Evidence for RNA or protein transport from somatic tissues to the male reproductive tract in mouse
Figures
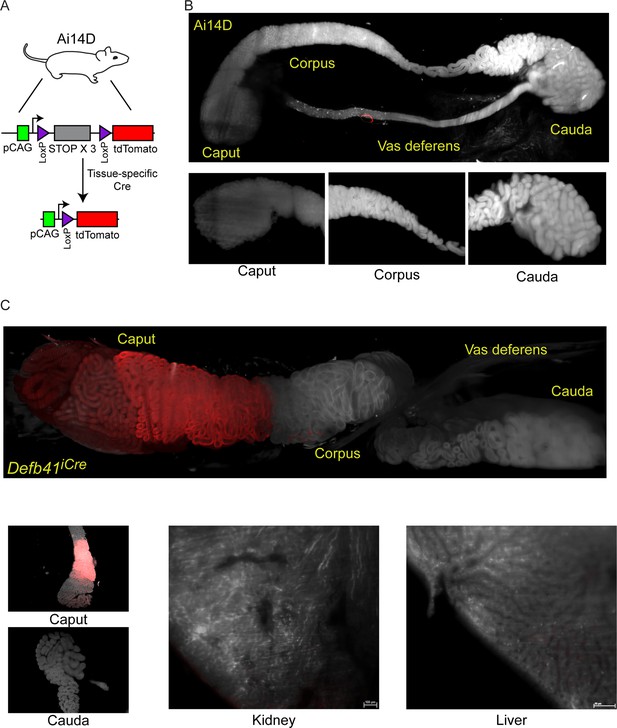
Analysis of Cre-dependent reporter gene expression in the murine epididymis.
(A) Schematic of the Cre-dependent reporter present in the Ai14D strain. Reporter cassette, inserted at the safe harbor Rosa26 locus, carries a strong pCAG promoter followed by a LoxP-flanked STOP cassette with three repeated in frame sequences. In the absence of Cre activity, transcription through this construct leads to transcriptional termination and no reporter activity. Following Cre-dependent excision of the LoxP cassette, the pCAG promoter is juxtaposed adjacent to a sequence encoding a tandem Tomato (tdTomato) fluorescent protein, resulting in robust expression of Tomato in tissues expressing Cre. (B) Lightsheet imaging of the mouse epididymis in the negative control Ai14D background. Following animal sacrifice, tissues were cleared for imaging according to a modified CLARITY protocol (Materials and methods). The entire epididymis, along with a large section of the vas deferens, was then imaged using the LaVision Biotec lightsheet microscope. Regions corresponding to the proximal (caput), middle (corpus), and distal (cauda) epididymis, and the vas deferens, are indicated on the image of the entire tissue sample. Insets show representative 2D slices of the indicated anatomical regions. (C) Positive control showing robust Tomato expression driven by the caput-specific Defb41 Cre driver (Björkgren et al., 2012). Panels show whole tissue image and representative sections as described in panel (B), along with representative sections of the kidney and liver, as indicated. See also Figure 1—figure supplement 1.

Histology of Ai14D; Defb41Cre double transgenic.
DAPI-stained histological section of the caput and corpus epididymis from an Ai14D; Defb41Cre animal, showing expected tdTomato expression specifically in the principal cells of caput epididymis. Top panel shows the entire caput and corpus, bottom panel shows a zoom-in from the distal caput.
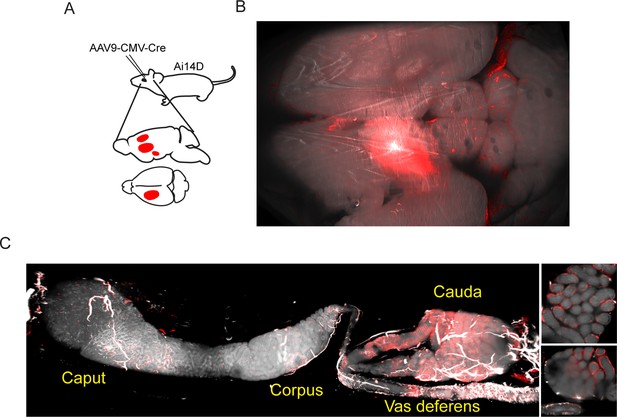
Adeno-associated viral (AAV)-mediated Cre expression in the brain drives reporter expression in the epididymis.
(A) Experimental schematic showing stereotactically guided injection of AAV into the brain of Ai14D reporter animals to drive localized Cre-Lox recombination in the brain. Fifteen or 25 days post injection (depending on the experiment), mice were sacrificed and subject to tissue clearing and analysis by lightsheet microscopy. (B) Tomato reporter activity in the brain of typical AAV-injected animal. Panel shows maximum intensity projection for the entire mouse brain, viewed from above. (C) Images of the epididymis, as in Figure 1B–C. Right panels show two distinct 2D slices of the cauda epididymis, highlighting the Tomato-positive rim surrounding the Tomato-negative lumen. See also Figure 2—figure supplements 1–2.
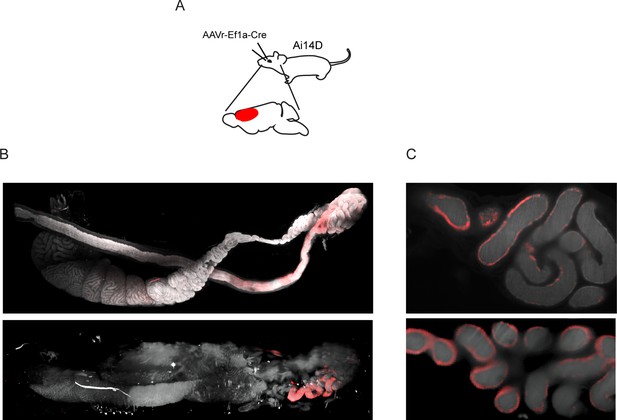
Additional examples of Tomato-positive epididymis samples from reporter animals subject to adeno-associated viral (AAV)-mediated Cre expression in the central nervous system (CNS).
(A) AAV injection scheme. As in Figure 2, but with rAAV2-retro-Eif1a-Cre injections into the prefrontal cortex. (B) Lightsheet images of the entire epididymis from two animals injected with rAAV2-retro-Eif1a-Cre in the prefrontal cortex. (C) Individual 2D cross sections from the cauda epididymis from the samples shown in panel (B).
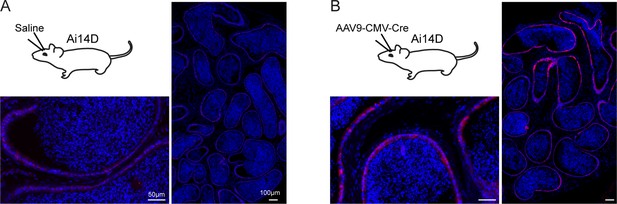
Fluorescence microscopy images of epididymis from reporter animals subject to adeno-associated viral ( AAV)-mediated Cre expression in the central nervous system (CNS).
(A) Microscopy images of the cauda epididymis from animals mock-injected with saline, as indicated in the accompanying schematic. (B) As in panel (A), but for animals injected with AAV9-CMV-Cre in the lateral somatosensory cortex, caudate putamen, and nucleus accumbens (see also Figure 2).
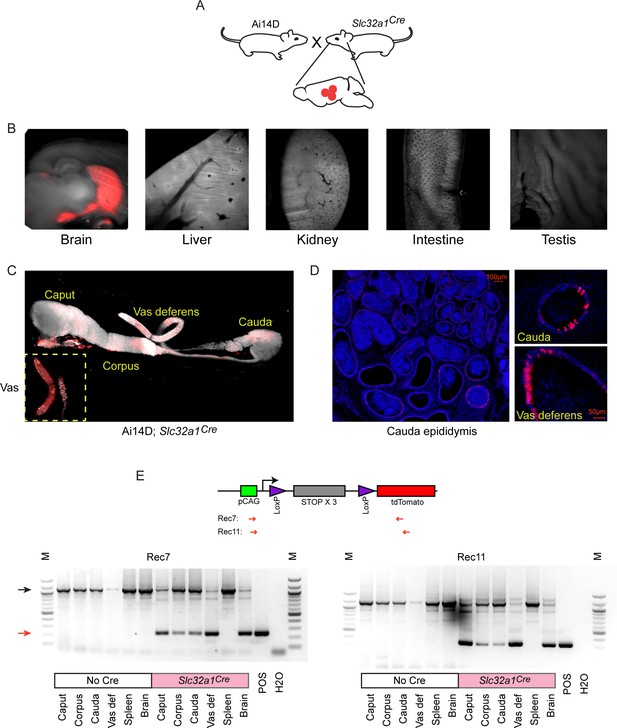
Cre expression in GABAergic neurons drives reporter expression in the epididymis.
(A) Schematic showing the cross that generated the double transgenic analyzed here, generated via crossing the Ai14D reporter with a strain bearing the Slc32a1Cre transgene (Materials and methods). (B) Lightsheet images of the indicated tissues in Ai14D; Slc32a1Cre double transgenic animals. (C) Lightsheet images of the epididymis in Ai14D; Slc32a1Cre double transgenic animals. (D) Two representative histology sections from Ai14D; Slc32a1Cre cauda epididymis and vas deferens, counterstained with DAPI, clearly showing Tomato-positive principal cells in the epithelium. See also Figure 3—figure supplement 1. (E) Recombination at the reporter locus in Ai14D; Slc32a1Cre tissues. Top: Schematic shows location of primers used on the PCR, with expected sizes following recombination of 248 bp (Rec7) and 332 bp (Rec11). Bottom: Each gel shows six tissues from a negative control Ai14D animal (no exposure to Cre), along with six tissues from the Ai14D; Slc32a1Cre double transgenic. POS: positive control tissue (liver from an Ai14D; AlbCre transgenic). Red arrow indicates the band arising from the recombined locus, with clear recombination observed in the brain, as well as all epididymal samples, in Ai14D;Slc32a1Cre animals.
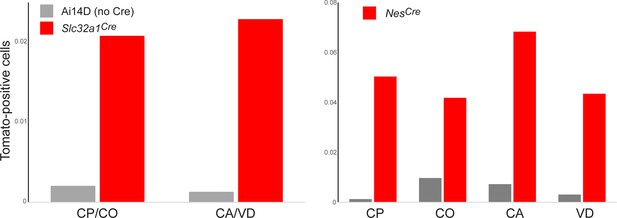
Flow cytometry analysis of Tomato-positive cells in the epididymis.
Bar charts show % Tomato-positive cells obtained from the indicated tissue samples, in negative control Ai14D animals (gray bars) or two double transgenic animals (red bars), as indicated. Both Slc32a1Cre and NesCre drove an increase in Tomato-positive cell numbers in the epididymis, consistent with our lightsheet, histology, and genotyping studies. To confirm the identity of Tomato-positive cells, we obtained ~10,000 Tomato-positive cells by FACS from several double transgenic animals, and carried out RNA-Seq, confirming the high level expression of principal cell markers expected from our histological studies in Figure 3D (Supplementary file 2).
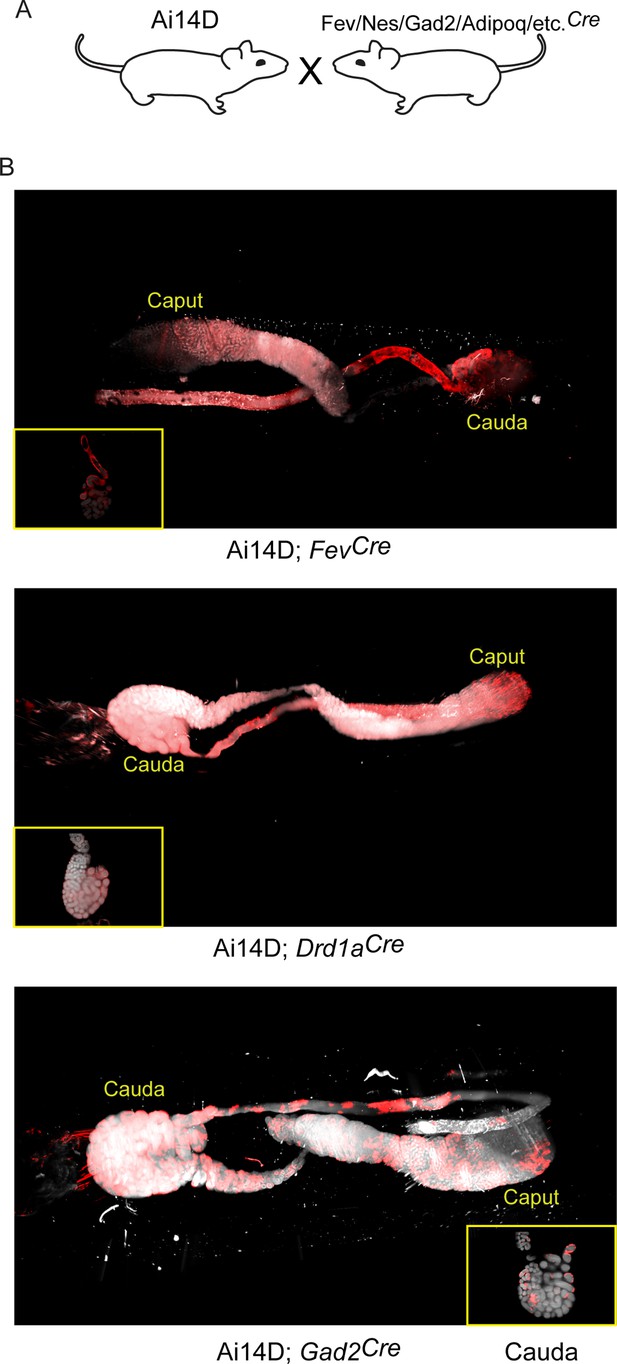
Multiple neuronal Cre drivers result in reporter activity in the epididymis.
(A) Schematic showing the cross that generated the double transgenics analyzed here, as in Figure 3A. (B) Representative lightsheet images of the epididymis from the indicated double transgenic animals. See also Figure 4—figure supplements 1–4.
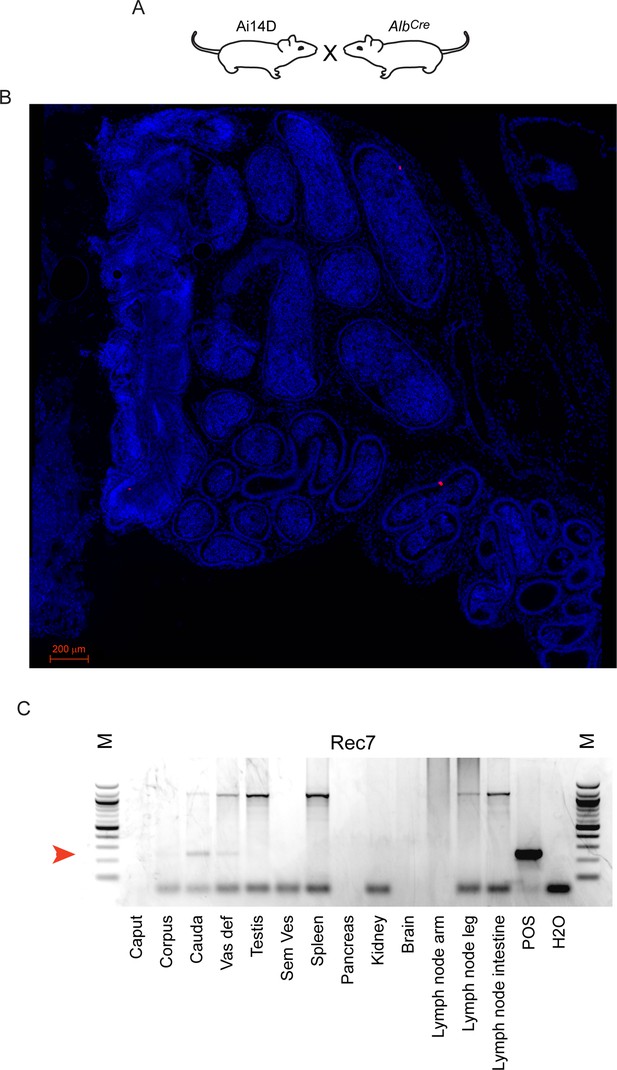
Minimal off-target recombination driven by AlbCre.
(A) Experimental schematic of mating strategy. (B) Histology of the cauda and corpus epididymis from Ai14D; AlbCre double transgenic animal. Across the entire section we find two isolated Tomato-positive cells in this animal. (C) PCR assay for recombination in the indicated tissues obtained from Ai14D; AlbCre double transgenic animals. A light recombination band is seen in the cauda epididymis, consistent with low level reporter activity in panel (B), while we find no evidence for any other off-target recombination.
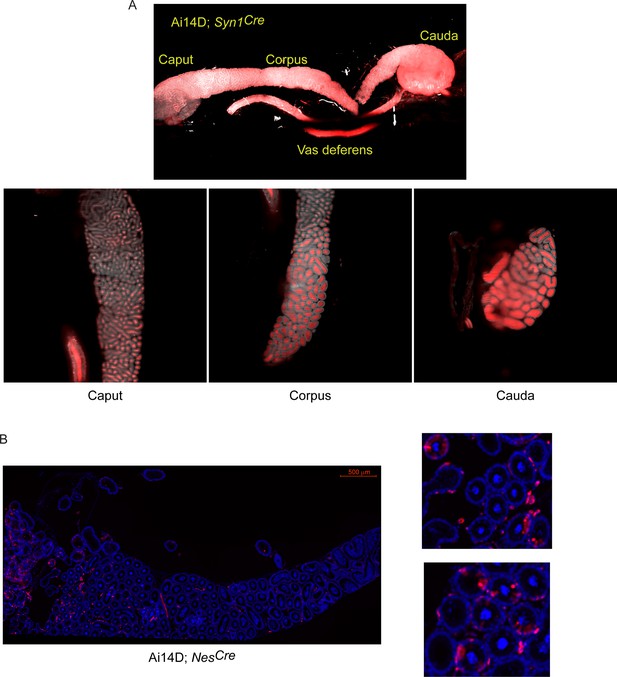
Tomato expression in epididymis samples from Ai14D animals carrying pan-neuronal Cre drivers.
(A) Ai14D; Syn1Cre double transgenic sample. Abundant Tomato signal here is clearly localized to the lumen of the epididymal tubule, consistent with Tomato expression in the male reproductive tract and potentially in the sperm, resulting from germline recombination in this animal. (B) Ai14D; NesCre double transgenic sample. Here, histology images shown widespread principal cell Tomato expression, similar to that observed in double transgenic Ai14D; Slc32a1Cre animals (Figure 3D), but with positive cells being found throughout all regions of the epididymis (in contrast to the cauda and vas deferens-enriched recombination in the Slc32a1Cre animals).
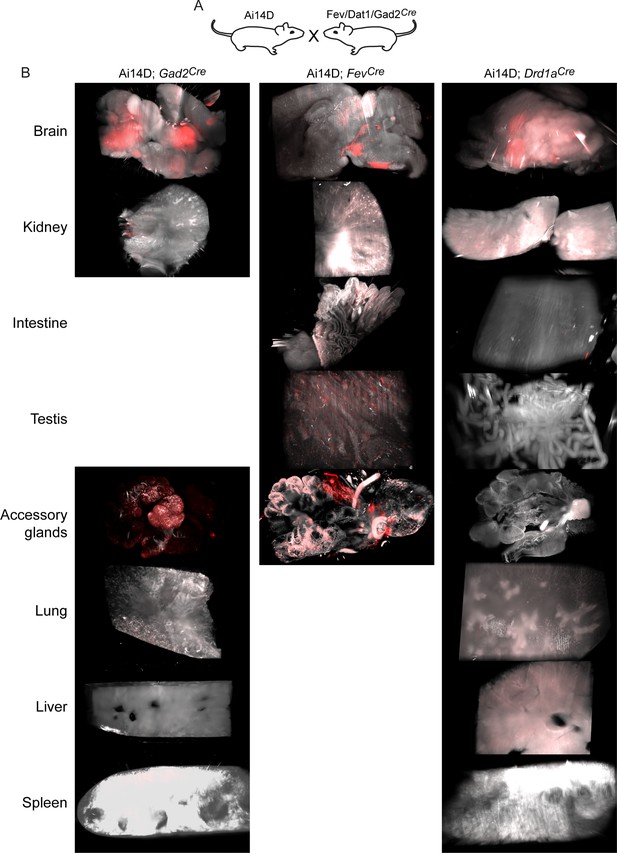
Examples of off-target Cre activity beyond the epididymis.
Lightsheet images of the indicated tissues showing Tomato expression in the various Ai14D; Cre double transgenic lines, as indicated.
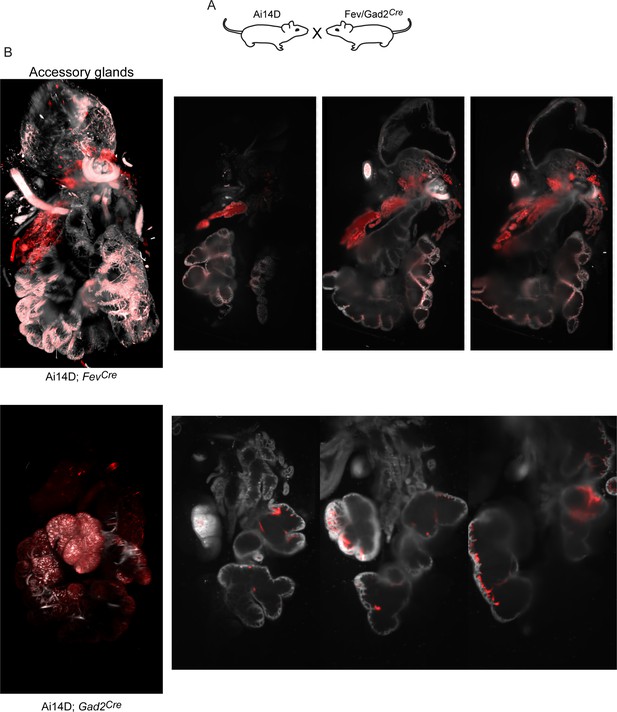
Off-target reporter activity in accessory glands.
Lightsheet images of the accessory glands are shown to the left, along with three representative sections in the right panels, for the two indicated Cre driver lines.
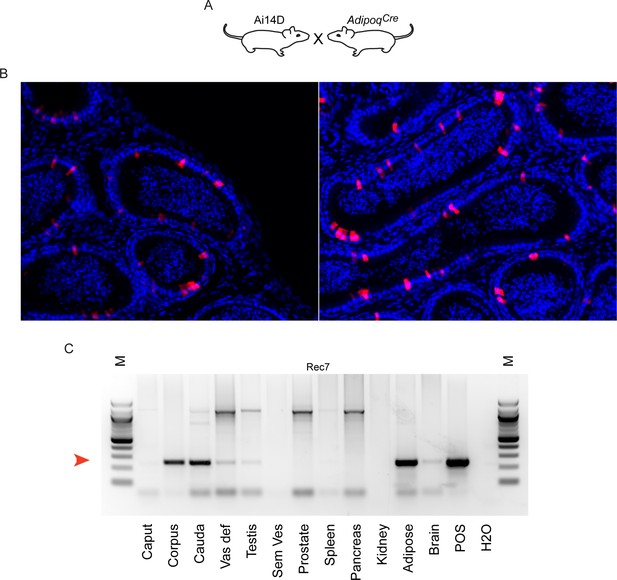
Reporter activity in AdipoqCre animals.
(A) Schematic, as in Figures 3A and 4A. (B) Histology from Ai14D; AdipoqCre animals, showing Tomato expression scattered throughout the epididymis epithelium. (C) PCR analysis of indicated tissues of Ai14D; AdipoqCre animals, revealing robust recombination in the epididymis. POS: positive control tissue. Red arrow indicates the band arising from the recombined locus. See also Figure 5—figure supplement 1.
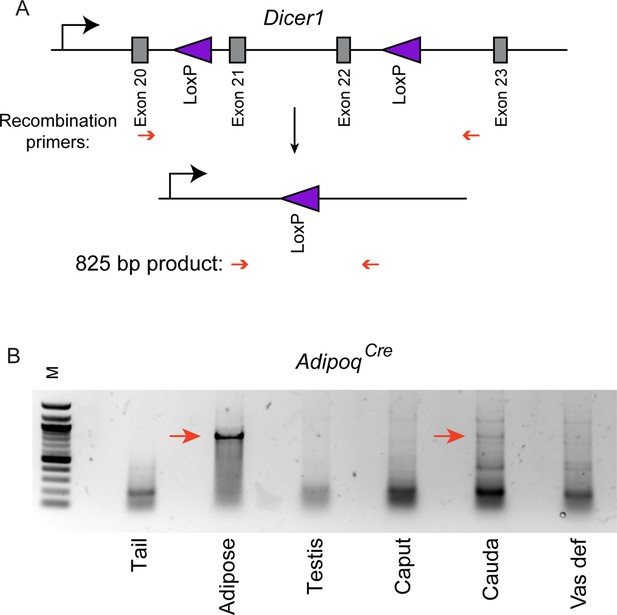
Adipoq Cre drives recombination of a second target locus in the epididymis.
(A) Schematic showing the genomic structure of the LoxP-flanked region of Dicer1. Primers used in (B) to assess recombination are indicated on the predicted recombination product. (B) Genotyping PCR for recombination in the indicated tissues obtained from AdipoqCre; Dicer1flx/flx double transgenic animals. Notable here is the expected recombination product in adipose tissue and no recombination in the tail tip negative control. Among reproductive tissues, we find robust recombination at this locus specifically in the cauda epididymis (and to a lesser extent in the vas deferens). To further confirm the identity of the recombination product we gel-purified this band and sequenced the resulting DNA, confirming that this product corresponds to the recombination product shown in (A).
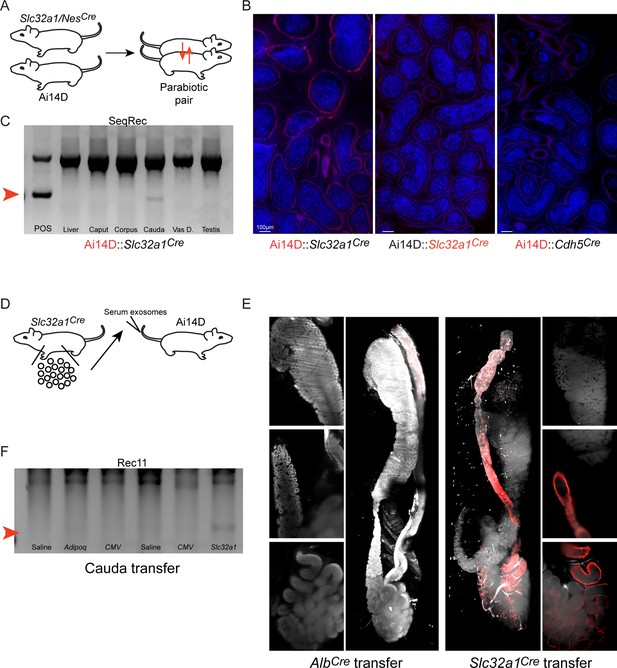
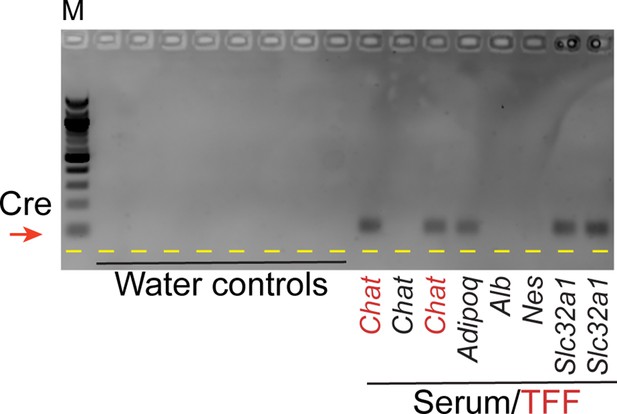
Cre activity present in the circulation can be transferred from a Cre driver to a reporter animal.
(A) Experimental schematic. Parabiosis surgery was used to link the circulatory systems of an Ai14D reporter animal and that of a Slc32a1Cre or NesCre animal. After 8–9 weeks together, animals were sacrificed and tissues from both animals were obtained for analysis. (B) Histology from the indicated sides (highlighted in red) of two Ai14D::Cre parabiotic pairs. We observe Tomato expression specifically on the reporter side, but not the Cre side, of Ai14D::Slc32a1Cre animals. (C) Genotyping PCR from the Ai14D side, and the Cre side, of the indicated parabiotic pairs, with faint recombination detected in the cauda epididymis of Ai14D::Slc32a1Cre animals. (D) Schematic showing serum/exosome transfer experiments. Note that these experiments were carried out with a wide range of injection schedules (Materials and methods), using both whole serum transfers and TFF-enriched exosome transfers for recipient animals. (E) Lightsheet images of Ai14D recipient following transfer of serum and exosomes from AlbCre donors (left panels), or from Slc32a1Cre donors (right panels). (F) Genotyping PCR from cauda epididymis samples obtained from Ai14D animals receiving serum or TFFs from various Cre donor lines. Red arrow indicates the band diagnostic of a recombined locus.
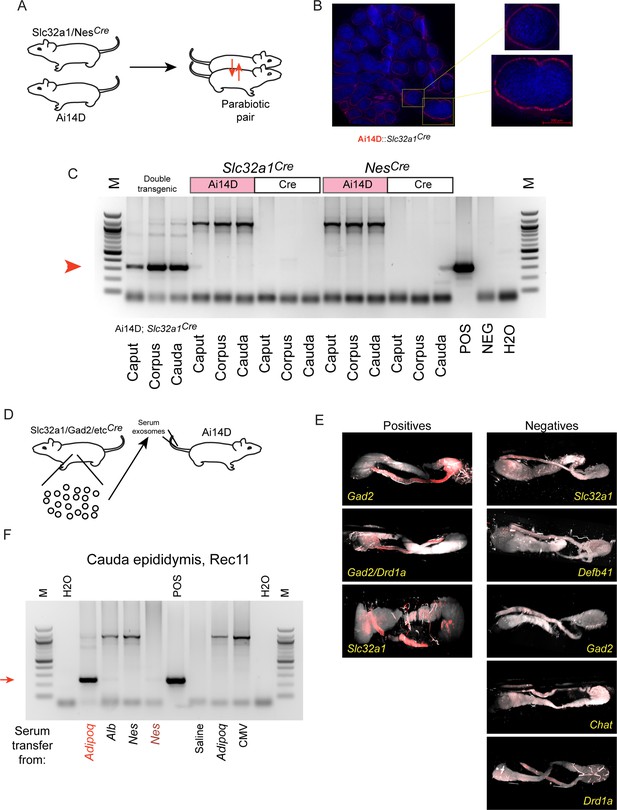
Inconsistent Cre transfer via parabiosis or serum transfer experiments.
(A) Parabiosis schematic. (B) Histology images of the cauda epididymis from the Ai14D recipient animal after 8 weeks in a parabiosis with a Slc32a1Cre donor. Zoom-in images show Tomato-positive epithelial cells in cauda epididymis. (C) Genotyping PCR for the indicated samples. The first three samples were gathered from the indicated double transgenic. For the two parabiotic animal pairs here, samples from the Ai14D recipient and the Cre donor were each assayed. (D) Serum/exosome transfer schematic. (E) Lightsheet images from eight serum/exosome recipients, with the Cre driver used as the serum/exosome donor indicated for each sample. Tomato-positive and -negative samples are organized separately. Note that one recipient received exosomes from both Gad2Cre donor and Drd1Cre donors on different injection days. (F) Genotyping PCR for recombination in the indicated serum/exosome recipients. Most recipients were negative for recombination, with strong recombination in one recipient of AdipoqCre exosomes, and very weak recombination observed in one recipient of NesCre serum and exosomes.
Additional files
-
Supplementary file 1
Oligonucleotides used in this study.
Table lists oligo sequences used for genotyping PCRs.
- https://cdn.elifesciences.org/articles/77733/elife-77733-supp1-v2.xlsx
-
Supplementary file 2
RNA-Seq from FACS-isolated Tomato-positive cells.
Table shows mRNA abundance (expressed as FPKM) for RNA-Seq libraries obtained from FACS-isolated Tomato-positive cells obtained from the cauda epididymis of Ai14D animals carrying either Defb41Cre or Slc32a1Cre, as indicated.
- https://cdn.elifesciences.org/articles/77733/elife-77733-supp2-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/77733/elife-77733-transrepform1-v2.docx
-
Source data 1
All source data files are original uncropped gel images used in the corresponding figures: Figure 3E, Figure 5C, Figure 6C, Figure 6F, Figure 4—figure supplement 1C, Figure 6—figure supplement 1C, Figure 6—figure supplement 1F, Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/77733/elife-77733-data1-v2.zip