HIF-1α induces glycolytic reprograming in tissue-resident alveolar macrophages to promote cell survival during acute lung injury
Figures
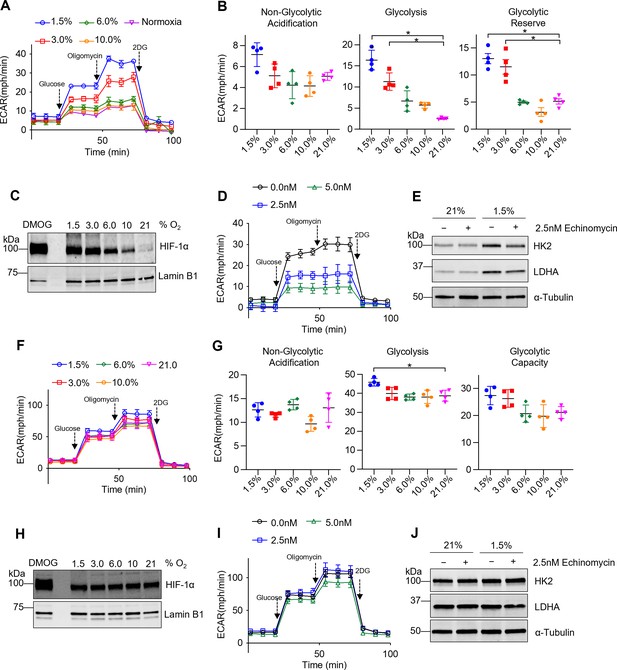
Tissue-resident alveolar macrophages (TR-AMs) exhibit hypoxia-inducible factor 1-alpha (HIF-1α) stabilization and develop a glycolytic phenotype in response to hypoxia, while bone marrow-derived macrophages (BMDMs) have limited metabolic adaptation to hypoxia.
TR-AMs (A–E) and BMDMs (F–J) were incubated overnight (16 hr) at varying O2 concentrations. (A) Using Seahorse XF24 analyzer, glycolysis was measured as extracellular acidification rate (ECAR). TR-AMs were sequentially treated with glucose, oligomycin (ATP synthase inhibitor), and 2-deoxyglucose (2-DG) (inhibitor of hexokinase 2, or glycolysis). (B) Interleaved scatter plots quantifying glycolytic parameters. Data represent at least three independent experiments (n = 4 separate wells per group). Glycolytic parameters were compared against 21% O2 and significance was determined by one-way ANOVA with Bonferroni correction. (C) Western blot analysis of nuclear extract to assess HIF-1α expression in TR-AMs treated with different concentrations of O2. DMOG served as a positive control. (D) Glycolysis stress test of TR-AMs under 1.5% O2 in combination with echinomycin (16 hr). (E) Western blot analysis of whole-cell lysates of TR-AMs treated with 21 or 1.5% O2 in combination with echinomycin (16 hr). (F) BMDM glycolysis measurements (ECAR) using Seahorse XF24 analyzer. (G) Interleaved scatter plots quantifying glycolytic parameters. Data represent at least three independent experiments (n = 4 separate wells per group). Glycolytic parameters were compared against 21% O2 and significance was determined by one-way ANOVA with Bonferroni correction. (H) Western blot analysis of nuclear extract to assess HIF-1α expression in BMDMs treated with different concentrations of O2. (I) Glycolysis stress test of BMDMs under 1.5% O2 in combination with echinomycin (16 hr). (J) Western blot analysis of whole-cell lysates of BMDMs treated with 21 or 1.5% O2 in combination with echinomycin (16 hr). All error bars denote mean ± SD. *p<0.05.
-
Figure 1—source data 1
The effect of different O2 concentrations on hypoxia-inducible factor 1-alpha HIF-1α expression in tissue-resident alveolar macrophages (TR-AMs).
Uncropped Western blot images of HIF-1α protein expression in TR-AMs under different concentrations of oxygen.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig1-data1-v2.zip
-
Figure 1—source data 2
The effect of echinomycin on glycolytic enzyme protein expression in tissue-resident alveolar macrophages (TR-AMs).
Uncropped Western blot images of HK2, LDHA, and α-tubulin in TR-AMs treated with echinomycin under normoxia or hypoxia.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig1-data2-v2.zip
-
Figure 1—source data 3
The effect of different O2 concentrations on hypoxia-inducible factor 1-alpha (HIF-1α) expression in bone marrow-derived macrophages (BMDMs).
Uncropped Western blot images of HIF-1α protein expression in BMDMs under different concentrations of oxygen.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig1-data3-v2.zip
-
Figure 1—source data 4
The effect of echinomycin on glycolytic enzyme protein expression in bone marrow-derived macrophages (BMDMs).
Uncropped Western blot images of HK2, LDHA, and α-tubulin in BMDMs treated with echinomycin under normoxia or hypoxia.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig1-data4-v2.zip
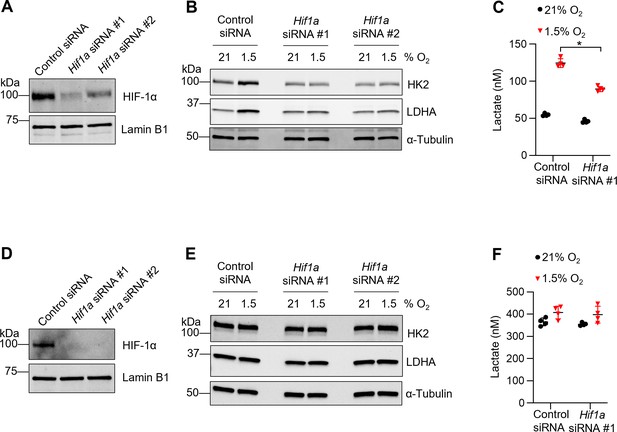
Knockdown of Hif1a diminishes hypoxia-induced glycolytic phenotype in tissue-resident alveolar macrophages (TR-AMs).
TR-AMs (A–C) and bone marrow-derived macrophages (BMDMs) (D–F) were transfected with Hif1a or control siRNA and subsequently incubated overnight (16 hr) at 21 or 1.5% O2.
Western blot analysis of nuclear extracts to assess successful Hif1a knockdown in (A) TR-AMs and (D) BMDMs under 1.5% O2. Western blot analysis of whole-cell extracts from (B) TR-AMs and (E) BMDMs. Extracellular lactate levels in (C) TR-AMs and (F) BMDMs incubated overnight (16 hr) at 21 or 1.5% O2. Significance was determined by two-way ANOVA with Bonferroni correction. All error bars denote mean ± SD. *p<0.05.
-
Figure 1—figure supplement 1—source data 1
Validation of Hif1a siRNA knockdown in tissue-resident alveolar macrophages (TR-AMs).
Uncropped Western blot images of hypoxia-inducible factor 1-alpha (HIF-1α) protein expression in TR-AMs treated with either control siRNA or two different Hif1a siRNAs.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig1-figsupp1-data1-v2.zip
-
Figure 1—figure supplement 1—source data 2
The effect of Hif1a siRNA knockdown on glycolytic enzyme protein expression in tissue-resident alveolar macrophages (TR-AMs) under normoxia and hypoxia.
Uncropped Western blot images of HK2, LDHA, and α-tubulin expression in TR-AMs treated with either control siRNA or two different Hif1a siRNAs under normoxia or hypoxia.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig1-figsupp1-data2-v2.zip
-
Figure 1—figure supplement 1—source data 3
Validation of Hif1a siRNA knockdown in bone marrow-derived macrophages (BMDMs).
Uncropped Western blot images of hypoxia-inducible factor 1-alpha (HIF-1α) protein expression in BMDMs treated with either control siRNA or two different Hif1a siRNAs.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig1-figsupp1-data3-v2.zip
-
Figure 1—figure supplement 1—source data 4
The effect of Hif1a siRNA knockdown on glycolytic enzyme protein expression in bone marrow-derived macrophages (BMDMs) under normoxia and hypoxia.
Uncropped Western blot images of HK2, LDHA, and α-tubulin expression in BMDMs treated with either control siRNA or two different Hif1a siRNAs under normoxia or hypoxia.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig1-figsupp1-data4-v2.zip
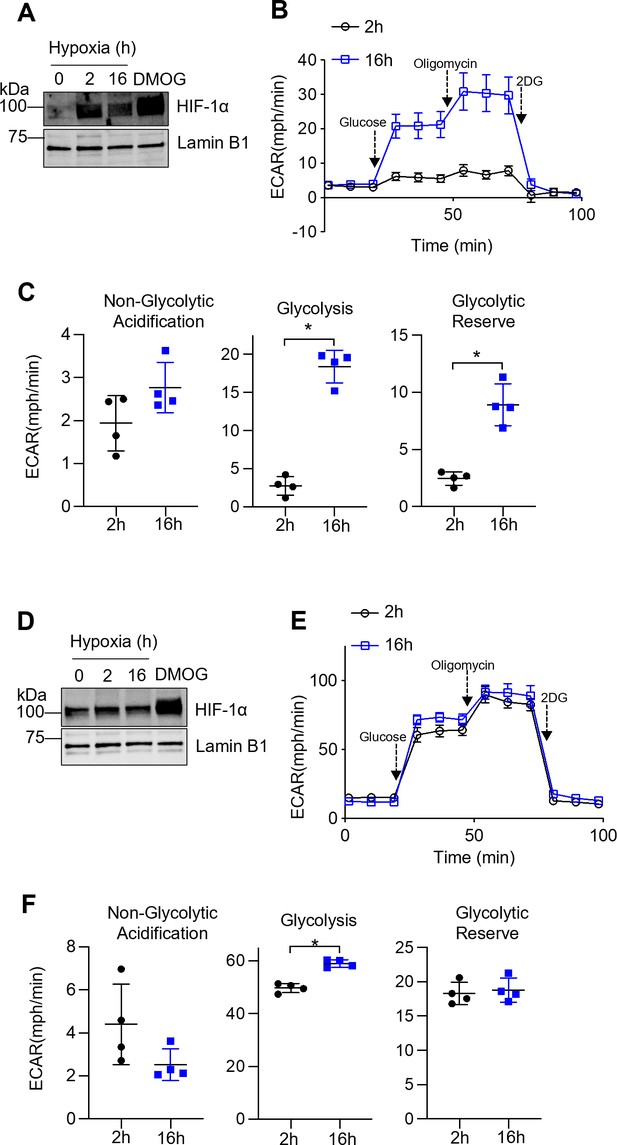
Prolonged but not short-term hypoxia induces glycolysis in tissue-resident alveolar macrophages (TR-AMs).
TR-AMs (A–C) or bone marrow-derived macrophages (BMDMs) (D–F) were incubated for 2 hr or overnight (16 hr) at 1.5% O2. (A) Western blot analysis of TR-AM (A) and BMDM (D) nuclear extracts to assess hypoxia-inducible factor 1-alpha (HIF-1α) protein expression. Using Seahorse XF24 technology, TR-AM (B) and BMDM (E) glycolysis was measured as extracellular acidification rate (ECAR). Interleaved scatter plots quantifying glycolytic parameters in TR-AMs (C) and BMDMs (F). Data represents at least three independent experiments (n = 4 separate wells per group). Significance was determined by unpaired, two-tailed t-test. *p<0.05.
-
Figure 1—figure supplement 2—source data 1
Expression of hypoxia-inducible factor 1-alpha (HIF-1α) protein in tissue-resident alveolar macrophages (TR-AMs) at different time points following exposure to hypoxia.
Uncropped Western blot images of HIF-1α protein expression in TR-AMs treated with hypoxia for 0, 2, or 16 hr or with DMOG.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig1-figsupp2-data1-v2.zip
-
Figure 1—figure supplement 2—source data 2
Expression of hypoxia-inducible factor 1-alpha (HIF-1α) protein in bone marrow-derived macrophages (BMDMs) at different time points following exposure to hypoxia.
Uncropped Western blot images of HIF-1α protein expression in BMDMs treated with hypoxia for 0, 2, or 16 hr or with DMOG.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig1-figsupp2-data2-v2.zip
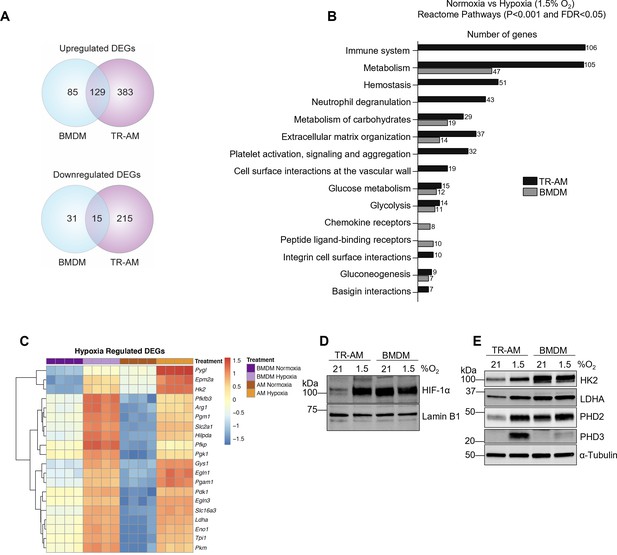
The hypoxia-induced transcriptomic response differs substantially between tissue-resident alveolar macrophages (TR-AMs) and bone marrow-derived macrophages (BMDMs).
TR-AMs and BMDMs were incubated overnight (16 hr) under normoxia (21.0% O2) or hypoxia (1.5% O2). (A) Venn diagrams show differentially expressed genes (DEGs) altered by hypoxia in TR-AMs (741 total DEGs), and BMDMs (260 total DEGs). DEGs were identified using DESeq2 at FC > 2 and false discovery rate (FDR)-adjusted p-value of <0.05. (B) Reactome pathway enrichment comparing number of genes in a given pathway altered by hypoxia in TR-AMs and BMDMs. (C) Heatmap representing the top 20 significant metabolic genes altered by hypoxia in both TR-AMs and BMDMs. (D) Western blot analysis of nuclear extracts to assess hypoxia-inducible factor 1-alpha (HIF-1α) protein expression. (E) Western blot analysis of whole cell extracts to assess glycolytic enzyme (HK2, LDH) and prolyl hydroxylase (PHD2, PHD3) protein expression.
-
Figure 2—source data 1
Read count data for hypoxia-regulated genes in tissue-resident alveolar macrophages (TR-AMs) and bone marrow-derived macrophages (BMDMs).
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Differences in hypoxia-inducible factor 1-alpha (HIF-1α) expression between tissue-resident alveolar macrophages (TR-AMs) and bone marrow-derived macrophages (BMDMs) under normoxia and hypoxia.
Uncropped Western blot images of HIF-1α expression in TR-AMs and BMDMs treated with normoxia or hypoxia.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig2-data2-v2.zip
-
Figure 2—source data 3
Differences in glycolytic enzyme and prolyl hydroxylase protein expression between tissue-resident alveolar macrophages (TR-AMs) and bone marrow-derived macrophages (BMDMs) under normoxia and hypoxia.
Uncropped Western blot images of HK2, LDHA, PHD2, PHD3, and α-tubulin expression in TR-AMs and BMDMs treated with normoxia or hypoxia.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig2-data3-v2.zip
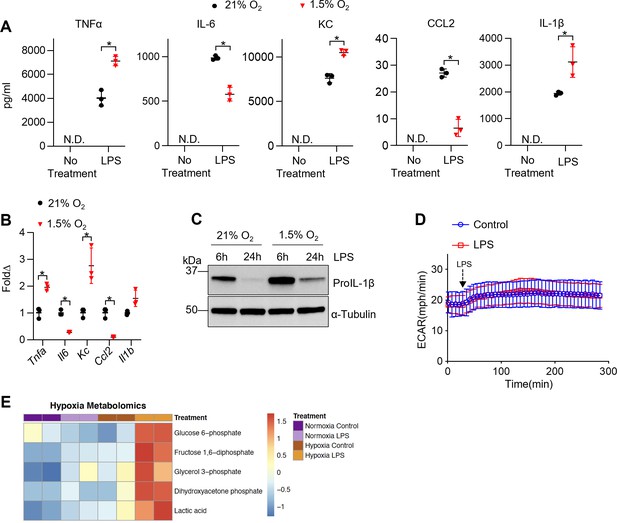
Hypoxia modulates tissue-resident alveolar macrophage (TR-AM) cytokine production and metabolic response to lipopolysaccharide (LPS).
TR-AMs were incubated overnight (16 hr) under 21 or 1.5% O2, then stimulated with 20 ng/ml LPS for 6 hr while maintaining pretreatment conditions. For IL-1β measurements, 5 mM ATP was added to TR-AMs for 30 min following 6 hr LPS treatment to activate caspase 1, ensuring IL-1β release. (A) We measured cytokine (TNFα, IL-6, KC, CCL2, and IL-1β) levels in media using ELISA. Data represent at least three independent experiments; n = 3 per group. Significance was determined by unpaired, two-tailed t-test. (B) qPCR was used to measure mRNA expression (Tnfa, Il6, Kc, Ccl2, and Il1b). Gene expression was normalized to corresponding gene ct values in 21% group and represented as fold change using the ∆∆ct method. Data represent at least three independent experiments; n = 3 per group. Significance was determined by unpaired, two-tailed t-test. (C) Western blot analysis of whole-cell extracts at 6 and 24 hr post LPS treatment. (D) Extracellular acidification rate (ECAR) was measured in following acute LPS injection (final concentration: 20 ng/ml) in TR-AMs conditioned in 1.5% O2. (E) Capillary electrophoresis-mass spectrometry (CE-MS) metabolite heatmap for glycolytic intermediates. All error bars denote mean ± SD. *p<0.05.
-
Figure 3—source data 1
Changes in lipopolysaccharide (LPS)-induced expression of proIL-1β protein in tissue-resident alveolar macrophages (TR-AMs) under normoxia and hypoxia.
Uncropped Western blot images of proIL-1β protein in TR-AMs treated with LPS for 6 or 24 hr under normoxia or hypoxia.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig3-data1-v2.zip
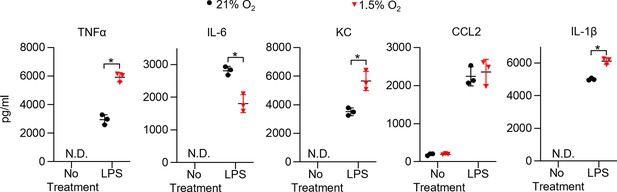
Hypoxia alters cytokine production in bone marrow-derived macrophages (BMDMs).
BMDMs were incubated overnight (16 hr) under normoxia or 1.5% O2, then stimulated with lipopolysaccharide (LPS) (20 ng/ml) for 6 hr while maintaining pretreatment conditions. For IL-1β, 5 mM ATP was added to BMDMs for 30 min following 6 hr of LPS treatment to activate caspase 1, ensuring IL-1β release. (A) Sandwich ELISA was used to measure secreted cytokine (TNFα, IL-6, KC, CCL2, and IL-1β). Data represent at least three independent experiments; n = 3 per group. Significance was determined by unpaired, two-tailed t-test. All error bars denote mean ± SD. *p<0.05.
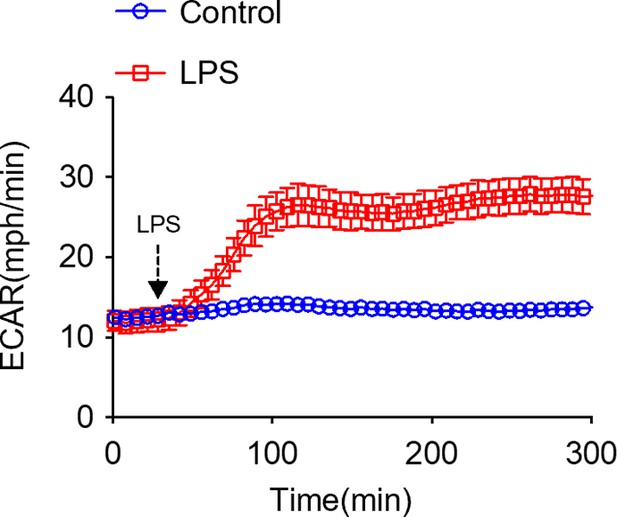
Lipopolysaccharide (LPS) induces an immediate increase in glycolysis in bone marrow-derived macrophages (BMDMs).
Extracellular acidification rate (ECAR) was measured in normoxic BMDMs following acute LPS injection (final concentration: 20 ng/ml).
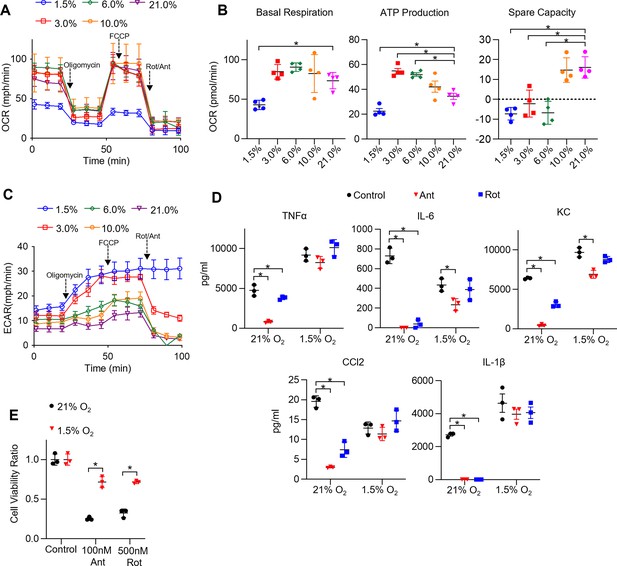
Hypoxia rescues ETC inhibitor-induced cell death and impaired cytokine production in tissue-resident alveolar macrophages (TR-AMs).
(A) Mitochondrial stress test to measure oxygen consumption rate (OCR) using Seahorse XF24 in TR-AMs, which were treated sequentially with oligomycin (ATP synthase inhibitor), FCCP (uncoupler), and rotenone (Rot)/antimycin A (Ant) (complex I and III inhibitors, respectively). (B) Interleaved scatter plots quantifying mitochondrial respiration parameters. Data represents at least three experiments (n = 4 separate wells per group). Mitochondrial parameters were compared against 21% O2 and significance was determined by one-way ANOVA with Bonferroni correction. (C) Extracellular acidification rate (ECAR) measurement during mitochondrial stress test to visualize TR-AMs’ ability to upregulate glycolysis in response to mitochondrial inhibition. (D) TR-AMs were incubated overnight (16 hr) under 21 or 1.5% O2, then stimulated with 20 ng/ml lipopolysaccharide (LPS) in the presence or absence of mitochondrial inhibitors (20 nM Ant or Rot) for 6 hr while maintaining pretreatment conditions. ELISA was used to measure secreted cytokine (TNFα, IL-6, KC, CCL2, and IL-1β) levels in media. ATP added to cells prior to collection for IL-1β assessment. Data represent at least three independent experiments; n = 3 per group. Significance was determined by one-way ANOVA with Bonferroni correction. (E) TR-AMs were cultured under 21 or 1.5% O2 for 6 hr, then treated with mitochondrial inhibitors (100 nM Ant or 500 nM Rot) overnight and a sulforhodamine B assay was performed to measure cytotoxicity. Graphs represent cell viability compared to control, 21% O2 group. Data represent at least three independent experiments (n = 3 per group). Significance was determined by two-way ANOVA with Bonferroni correction. All error bars denote mean ± SD. *p<0.05.
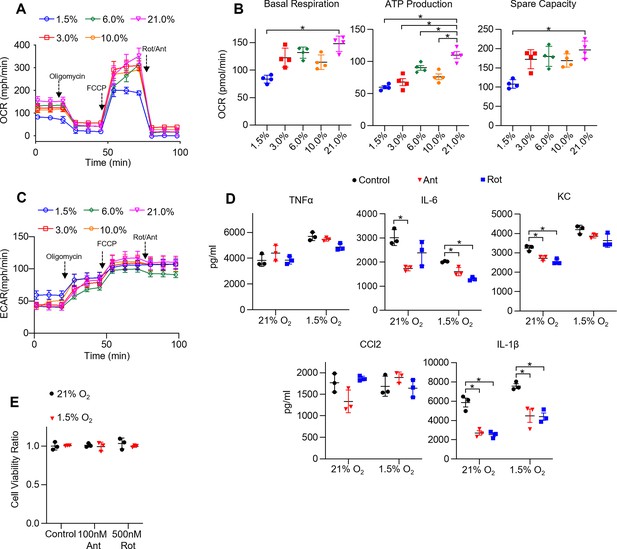
The effect of hypoxia on bone marrow-derived macrophage (BMDM) mitochondrial function, cytokine production, and cell viability under ETC inhibition.
(A) Mitochondrial stress test to measure oxygen consumption rate (OCR) using Seahorse XF24 in BMDMs. (B) Interleaved scatter plots quantifying mitochondrial respiration parameters. Data represents at least three experiments (n = 4 separate wells per group). Mitochondrial parameters were compared against 21% O2 and significance was determined by one-way ANOVA with Bonferroni correction. (C) Extracellular acidification rate (ECAR) measurement during mitochondrial stress test. (D) BMDMs were incubated overnight (16 hr) under 21 or 1.5% O2, then stimulated with 20 ng/ml lipopolysaccharide (LPS) in the presence or absence of mitochondrial inhibitors (20 nM antimycin A [Ant] or rotenone [Rot]) for 6 hr while maintaining pretreatment conditions. ELISA was used to measure secreted cytokine (TNFα, IL-6, KC, CCL2, and IL-1β) levels in media. ATP added to cells prior to collection for IL-1β assessment. Data represent at least three independent experiments; n = 3 per group. Significance was determined by one-way ANOVA with Bonferroni correction. (E) BMDMs were cultured under 21 or 1.5% O2 for 6 hr, then treated with mitochondrial inhibitors (100 nM Ant or 500 nM Rot) overnight and a sulforhodamine B assay was performed to measure cytotoxicity. Graphs represent cell viability compared to control, 21% O2 group. Data represent at least three independent experiments (n = 3 per group). Significance was determined by two-way ANOVA with Bonferroni correction. All error bars denote mean ± SD. *p<0.05.
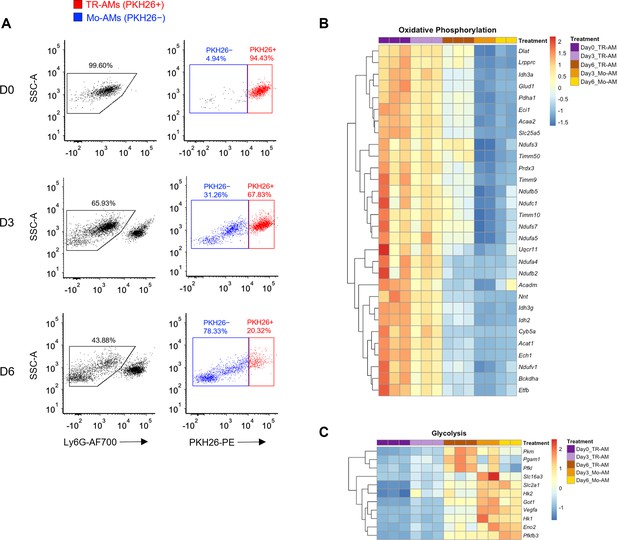
Tissue-resident alveolar macrophage (TR-AM) survival correlates with a shift to glycolytic metabolism during influenza-induced acute lung injury.
(A) FACS plots of bronchoalveolar lavage fluid (BALF) samples collected from C57BL/6 mice infected with PR8 (100 PFU) at baseline (D0), 3 days (D3), and 6 days (D6) post infection. First, debris, red blood cells, and lymphocytes were eliminated based on size (forward scatter signal [FSC]) and granularity (side scatter signal [SSC]). Samples were first gated on single cells based on the SSC/FSC, and then live cells were selected (SYTOX Green−). Ly6G− used to exclude neutrophils. TR-AMs were identified as being PKH26+, and nonresident/infiltrating monocyte-derived alveolar macrophages (Mo-AMs) were PKH26−. Gene expression heatmaps representing (B) oxidative phosphorylation and (C) glycolytic gene expression. Heatmaps were generated through differentially expressed gene (DEG) analysis of UniProt oxidative phosphorylation and glycolysis gene sets for FAC TR-AMs (PKH+; n = 3/group) and Mo-AMs (PKH26−; n = 2/group) over the infection time course.
-
Figure 5—source data 1
Read counts data for genes of oxidative phosphorylation in tissue-resident alveolar macrophages (TR-AMs) and monocyte-derived alveolar macrophages (Mo-AMs).
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig5-data1-v2.csv
-
Figure 5—source data 2
Read counts data for genes of glycolysis in tissue-resident alveolar macrophages (TR-AMs) and monocyte-derived alveolar macrophages (Mo-AMs).
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig5-data2-v2.csv
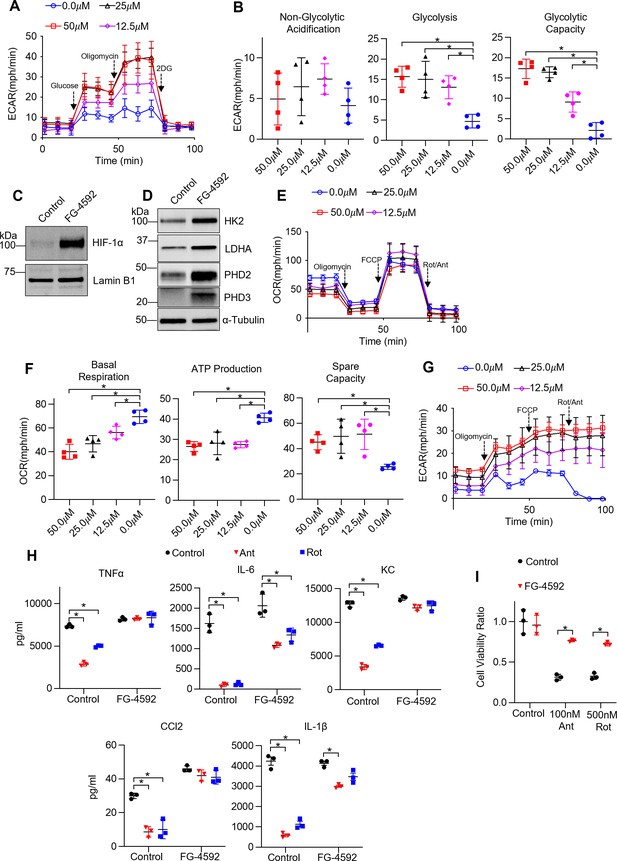
Non-hypoxic stabilization of hypoxia-inducible factor 1-alpha (HIF-1α) induces glycolysis and rescues ETC inhibitor-induced reduction in cytokine production and cell death in tissue-resident alveolar macrophages (TR-AMs).
TR-AMs were treated (16 hr) overnight ±FG-4592 (25.0 μM when not stated otherwise). (A) Glycolysis was measured as extracellular acidification rate (ECAR). (B) Quantification of glycolytic parameters. Data represent at least three independent experiments (n = 4 separate wells per group). Glycolytic parameters compared to control group (0.0 μM) and significance was determined by one-way ANOVA with Bonferroni correction. (C) Western blot analysis of nuclear extract for HIF1α expression and (D) whole cell lysate for glycolytic enzyme and prolyl hydroxylase expression. (E) Mitochondrial stress test to measure oxygen consumption rate (OCR). (F) Quantification of mitochondrial respiration parameters. Data represents at least three experiments (n = 4 separate wells per group). Mitochondrial parameters were compared to control group (0.0 μM) and significance was determined by one-way ANOVA with Bonferroni correction. (G) ECAR measurement during mitochondrial stress test. (H) TR-AMs were pretreated overnight (16 hr) with 0.0 μM (no treatment) or 25.0 μM FG-4592, then stimulated with 20 ng/ml lipopolysaccharide (LPS) in the presence or absence of mitochondrial inhibitors (20 nM antimycin A [Ant] or rotenone [Rot]) for 6 hr while maintaining pretreatment conditions. Sandwich ELISA was used to measure secreted cytokine (TNFα, IL-6, KC, and CCL-2). Data represents at least three independent experiments; n = 3 per group. (I) TR-AMs were treated with FG-4592 for 6 hr, then treated with mitochondrial inhibitors (100 nM Ant or 500 nM Rot) overnight and a sulforhodamine B assay was performed to measure cytotoxicity. Bar graphs represent cytotoxicity compared to control, 0.0 µM group. Data represents at least three independent experiments (n = 3 per group). Significance was determined by two-way ANOVA with Bonferroni correction. All error bars denote mean ± SD. *p<0.05.
-
Figure 6—source data 1
The effect of FG-4592 on hypoxia-inducible factor 1-alpha (HIF-1α) expression in tissue-resident alveolar macrophages (TR-AMs).
Uncropped Western blot images of HIF-1α in TR-AMs treated with FG-4592 or control vehicle.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig6-data1-v2.zip
-
Figure 6—source data 2
The effect of FG-4592 on glycolytic enzyme and prolyl hydroxylase protein expression in tissue-resident alveolar macrophages (TR-AMs).
Uncropped Western blot images of HK2, LDHA, PHD2, PHD3, and α-tubulin in TR-AMs treated with FG-4592 or control vehicle.
- https://cdn.elifesciences.org/articles/77457/elife-77457-fig6-data2-v2.zip
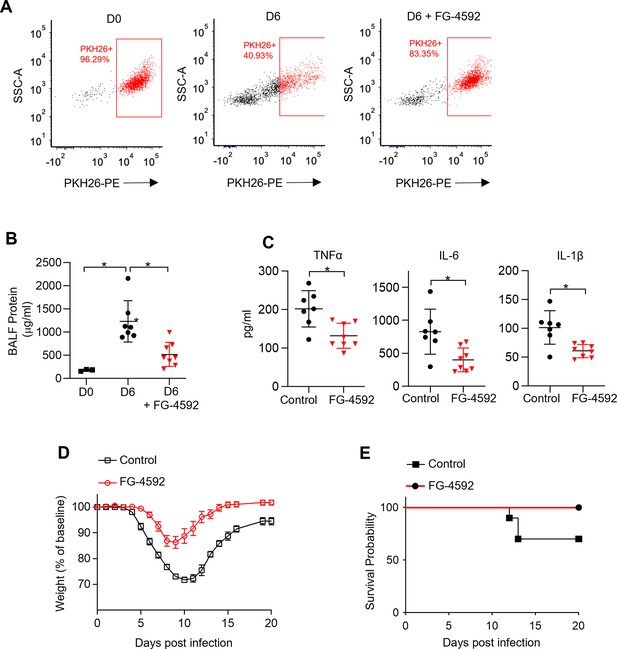
Non-hypoxic stabilization of hypoxia-inducible factor 1-alpha (HIF-1α) increases tissue-resident alveolar macrophage (TR-AM) survival and improves outcomes in influenza-induced acute lung injury.
We intratracheally infected C57BL/6 mice with PR8 (100 PFU) and collected bronchoalveolar lavage fluid (BALF) on day 0 (D0) (uninfected) and day 6 (D6) post infection. Mice also received either the HIF-1α stabilizer (FG-4592) or vehicle control on D0. (A) Representative FACS plot of BALF macrophages. (B) BALF protein concentration. (C) BALF proinflammatory cytokine levels at D6. BALF data generated from two separate experiments (n = 7 mice/control group and n = 8 mice/FG-4592 group). BALF data significance was determined by unpaired, two-tailed t-test. (D ,E) C57BL/6 mice infected with PR8 (200 PFU) (10 mice/group). (D) Weight loss represented as percentage and normalized to D0. (E) Survival curve. All error bars denote mean ± SD. *p<0.05.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL/6J | Jackson Laboratory | Stock no. 000664 | 6–8 weeks |
| Strain, strain background (influenza A virus) | A/PR8/34 (H1N1) | BEI Resources, NIAID, NIH | NR-348 | |
| Antibody | Anti-HK2 (rabbit monoclonal) | Cell Signaling Technology | Cat# C64G5 | WB (1:1000) |
| Antibody | Anti-LDHA (rabbit polyclonal) | Cell Signaling Technology | Cat# 2012S | WB (1:1000) |
| Antibody | Anti-PHD2/EGLN1 (rabbit monoclonal) | Cell Signaling Technology | Cat# 4835 | WB (1:1000) |
| Antibody | Anti- PHD3/EGLN3 (rabbit polyclonal) | Novus Biologicals | Cat# NB100-303 | WB (1:1000) |
| Antibody | Anti-IL-1β (mouse monoclonal) | Cell Signaling Technology | Cat# 12242 | WB (1:1000) |
| Antibody | Anti-Lamin B1 (rabbit polyclonal) | ProteinTech | Cat# 12987-1-AP | WB (1:1000) |
| Antibody | Anti-HIF-1α (rabbit polyclonal) | Cayman Chemical | Cat# 10006421 | WB (1:500) |
| Antibody | Anti-α-Tubulin (mouse monoclonal) | Sigma | Cat# T6074 | WB (1:20,000) |
| Antibody | Anti-rabbit IgG, HRP-linked Antibody (goat polyclonal) | Cell Signaling Technology | Cat# 7074 | WB (1:2500) |
| Antibody | Anti-mouse IgG, HRP-linked Antibody (horse polyclonal) | Cell Signaling Technology | Cat# 7076 | WB (1:2500) |
| Antibody | CD16/CD32 (FcBlock) (rat monoclonal) | BD Biosciences | Clone 2.4G2; Cat# 553141 | Flow cytometry (1:50) |
| Antibody | Alexa Fluor 700 anti-mouse Ly-6G (rat monoclonal) | BioLegend | Clone 1A8; Cat# 553141 | Flow cytometry (1:250) |
| Chemical compound, drug | FG-4592 (roxadustat) | Cayman Chemical | Cat# 15294 | |
| Chemical compound, drug | Recombinant mouse M-CSF | BioLegend | 576406 | |
| Chemical compound, drug | Oligomycin | Fisher Scientific | 49-545-510MG | |
| Chemical compound, drug | FCCP | MilliporeSigma | C2920 | |
| Chemical compound, drug | Antimycin A | MilliporeSigma | A8674 | |
| Chemical compound, drug | Rotenone | MilliporeSigma | R8875 | |
| Chemical compound, drug | Lipopolysaccharide | Santa Cruz | sc-3535 | |
| Commercial assay or kit | Mouse IL-6 DuoSet ELISA | R&D Systems | DY406 | |
| Commercial assay or kit | Mouse TNF-α DuoSet ELISA | R&D Systems | DY410 | |
| Commercial assay or kit | Mouse KC DuoSet ELISA | R&D Systems | DY453 | |
| Commercial assay or kit | Mouse CCL2 DuoSet ELISA | R&D Systems | DY479 | |
| Commercial assay or kit | Mouse IL-1β alpha DuoSet ELISA | R&D Systems | DY401 | |
| Commercial assay or kit | Lactate Assay Kit | MilliporeSigma | MAK064-1KT | |
| Commercial assay or kit | Mouse Macrophage Nucleofector Kit | Lonza | VPA-1009 | |
| Commercial assay or kit | Seahorse XFe24 FluxPak | Agilent | 102340-100 | |
| Commercial assay or kit | NE-PER Nuclear and Cytoplasmic Extraction Reagents | Thermo Fisher | Cat# 78833 | |
| Other | PKH26 Cell Linker Dye for Phagocytic Cell Labeling | MilliporeSigma | Cat# PKH26PCL-1KT | Dye to distinguish between TR-AMs and Mo-AMs |
| Other | SYTOX Green Nucleic Acid Stain | Thermo Fisher | Cat# S7020 | Stain to distinguish between live and dead cells. |
| Sequence-based reagent | Rpl19_F | This paper | PCR primers | CCGACGAAAGGGTATGCTCA |
| Sequence-based reagent | Rpl19_R | This paper | PCR primers | GACCTTCTTTTTCCCGCAGC |
| Sequence-based reagent | Il6_F | This paper | PCR primers | TTCCATCCAGTT GCCTTCTTGG |
| Sequence-based reagent | Il6_R | This paper | PCR primers | TTCCTATTTCCA CGATTTCCCAG |
| Sequence-based reagent | Tnfa_F | This paper | PCR primers | AGGGGATTAT GGCTCAGGGT |
| Sequence-based reagent | Tnfa_R | This paper | PCR primers | CCACAGTCCAGGTCACTGTC |
| Sequence-based reagent | Il1b_F | This paper | PCR primers | GCCACCTTTT GACAGTGATGAG |
| Sequence-based reagent | Il1b_R | This paper | PCR primers | GACAGCCCA GGTCAAAGGTT |
| Sequence-based reagent | Kc_F | This paper | PCR primers | AGACCATGGC TGGGATTCAC |
| Sequence-based reagent | Kc_R | This paper | PCR primers | ATGGTGGCTATGACTTCGGT |
| Sequence-based reagent | Ccl2_F | This paper | PCR primers | CTGTAGTTTTT GTCACCAAGCTCA |
| Sequence-based reagent | Ccl2_R | This paper | PCR primers | GTGCTGAAGA CCTTAGCCCA |
| Sequence-based reagent | Non-targeting (control) siRNA | Dharmacon | D-001810-01 | |
| Sequence-based reagent | Hif1a #1; J-040638-06 | Dharmacon | J-040638-06 | |
| Sequence-based reagent | Hif1a #2; J-040638-07 | Dharmacon | J-040638-07 | |
| Software, algorithm | FastQC | Babraham Institute | RRID:SCR_014583 | |
| Software, algorithm | STAR | PMID:23104886 | RRID:SCR_015899 | |
| Software, algorithm | DESeq2 | Bioconductor | RRID:SCR_015687 | |
| Software, algorithm | Reactome Cytoscape Plugin | PMID:14597658 | RRID:SCR_003032 | |
| Software, algorithm | Prism 9 | GraphPad | RRID:SCR_002798 |





