) for 3 hr in vitro, then intravenously injected to C57BL/6 mice. In the IAA+ treatment group, another IAA injection (200 mg/kg, ip) at time of parasite infection was applied for further DHHC2 depletion in vivo. From 12–192 hr postinfection, the parasitemia in mice infected with Tir1 and mAID::dhhc2 was monitored by blood smear every 12 hr. (B) Tir1 and mAID::dhhc2 parasite proliferation in mice (n=3 per group) at each treatment group in (A), two-tailed t-test, **p<0.01, ns, not significant. (C) Schematic of the schizont development from trophozoite in vitro. Purified early stage parasites including ring and trophozoite were cultured with vehicle, IAA, or 2-BP for 12 hr for schizont development. (D) Giemsa staining of the schizonts developed from Tir1 and mAID::dhhc2 parasites treated with vehicle, IAA, or 2-BP illustrated in (C). Scale bar = 5 μm. (E) Quantification of mature schizonts in (D). Purified early stage parasites including ring and trophozoite were treated with IAA or 2-BP for 12 hr. Values are means ± SEM (n=3 biological replicates), two-tailed t-test, **p<0.01, ***p<0.001. (F) Costaining of the mAID::dhhc2 schizonts in (C) with antibody against merozoite surface protein MSP1 and Hoechst 33342. Scale bar = 5 μm. (G) Quantification of Hoechst signal in schizonts indicating nuclear DNA contents in (F). More than 30 schizonts were analyzed in each group. Signal in ring stage parasites serves as a control. Values were means ± SD, Mann–Whitney U test applied, ns, not significant. (H) Immunoblot of MSP1 in the schizonts developed from the mAID::dhhc2 parasites treated with vehicle and IAA. Precursor form (~200 kD) and mature form (~42 kD) of MSP1 were shown. Histone H3 used as a loading control. (I) Representative images of transmission electron microscopy (TEM) of schizonts developed from the mAID::dhhc2 parasites treated with vehicle, IAA, or 2-BP. Purified early stage parasites including ring and trophozoite were treated with IAA or 2-BP for 12 hr. Right panels indicate three examples of representative daughter cell pellicle including plasma membrane (PM) and inner membrane complex (IMC). (J) Quantification of fully segmented merozoites in schizonts in (I). More than 30 schizonts were analyzed in each group. Values were shown as means ± SD, Mann–Whitney U test, ****p<0.0001.
-
Figure 5—source data 1
(related to Figure 5B) Tir1 and mAID-dhhc2 parasite proliferation in mice.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig5-data1-v3.xls
-
Figure 5—source data 2
(related to Figure 5E) Quantification of mature schizonts.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig5-data2-v3.xls
-
Figure 5—source data 3
(related to Figure 5G) Quantification of Hoechst signal in schizonts.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig5-data3-v3.xls
-
Figure 5—source data 4
(related to Figure 5H) Immunoblot of MSP1 in the schizonts developed from the mAID::dhhc2 parasites treated with vehicle and IAA.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig5-data4-v3.xls
-
Figure 5—source data 5
(related to Figure 5J) Quantification of fully segmented merozoites in schizonts.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig5-data5-v3.xls
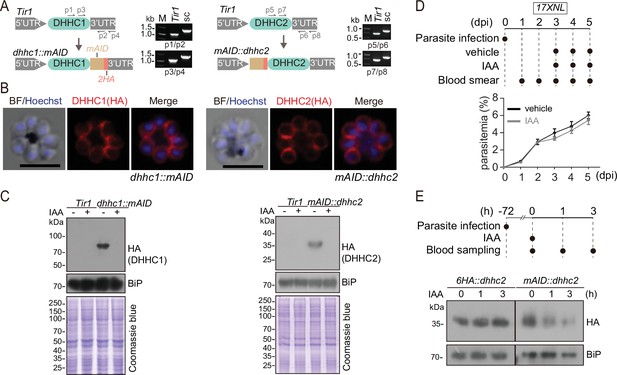
Generation of the modified strains with endogenous DHHC2 and DHHC2 tagged with a mAID motif for induced protein degradation.
(A) Schematic of generation of modified strains with endogenous DHHC1 and DHHC2 tagged with a mAID motif. The mAID::2 HA motif was inserted to the C-terminus of endogenous DHHC1 and the N-terminus of endogenous DHHC2, respectively, in the parental parasite Tir1 using the CRISPR-Cas9 method, generating the strains dhhc1::mAID and mAID::dhhc2. Diagnostic PCR results confirming correct modification were shown at the right panels. mAID: mini auxin-inducible degron. (B) Immunofluorescence assays (IFA) of the fusion proteins DHHC1::mAID (left panel) and mAID::DHHC2 (right panel) in the schizonts of dhhc1::mAID and mAID::dhhc2 parasites. Scale bar = 5 μm. (C) Immunoblot of fusion proteins DHHC1::mAID (left panel) and mAID::DHHC2 (right panel) in the schizonts of the Tir1, dhhc1::mAID and mAID::dhhc2 parasites treated with vehicle or indole-3-acetic acid (IAA) (1 mM) for 3 hr. BiP and Coomassie blue staining were used as the loading control. (D) Proliferation assessment of wildtype (WT) parasite in mice treated with IAA. Upper panel indicates the experimental design. C57BL/6 mice with ~2–3% parasitemia of WT parasite (17XNL) were injected intraperitoneally with 200 mg/kg/day IAA or vehicle each day for 3 days (days 3–5). Parasitemia was monitored by Giemsa staining of blood smear at indicated time. (E) IAA-induced degradation assessment of parasite mAID::DHHC2 protein in mice. Upper panel indicates the experimental design. C57BL/6 mice with ~10% parasitemia of mAID::dhhc2 or 6 HA::dhhc2 parasites were injected intraperitoneally with IAA (200 mg/kg) for one time and the parasite-infected red blood cells were collected for detecting the protein abundance by immunoblot at different time post IAA injection. BiP was used as a loading control.
-
Figure 5—figure supplement 1—source data 1
(related to Figure 5—figure supplement 1A) Diagnostic PCR of dhhc1::maid, mid::dhhc2 parasite lines.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig5-figsupp1-data1-v3.xls
-
Figure 5—figure supplement 1—source data 2
(related to Figure 5—figure supplement 1C) Immunoblot of fusion proteins DHHC1::mAID (left panel) and mAID::DHHC2 (right panel) in the schizonts of the Tir1, dhhc1::mAID and mAID::dhhc2 parasites treated with vehicle or IAA (1 mM) for 3 hr.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig5-figsupp1-data2-v3.xls
-
Figure 5—figure supplement 1—source data 3
(related to Figure 5—figure supplement 1D) Proliferation assessment of wildtype parasite in mice treated with IAA.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig5-figsupp1-data3-v3.xls
-
Figure 5—figure supplement 1—source data 4
(related to Figure 5—figure supplement 1E) IAA-induced degradation assessment of parasite mAID::DHHC2 protein in mice.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig5-figsupp1-data4-v3.xls
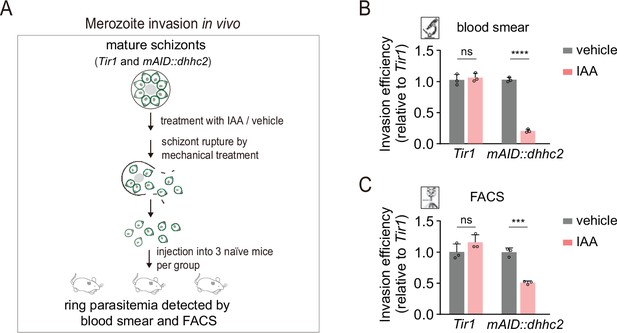
DHHC2 depletion impaired merozoite invasion.
(A) Schematic of merozoite invasion in the mouse. Purified mature schizonts of Tir1 and mAID::dhhc2 parasites were pretreated with indole-3-acetic acid (IAA) for 3 hr for protein depletion. Followed by mechanical treatment of schizont for rupture, the released merozoites were collected and injected intravenously into three naïve mice per condition. 20 min postinjection, the ability of merozoite invasion was evaluated by counting the parasitemia (ring stage parasite) using blood smear and flow cytometry. (B, C) Histogram showing the invasion efficiency of merozoite in the mice acquired by blood smear (B) and flow cytometry (C). Invasion efficiency of vehicle-treated group was set as 1.0. Values were means ± SD, two-tailed t-test, ***p<0.001, ****p<0.0001, ns: not significant.
-
Figure 5—figure supplement 2—source data 1
(related to Figure 5—figure supplement 2B) The invasion efficiency of merozoite in the mice evaluated by blood smear.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig5-figsupp2-data1-v3.xls
-
Figure 5—figure supplement 2—source data 2
(related to Figure 5—figure supplement 2C) The invasion efficiency of merozoite in the mice evaluated by flow cytometry.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig5-figsupp2-data2-v3.xls
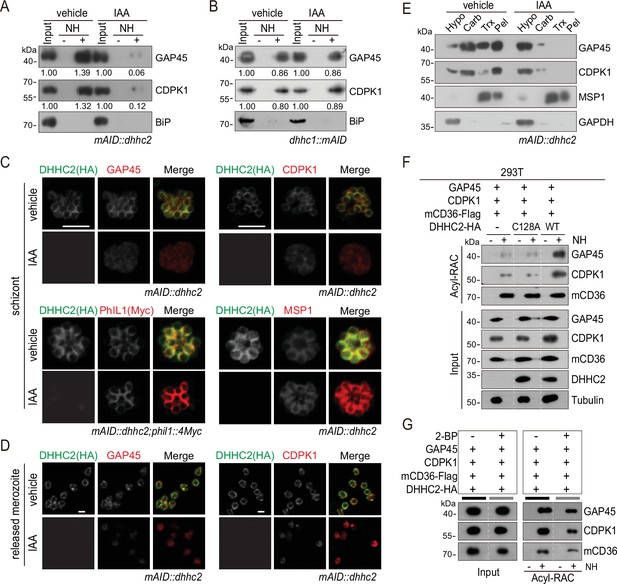
DHHC2 palmitoylates GAP45 and CDPK1 in the schizonts.
(A) Acyl-RAC method detecting palmitoylation of GAP45 and CDPK1 in the mAID::dhhc2 schizonts treated with vehicle or indole-3-acetic acid (IAA). BiP served as a loading control. Representative of two independent replicates. (B) Acyl-RAC method detecting palmitoylation of GAP45 and CDPK1 in the dhhc1::mAID schizonts treated with vehicle or IAA. BiP served as a loading control. Representative of two independent replicates. (C) Immunofluorescence assays (IFA) analysis of GAP45, CDPK1, PhIL1, and MSP1 expression in schizonts treated with vehicle or IAA. Purified mature schizonts were treated with IAA for 12 hr. Parasites mAID::dhhc2 and mAID::dhhc2;phil1::4Myc used are indicated. PhIL1 is an inner membrane complex (IMC) protein without palmitoylation. MSP1 is a plasma membrane protein. Scale bar = 5 μm. (D) IFA analysis of GAP45 and CDPK1 in released mAID::dhhc2 merozoites treated with vehicle or IAA. Scale bar = 5 μm. (E) Solubility assay detected membrane association of GAP45, CDPK1, and MSP1 in the mAID::dhhc2 schizonts treated with vehicle or IAA. PM protein MSP1 and cytoplasmic protein GAPDH are set as control. Representative of two independent replicates. (F) Palmitoylation analysis of GAP45 and CDPK1 ectopically expressed in human cells. Human embryonic kidney 293T cells were cotransfected with plasmids coding for the HA-tagged and human codon-optimized DHHC2 (WT) or its catalytic-deficient mutant (C128A), along with the GAP45 and CDPK1. Flag-tagged mouse CD36 (mCD36-Flag) was also cotransfected and serves as a control of evidenced palmitoylated protein. Tubulin served as a loading control. Representative of two independent replicates. (G) Palmitoylation analysis of GAP45 and CDPK1 ectopically expressed in human HEK293T cells treated with or without 2-BP. The cells were cotransfected with a HA-tagged and human codon-optimized DHHC2 and a flag-tagged mouse CD36 (mCD36-Flag). Representative of two independent replicates.
-
Figure 6—source data 1
(related to Figure 6A) Acyl-RAC method detecting palmitoylation of GAP45 and CDPK1 in the mAID::dhhc2 schizonts treated with vehicle or IAA.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig6-data1-v3.xls
-
Figure 6—source data 2
(related to Figure 6B) Acyl-RAC method detecting palmitoylation of GAP45 and CDPK1 in the dhhc1::mAID schizonts treated with vehicle or IAA.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig6-data2-v3.xls
-
Figure 6—source data 3
(related to Figure 6E) Solubility assay detected membrane association of GAP45, CDPK1, and MSP1 in the mAID::dhhc2 schizonts treated with vehicle or IAA.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig6-data3-v3.xls
-
Figure 6—source data 4
(related to Figure 6F) Palmitoylation analysis of GAP45 and CDPK1 ectopically expressed in human cells.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig6-data4-v3.xls
-
Figure 6—source data 5
(related to Figure 6G) Palmitoylation analysis of GAP45 and CDPK1 ectopically expressed in human HEK293T cells treated with or without 2BP.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig6-data5-v3.xls
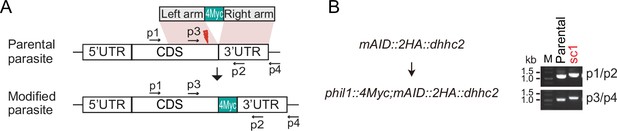
Genotyping of genetically modified parasites.
(A) Schematic of the phil1 gene tagging with a 4Myc at the C-terminus in the mAID::dhhc2 parasite using the CRISPR-Cas9 method. (B) Diagnostic PCR of parasite single clone (sc).
-
Figure 6—figure supplement 1—source data 1
(related to Figure 6—figure supplement 1B) Diagnostic PCR of phil1::4Myc; mid::dhhc2 parasite line.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig6-figsupp1-data1-v3.xls
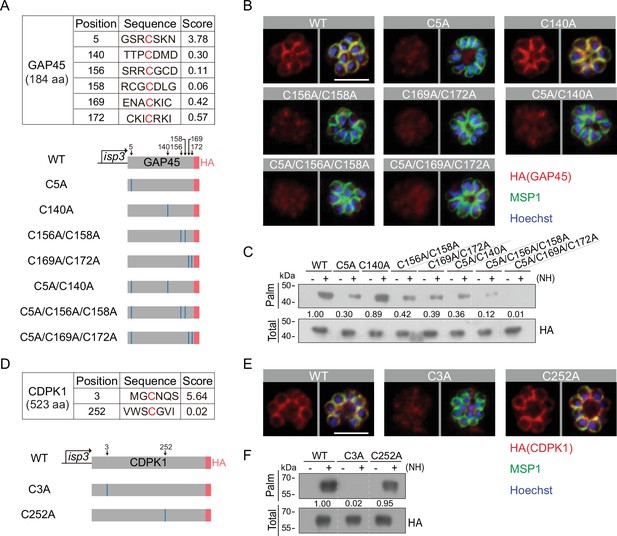
Residues for palmitoylation in GAP45 and CDPK1.
(A) Six cysteine predicated for palmitoylation in GAP45 (upper panel). The score of the predicted palmitoylation sites was provided by the CSS-Palm software. Lower panels indicate schematic of constructs expressing HA-tagged GAP45, each with a cysteine to alanine replacement in single, double, or triple residues (C5A, C140A, C156A/C158A, C169A/C172A, C5A/C140A, C5A/C156A/C158A, and C5A/C169A/C172A). These constructs were episomally expressed in the schizonts. (B) Immunofluorescence assays (IFA) of GAP45::HA and seven mutant proteins episomally expressed in schizonts. Scale bar = 5 μm. Representative of three independent repeats. (C) Palmitoylation analysis of GAP45::HA and seven mutant proteins episomally expressed in schizonts. Representative of two independent repeats. (D) Two cysteine predicated for palmitoylation in CDPK1 (upper panel). The score of the predicted palmitoylation sites was provided by the CSS-Palm software. Lower panels indicate schematic of constructs expressing HA-tagged CDPK1, each with a cysteine to alanine replacement in single residue (C3A and C252A). These constructs were episomally expressed in the schizonts. (E) IFA of CDPK1::HA and two mutant proteins episomally expressed in schizonts. Scale bar = 5 μm. Representative of three independent repeats. (F) Palmitoylation analysis of CDPK1::HA and two mutant proteins episomally expressed in schizonts. Representative of two independent repeats. HA: hemagglutinin;WT: wild type.
-
Figure 7—source data 1
(related to Figure 7C) Palmitoylation analysis of GAP45::HA and seven mutant proteins episomally expressed in schizonts.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig7-data1-v3.xls
-
Figure 7—source data 2
(related to Figure 7F) Palmitoylation analysis of CDPK1::HA and two mutant proteins episomally expressed in schizonts.
- https://cdn.elifesciences.org/articles/77447/elife-77447-fig7-data2-v3.xls
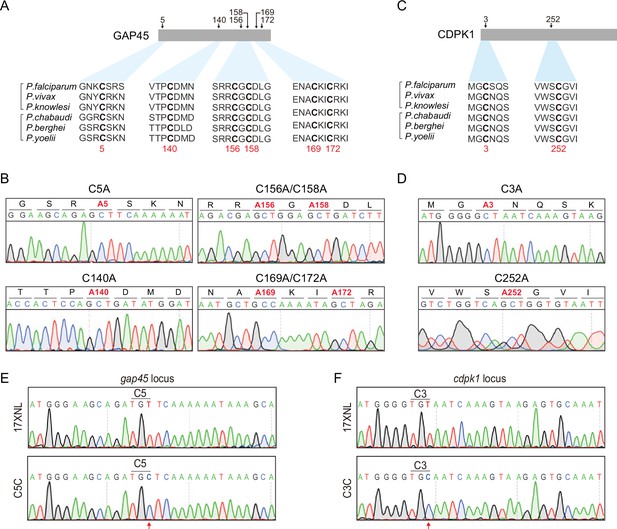
Potential cysteine(s) in GAP45 and CDPK1 for palmitoylation and protein mutants with cysteine replacement to validate the essentiality.
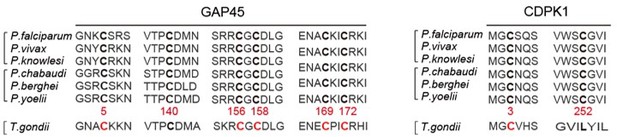
(A) Amino acid sequence of GAP45 from P. falciparum (PF3D7_1222700), P. vivax (PVX_123765), P. knowlesi (PKNH_1441800), P. chabaudi (PCHAS_1439600), P. berghei (PBANKA_1437600), and P. yoelii (PY17X_1440100) were aligned. Six conserved cysteine (highlighted in bold) predicated for palmitoylation was shown. (B) DNA sequencing confirmation of the constructs expressing hemagglutinin (HA)-tagged GAP45, each with a cysteine to alanine replacement in single or double residues (C5A, C140A, C156A/C158A, and C169A/C172A). These constructs were episomally expressed in the schizonts. (C) Amino acid sequence of CDPK1 from P. falciparum (PF3D7_0217500), P. vivax (PVX_002665), P. knowlesi (PKNH_0403400), P. chabaudi (PCHAS_0316300), P. berghei (PBANKA_0314200), and P. yoelii (PY17X_0314700) were aligned. Two conserved cysteine (highlighted in bold) predicated for palmitoylation was shown. (D) DNA sequencing confirmation of the constructs expressing HA-tagged CDPK1, each with a cysteine to alanine replacement (C3A and C252A). These constructs were episomally expressed in the schizonts. (E) DNA sequencing confirmation of nucleotide replacement in the gap45 locus of the GAP45 C5C parasite clone. Top panel shows the nucleotide sequence from the parasite 17XNL strain; the bottom panel shows the replaced nucleotide (silent mutation) in the clone GAP45 C5C. (F) DNA sequencing confirmation of nucleotide replacement in the gcdpk1 locus of CDPK1 C3C parasite clone. Top panel shows the nucleotide sequence from the parasite 17XNL strain; the bottom panel shows the replaced nucleotide (silent mutation) in the clone CDPK1 C3C.
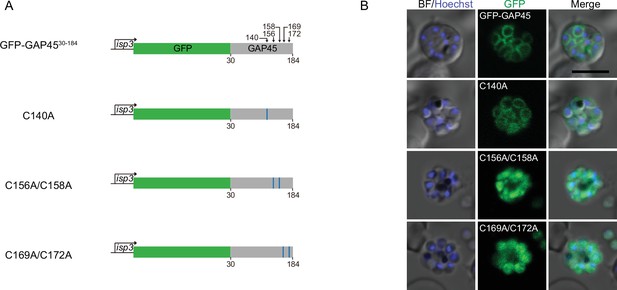
Potential cysteine(s) in GAP45 and CDPK1 for palmitoylation and protein mutants with cysteine replacement to validate the essentiality.
(A) Schematic of constructs expressing GFP-tagged C-terminal fragment (30–184 aa) of GAP45 and three derived mutants (C140A, C156A/C158A, and C169A/C172A). These constructs were driven by the promoter of gene isp3 and episomally expressed in the schizonts. (B) Fluorescence of GFP-tagged GAP45 fragment and mutants in (A). Scale bar = 5 μm.
Additional files
-
Supplementary file 1
List of IMC protein candidates identified in this study.
- https://cdn.elifesciences.org/articles/77447/elife-77447-supp1-v3.xlsx
-
Supplementary file 2
List of 188 filtered-out proteins by transcriptional profile.
- https://cdn.elifesciences.org/articles/77447/elife-77447-supp2-v3.xlsx
-
Supplementary file 3
Primers and oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/77447/elife-77447-supp3-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/77447/elife-77447-transrepform1-v3.docx