Retinoic acid-induced protein 14 controls dendritic spine dynamics associated with depressive-like behaviors
Figures
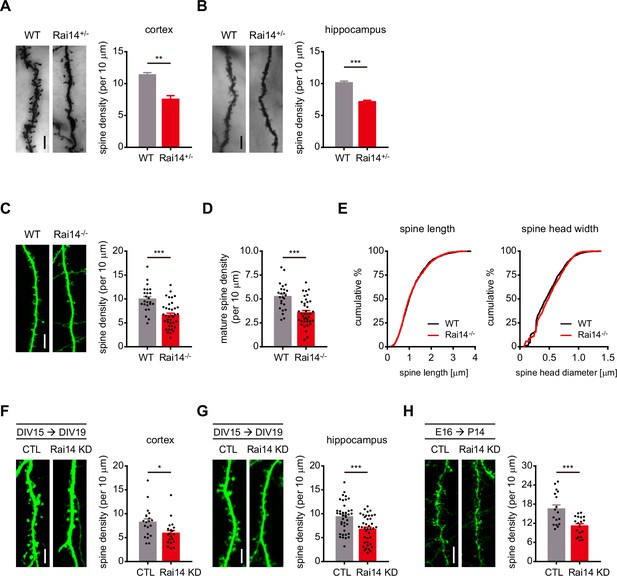
Rai14-depleted neurons exhibit decreased dendritic spine density.
(A) Golgi-stained basal dendrites of cortical layer II/ III pyramidal neurons from adult wild-type (WT) and Rai14+/- mouse brains. Representative images (left) and quantitative analysis of the dendritic spine density (right) are shown (n = 4 for each group, 7–11 neurons for each mouse were analyzed). (B) Golgi-stained basal dendrites of hippocampal CA1 pyramidal neurons from adult WT and Rai14+/- mouse brains. Representative images (left) and quantitative analysis of the dendritic spine density (right) are shown. (n = 4 for each group, 8–11 neurons for each mouse were analyzed). (C–E) Dendritic spine analysis of WT and Rai14-/- primary cultured hippocampal pyramidal neurons (DIV19) derived from WT and Rai14-/- embryos. GFP-empty vector was transfected to analyze neuronal morphology. (C) Representative images (left) and quantitative analysis of the dendritic spine density (right) are shown. (n = 24 neurons for WT, 38 neurons for Rai14-/- from three separate experiments). (D) Quantification of mature spine density of the dendritic segments shown in (C). For spine type classification criteria, please see Materials and methods. (E) Cumulative probability plot of the spine length (left, n = 2165 spines from WT and 2207 spines from Rai14-/- neurons) and the maximal diameter of spine head width (right, n = 1131 mature spines from wild type, and 1210 mature spines from Rai14-/- neurons). (F–G) Spine density analysis of primary cultured cortical (F) and hippocampal (G) pyramidal neurons expressing scrambled shRNA (CTL) or Rai14 shRNA (Rai14 KD). Neurons were transfected at DIV15, and fixed and analyzed at DIV19–20. (F) Representative images of dendritic segment from cortical neurons (left) and quantitative analysis of the dendritic spine density (right) are shown (n = 20 neurons for each group from three independent cultures). (G) Representative images of dendritic segment from hippocampal neurons (left) and quantitative analysis of the dendritic spine density (right) are shown (n = 41 neurons for CTL, 40 neurons for Rai14 KD from 4 independent cultures). (H) Spine density analysis of cortical layer II/ III pyramidal neurons expressing scrambled shRNA (CTL) or Rai14 shRNA (Rai14 KD) from mouse brains. Embryos were electroporated in utero with scrambled or Rai14 shRNA at E16, and brains were analyzed at P14. Representative images (left) and quantitative analysis of the dendritic spine density (right) are shown (n = 17 neurons from 3 mice for CTL, 18 neurons from 3 mice for Rai14 KD). Scale bars represent 5 μm. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 determined by student’s t-test for (A), (B), (C), (D), (F), (G) and (H). Kolmogorov-Smirnov test was used for (E). All experiments were repeated at least three times. See also Figure 1—figure supplement 1 and Figure 1—source data 1.
-
Figure 1—source data 1
Values for dendritic spine density analysis in Rai14-deficient groups.
- https://cdn.elifesciences.org/articles/77755/elife-77755-fig1-data1-v2.xlsx
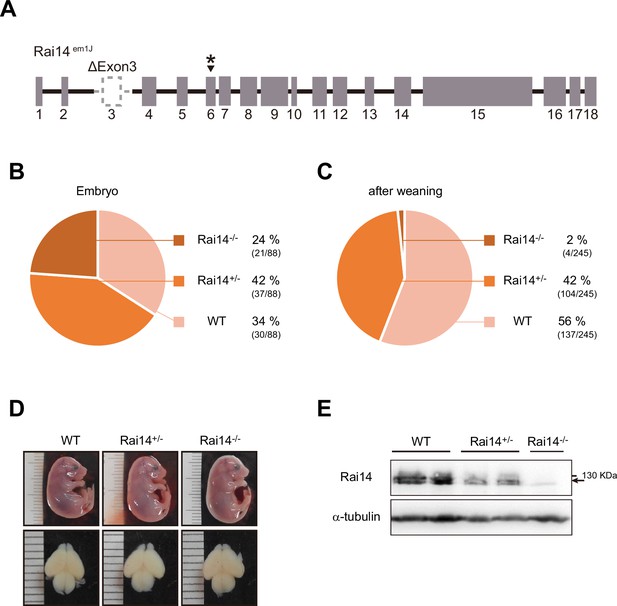
Loss of Rai14 causes perinatal lethality.
(A) Knockout scheme of Rai14-/- mouse (Rai14em1(IMPC)J, The Jackson Laboratory). Cas9 RNA and four guide sequences results in the deletion of exon 3 and 336 bp of flanking intronic sequences including the splice acceptor and donor, followed by a change in amino acid sequence after residue 12 and early truncation 59 amino acid later (asterisk) (MGI Ref ID: J:188991). (B) Genotype distribution of E17.5–E18 embryos derived from the timed breeding of Rai14+/- mice. Embryos from the timed mating were isolated at E17.5–E18, and genotyped by PCR from the arms, legs, and tail snips. (C) Genotype distribution of pups derived from the timed breeding of Rai14+/- mice. Pups were separated from dams at P21–P28 and genotyped by PCR from the tail snips. (D) Morphology of Rai14-deificient (Rai14+/- and Rai14-/-) and wild-type (WT) littermate embryos at E18.5 (upper) and embryonic brains (lower). (E) A western blot of endogenous Rai14 in brain lysates of Rai14-deificient (Rai14+/- and Rai14-/-) and WT littermate embryos.
-
Figure 1—figure supplement 1—source data 1
Uncropped western blot images with relevant bands labeled.
- https://cdn.elifesciences.org/articles/77755/elife-77755-fig1-figsupp1-data1-v2.pdf
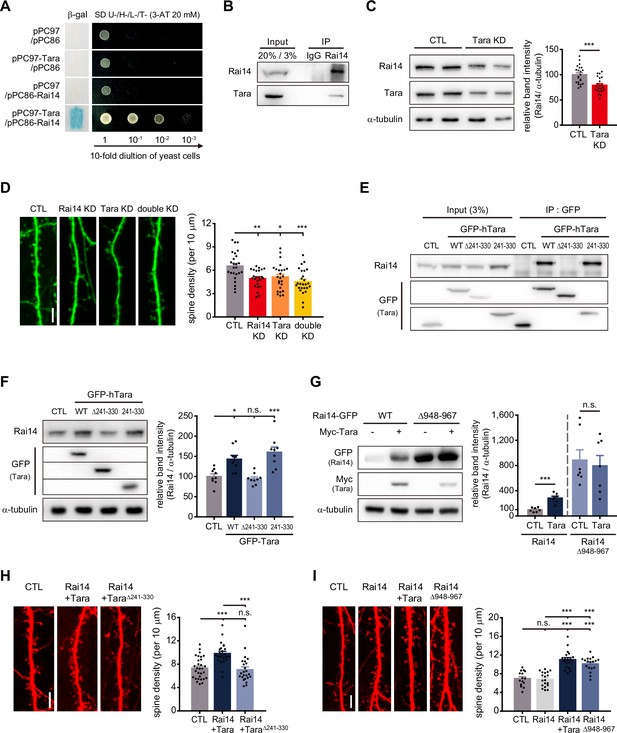
Tara-mediated stabilization of Rai14 up-regulates dendritic spine density.
(A) Yeast two-hybrid assay of Rai14 and Tara. pPC97-Tara and pPC86-Rai14 co-transformants were analyzed byβ-galactosidase activity assay using X-gal as substrate (left) and growth on minimal media in decreasing concentrations of yeast (right). (B) Co-immunoprecipitation of endogenous Rai14 and Tara from P14 mouse brain lysates. (C) Down-regulation of Rai14 by Tara KD. Western blot image of endogenous Rai14 from HEK293 cell lysates transfected with scrambled shRNA (CTL) or Tara shRNA (Tara KD) (left) and relative Rai14 band intensity normalized to α-tubulin (right) are shown (n = 22 for CTL, 22 for Tara KD). (D) Spine density analysis of Tara and/or Rai14 KD conditions. Representative images of dendritic segments from DIV19 primary cultured hippocampal pyramidal neurons expressing indicated shRNA(s) (left) and quantification of the dendritic spine density (right) are shown (n = 25 neurons for CTL, 24 neurons for Rai14 KD, 25 neurons for Tara KD, and 26 neurons for double KD). (E) Localization of Tara region for interaction with Rai14. Co-immunoprecipitation of endogenous Rai14 with Tara deletion mutants was carried out in HEK293 cells. CTL: GFP-empty vector. (F) Up-regulation of Rai14 by Tara interaction. Western blot image of endogenous Rai14 from HEK293 cell lysates transfected with indicated plasmids (left), and relative Rai14 band intensity normalized to α-tubulin (right) are shown (n = 9). CTL: GFP-empty vector (G) Stabilization of Rai14 by deletion of C-terminal tip. Western blot image of Rai14 from HEK293 cell lysates transfected with indicated plasmids (left) and relative Rai14 band intensity normalized to α-tubulin (right) are shown (n = 7). (H) Regulation of spine density by Tara-Rai14 interaction. Representative images of dendritic segments from DIV17–19 primary cultured hippocampal pyramidal neurons expressing indicated plasmids (left) and quantification of the dendritic spine density (right) are shown (n = 30 neurons for CTL, 24 neurons for Rai14 +Tara and Rai14 +TaraΔ241–330). (I) Regulation of spine density by Rai14 stabilization. Representative images of dendritic segments from DIV17–19 primary cultured hippocampal pyramidal neurons expressing indicated plasmids (left) and quantification of the dendritic spine density (right) are shown (n = 15 neurons for CTL, 19 neurons for Rai14 and Rai14 +Tara, and 18 neurons for Rai14Δ948–967). Scale bars represent 5 μm. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 from student’s t-test for (C), (G) and one-way ANOVA with Bonferroni’s multiple comparison test for (D), (F), (H), and (I). Experiments were repeated at least three times. See also Figure 2—figure supplements 1 and 2, and 3, and Figure 2—source data 1.
-
Figure 2—source data 1
Quantification on Rai14 expression and spine density in association with Tara.
- https://cdn.elifesciences.org/articles/77755/elife-77755-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Uncropped western blot images with relevant bands labeled.
- https://cdn.elifesciences.org/articles/77755/elife-77755-fig2-data2-v2.pdf
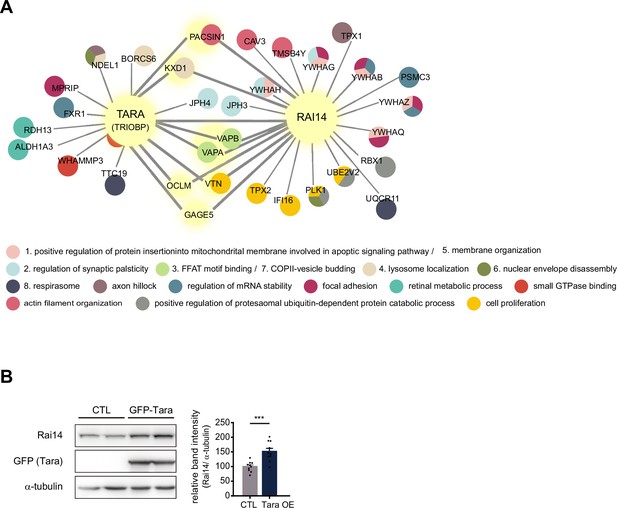
Rai14 and Tara form a complex.
(A) Rai14 and TRIOBP (Tara) protein interactome image generated from BioPlex (Huttlin et al., 2017; Schweppe et al., 2018) (Modified for clear representation). Shared interactors are indicated by light yellow nodes and bold lines. Secondary network Gene Ontology annotations with significant adjusted p-values are listed below, and GO terms associated with the protein are indicated by designated colors in the circle. (B) Increased Rai14 protein level by Tara overexpression. A western blot image of endogenous Rai14 in the lysate of HEK293 cell transfected with GFP-Tara. Relative intensity of the Rai14 band intensity normalized against α-tubulin is shown (n = 10). Data are plotted as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 from student’s t-test.
-
Figure 2—figure supplement 1—source data 1
Uncropped western blot images with relevant bands labelled.
- https://cdn.elifesciences.org/articles/77755/elife-77755-fig2-figsupp1-data1-v2.pdf
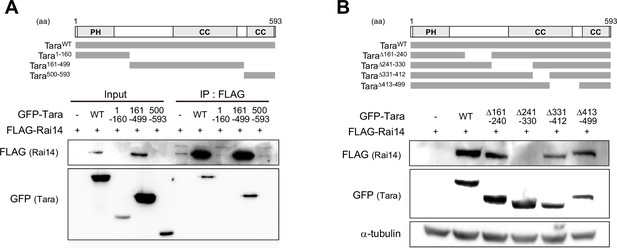
Domain mapping of Tara for interaction with Rai14.
(A) Mapping of Tara domain involved in Rai14 interaction. Upper, The diagram of human Tara deletion mutants: Tara1–160, Tara161–499, Tara500–593. PH: Pleckstrin homology domain. CC: coiled-coil domain. Lower, Co-immunoprecipitation of FLAG-Rai14 with Tara deletion mutants in HEK293 cells. IP with anti-FLAG antibody. The Tara mutant fragment containing amino acid residues 161–499 interacted with FLAG-Rai14. (B) Mapping of Tara domain involved in Rai14 upregulation. Upper, The diagram of the domain structure of human Tara and its deletion mutants: TaraΔ161–240, TaraΔ241–330, TaraΔ331–412, and TaraΔ413–499. Lower, Western blots using HEK293 cell lysates transfected with FLAG-Rai14 and indicated Tara constructs. The Tara mutant lacking amino acid residues 241–330 failed to upregulate FLAG-Rai14.
-
Figure 2—figure supplement 2—source data 1
Uncropped western blot images with relevant bands labeled.
- https://cdn.elifesciences.org/articles/77755/elife-77755-fig2-figsupp2-data1-v2.pdf
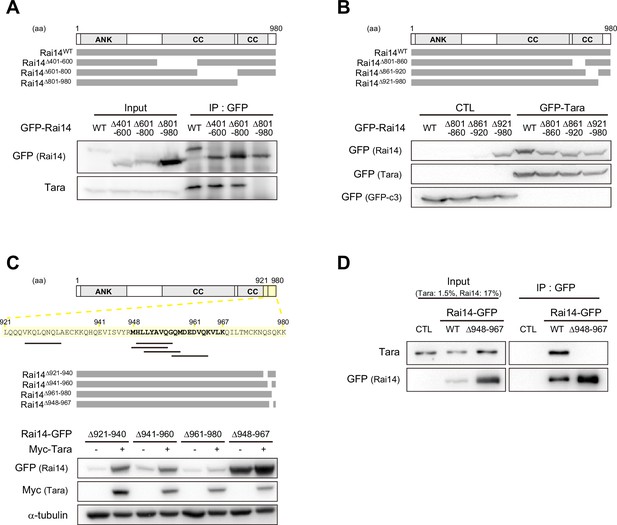
Mapping of Rai14 domain involved in Tara-mediated Rai14 upregulation.
(A) Mapping of Rai14 domain involved in Tara interaction. Upper, The diagram of the domain structure of human Rai14 and its deletion mutants: Rai14Δ401–600, Rai14Δ601–800, and Rai14Δ801–980. ANK: ankyrin repeat domain, CC: coiled-coil domain. Lower, Co-immunoprecipitation of endogenous Tara with Rai14 deletion mutants in HEK293 cells. IP with anti-GFP antibody. The Rai14 mutant lacking amino acid residues 801–900 lost its interaction with Tara. (B–C) Mapping of Rai14 domain involved in Tara-mediated Rai14 upregulation. (B) Upper, The diagram of the domain structure of human Rai14 and its deletion mutants: Rai14Δ801–860, Rai14Δ861–920, and Rai14Δ921–980. Lower, Western blots using HEK293 cell lysates transfected with indicated Rai14 constructs with or without GFP-Tara. CTL: GFP-empty vector. The Rai14 mutant lacking 921–980 amino acid sequence showed upregulated protein level even if Tara was not co-expressed. (C) Upper, The diagram of the domain structure of human Rai14 and its deletion mutants: Rai14Δ921–940, Rai14Δ941–960, Rai14Δ961–980, and Rai14Δ948–967. Amino acid residues 921–980 are highlighted with light yellow box. Black lines under the sequence represent predicted protease cleavage sites (Kumar et al., 2020; Li et al., 2020). Lower, Western blots using HEK293 cell lysates transfected with indicated Rai14 constructs with or without Myc-Tara. The Rai14 mutant lacking amino acid residues 948–967 showed upregulated protein level even if Tara was not co-expressed. (D) Co-immunoprecipitation of endogenous Tara with Rai14-GFP or Rai14Δ948–967-GFP in HEK293 cells. IP with anti-GFP antibody. CTL: GFP-empty vector. Deletion of amino acid residues 948–967 completely abolished the interaction of Rai14 with Tara.
-
Figure 2—figure supplement 3—source data 1
Uncropped western blot images with relevant bands labeled.
- https://cdn.elifesciences.org/articles/77755/elife-77755-fig2-figsupp3-data1-v2.pdf
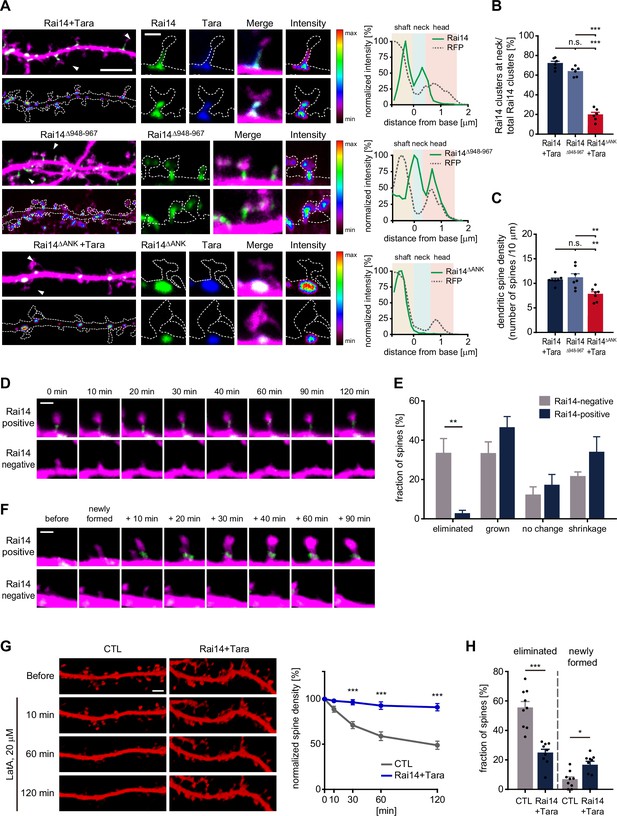
Tara-Rai14 complex accumulates at the neck of dendritic spines and protects spines from elimination.
(A–B) Localization analyses of Rai14 at the dendritic spine. (A) Dendritic segments of DIV17–18 primary cultured hippocampal pyramidal neurons transfected with indicated Rai14-GFP and/or FLAG-Tara constructs are shown with an intensity heat map of Rai14 and Rai14 mutants (left, green: Rai14WT/mut-GFP, blue: FLAG-Tara, magenta: RFP). Spines indicated by white arrowhead are shown in higher magnification with an intensity heat map of Rai14 and Rai14 mutants (middle). Representative intensity profiles of Rai14 and Rai14 mutants in the indicated spines are also shown (right, RFP: a morphology marker). Scale bar represents 5 μm for dendritic segments and 1 μm for magnified spine images. The contours of the dendritic shaft and spines are outlined by dashed lines. (B) Fraction of Rai14 clusters at spine neck relative to total Rai14 clusters within the designated dendritic segments. (n = 6 neurons) (C) Impact of stabilized (Rai14Δ948–967) or mislocalized forms of Rai14 (Rai14ΔANK) expression on dendritic spine density of primary hippocampal pyramidal neurons (n = 7 neurons, DIV17–18). (D–E) Spine dynamics of dendritic spines with or without Rai14 from time-lapse imaging on DIV15–17 primary cultured hippocampal pyramidal neurons expressing Rai14-GFP, FLAG-Tara, and RFP. Rai14-positive spines: spines containing Rai14-GFP clusters within their neck at time 0 min, Rai14-negative spines: spines that does not contain Rai14-GFP clusters within their neck at time 0 min. (D) Representative images of a stable Rai14-positive spine (upper) and an eliminated Rai14-negative spine (lower). Scale bar represents 2 μm. (E) Quantification on the dynamics of Rai14-positive and Rai14-negative spines at 120 min compared to 0 min. (n = 5 neurons) (F) Representative images of newly formed dendritic spines in which Rai14-GFP recruited (upper, Rai14-positive) or not (lower, Rai14-negative) at the spine neck. (G–H) Impact of Rai14 and Tara expression on spine maintenance upon latrunculin A (LatA) treatment. (G) Representative images of hippocampal dendritic segments (left, morphology marker: RFP-LifeAct) and normalized spine density at indicated time points after LatA treatment (right, 20 μΜ) are shown (n = 9 neurons, DIV17–18). Each spine density after LatA treatment was normalized to the spine density before LatA treatment. Scale bar represents 5 μm. (H) Fractions of the eliminated spines and newly formed spines at 120 min time point after LatA treatment. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 determined by one-way ANOVA for (B) and (C), student’s t-test for (H), and two-way ANOVA with Bonferroni’s multiple comparison test for (E) and (G). All experiments were repeated at least three times. See also Figure 3—figure supplement 1, and Figure 3—source data 1.
-
Figure 3—source data 1
Source data for Rai14 localization and dendritic spine dynamics.
- https://cdn.elifesciences.org/articles/77755/elife-77755-fig3-data1-v2.xlsx
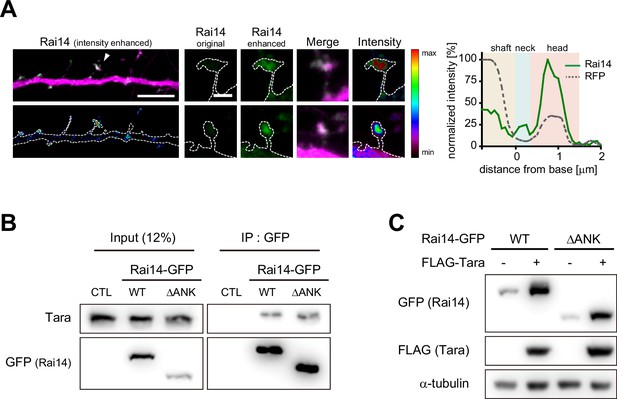
Characterization of Rai14ΔANK protein.
(A) Localization of Rai14-GFP at the dendritic spines. Dendritic segments of DIV17–18 hippocampal pyramidal neurons transfected with Rai14-GFP (left, upper) are shown with an intensity heat map of Rai14 (left, lower). Spines indicated by white arrowheads are shown in higher magnification with an intensity heat map of Rai14-GFP (middle). Representative intensity profile of Rai14 in the indicated spine is also shown (right, RFP: a morphology marker). Scale bar represents 5 μm for dendritic segments and 1 μm for magnified spine images. The contours of the dendritic shaft and spines are outlined by dashed lines. (B) Co-immunoprecipitation of endogenous Tara with Rai14-GFP or Rai14ΔANK-GFP in HEK293 cell. CTL: GFP-empty vector. ANK: ankyrin repeat domains (amino acid residues 17–252). The interaction of Rai14 with Tara was not compromised by deletion of ankyrin repeat domain of Rai14. (C) Representative western blot image of Rai14-GFP or Rai14ΔANK-GFP from HEK293 cell lysates co-transfected with/without FLAG-Tara. ANK: ankyrin repeat domains. The Rai14 mutant lacking ankyrin repeat domain was upregulated upon increased Tara expression.
-
Figure 3—figure supplement 1—source data 1
Uncropped western blot images with relevant bands labeled.
- https://cdn.elifesciences.org/articles/77755/elife-77755-fig3-figsupp1-data1-v2.pdf
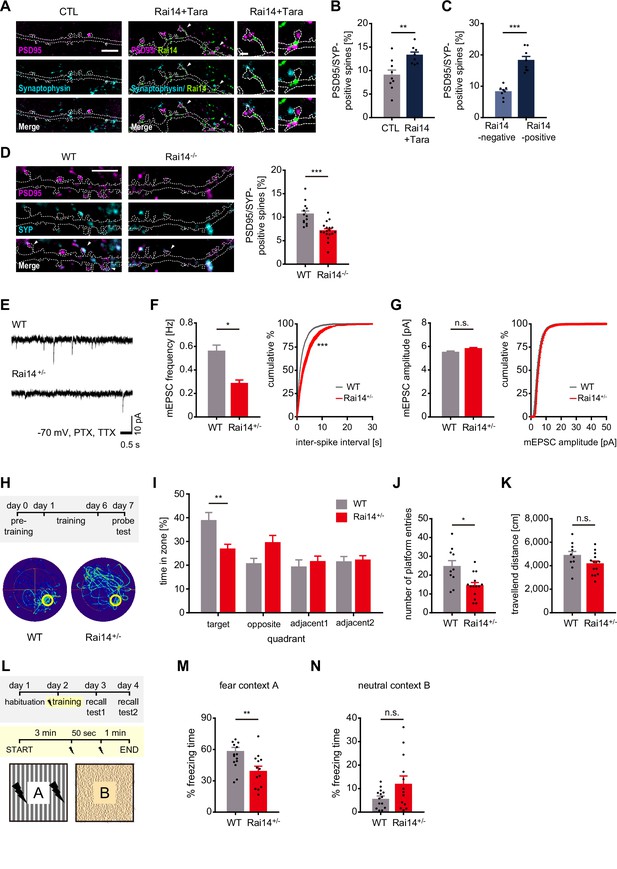
Rai14 affects functional synaptic activity.
(A–C) Enhanced synapses in the hippocampal pyramidal neurons over-expressing Rai14 and Tara. (A) Representative images of DIV17–18 dendritic segments and spines are shown (magenta: PSD95, cyan: Synaptophysin, green: Rai14-GFP). Spines indicated with white arrowheads are shown in higher magnification. Scale bar represents 5 μm for dendritic segment image (left) and 1 μm for magnified spine images (right). Dashed lines indicate the contours of the dendritic shaft and spines. (B) Fractions of synapse-bearing spines (n = 9 neurons for CTL, 8 neurons for Rai14 +Tara). SYP: Synaptophysin. The fraction of synaptic clusters co-localized with dendritic spines relative to entire spines was analyzed. (C) Fractions of synapse-bearing spines in Rai14-positive and Rai14-negative spines in hippocampal neurons expressing Rai14 and Tara (n = 8 neurons). SYP: Synaptophysin, Rai14-positive spines: spines containing Rai14-GFP clusters within their neck, Rai14-negative spines: spines without Rai14-GFP within their neck. (D) Decreased synapse number in the DIV18–20 hippocampal Rai14-/- pyramidal neurons. The fraction of synaptic clusters co-localized with dendritic spines relative to entire spines was analyzed. Representative images of dendritic segments (left,) and fractions of synapse-bearing spines (right, n = 13 neurons for WT, 18 neurons for Rai14-/-). Dashed lines: contours of the dendritic shaft and spines. Scale bar: 5 μm. magenta: PSD95, cyan: SYP (synaptophysin) (E–G) miniature excitatory postsynaptic currents (mEPSCs) recorded from principal hippocampal CA1 pyramidal neurons of WT and Rai14+/- mice. mEPSCs were recorded at –70 mV holding potential in the presence of picrotoxin (PTX) and tetrodotoxin (TTX). (E) Representative mEPSC traces. Scale bars represent 0.5 s and 10 pA. (F) Left, Average mEPSC frequency of principal hippocampal neurons from WT and Rai14+/- mice. Right, Cumulative probability distributions of mEPSC inter-spike intervals (n = 3 for each group, 10–12 neurons for each mouse were analyzed). (G) Average (left) and cumulative probability distributions (right) of mEPSC amplitude in neurons analyzed in (F). (H–K) Morris water maze test. Performance was assessed by comparing 11- to 12-week-old male WT and Rai14+/- mice (n = 10 for WT, 14 for Rai14+/-). (H) Experimental scheme of Morris water maze test (upper) and representative trajectories of WT and Rai14+/- mice during the probe test (lower). Pre-training: training with visible platform (5 trials/ day, on day 0), training: training with hidden platform (5 trials/ day, on day 1–day 6), probe test: test with platform removed (5 min/ test, on day 7). The platform is indicated with a yellow circle. (I) Permanence time of WT and Rai14+/- mice in indicated quadrants during the probe test. (J) Number of platform entries during the probe test. (K) Total traveled distance during the probe test. (L–N) Contextual fear conditioning test (n = 14 for WT, 13 for Rai14+/-, 11–12 week old). (L) Experimental scheme of the contextual fear conditioning test. In fear context A, two electric foot-shocks (0.4 mA for 1 s) were delivered with a 50 s interval. (M) Mean fractions of freezing time in the fear context (N) Mean percentage of freezing time in the neutral context. Error bars indicate the mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 determined by student’s t-test for (B), (C), (D), (J), (K), (M), and (N), two-way ANOVA with Bonferroni’s multiple comparison test for (I). Unpaired t-test with Welch’s correction was used for bar graphs, and Kolmogorov-Smirnov test was used for cumulative graphs in (F) and (G). All experiments were repeated at least three times. See also Figure 4—figure supplement 1, and 2, and Figure 4—source data 1.
-
Figure 4—source data 1
Source data for synapse number and synaptic function in Rai14-deficient groups.
- https://cdn.elifesciences.org/articles/77755/elife-77755-fig4-data1-v2.xlsx
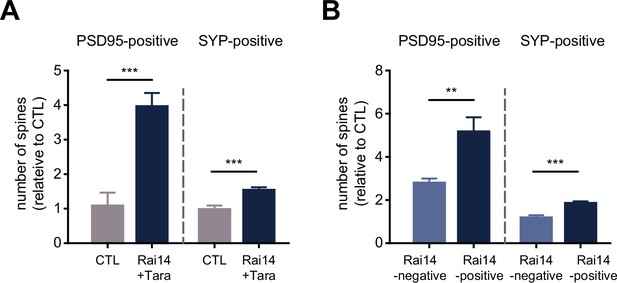
Spine analyses by pre- and postsynaptic markers.
(A) The fractions of PSD95-bearing and synaptophysin-positive spines (n = 8 neurons for CTL, 8 neurons for Rai14 +Tara). SYP: synaptophysin. The numbers of PSD95 /SYP-positive spines are normalized to those of CTL. (B) The fractions of PSD95-bearing and synaptophysin-positive spines in the Rai14-positive or Rai14-negative spine groups (n = 8 neurons). SYP: synaptophysin, Rai14-positive spines: spines containing Rai14-GFP clusters within their neck, Rai14-negative spines: spines without Rai14-GFP within their neck. The numbers of PSD95 /SYP-positive spines are normalized to those of CTL. Data are plotted as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 from student’s t-test.
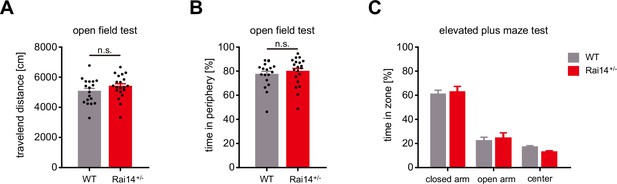
Anxiety-related behavioral tests of Rai14+/- mice.
(A–B) Open-field test. Performance was assessed by comparing 10- to 12-week-old male WT and Rai14+/- mice (n = 18 mice for WT, 20 mice for Rai14+/-). (A) Total travelled distance of WT and Rai14+/- mice in the open field test. (B) The fraction of time spent in periphery in the open field test. (C) Elevated plus maze (EPM) test. The fraction of time spent in closed arms, open arms, and center during EPM test. Performance was assessed by comparing 10- to 12-week-old male WT and Rai14+/- mice (n = 14 for WT, 19 for Rai14+/-). Data are plotted as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 from student’s t-test (A), (B), and one-way ANOVA (C).
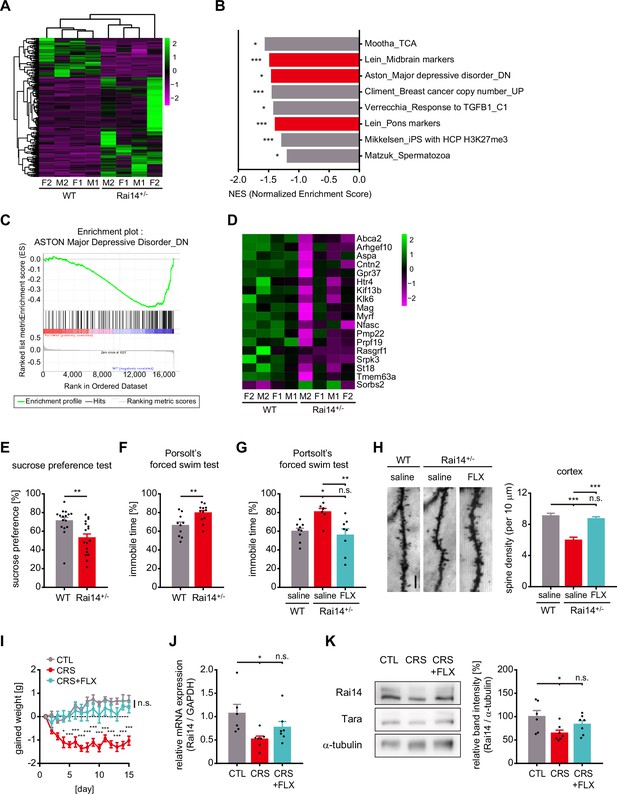
Rai14-deficient mice exhibit depressive-like behaviors associated with stress.
(A–D) RNA sequencing and gene set enrichment analysis (GSEA) on whole brains of 9-week-old Rai14+/- and littermate controls. (A) Heat map of the one-way hierarchical clustering for gene expression value (log2 based normalized). 273 genes showing |fold change| ≥ 2 and raw p-value < 0.05. Green: higher expression, magenta: lower expression, F: female, M: male (n = 4 mice, 2 females + 2 males). (B) GSEA results using curated chemical and genetic perturbations (CGP) gene set collection from MSigDB. Significant gene sets (nominal p-value < 0.05) negatively enriched in Rai14+/- mouse brains are listed in the order of normalized enrichment score (NES), and gene sets associated with the nervous system are indicated with red color. *p < 0.05, **p < 0.01, and ***p < 0.001. (C) The enrichment plot of the genes in the gene set ‘Aston_Major depressive disorder_DN’ generated from GSEA (Mootha et al., 2003; Subramanian et al., 2005). Upper: Profile of running enrichment score. Lower: Positions of the gene set members on the ranked ordered list. Green line: enrichment profile, black line: hits of gene set members, red zone: upregulated in Rai14+/- brain, blue zone: downregulated in Rai14+/- brain. (D) Heat map representation of transcripts included both in the ‘ASTON-Major depressive disorder_DN’ gene set and significant DEGs in Rai14+/- mouse brains. Green: higher expression, magenta: lower expression, F: female, M: male. (E) Sucrose preference test. Ten- to 12-week-old male WT and Rai14+/- mice were individually housed and given a free choice between 2% sucrose solution and plain water (n = 16 for WT, 17 for Rai14+/-). (F) Porsolt’s forced swim test. Performance was assessed by comparing 10- to 12week old male WT and Rai14+/- mice (n = 10 for WT, 12 for Rai14+/-). The fractions of immobile time are shown. (G) Porsolt’s forced swim test upon anti-depressant administration. Fluoxetine (FLX, 10 mg/ kg) or saline were treated for 15 days ahead of the test (n = 9 for WT-saline, 7 for Rai14+/--saline, and 8 for Rai14+/--FLX) (H) Effects of fluoxetine (FLX) on dendritic spine density. FLX (10 mg/ kg) or saline was treated for 15 days ahead of the sampling. Representative images of Golgi-stained dendrites of cortical layer II/ III pyramidal neurons (left) and quantitative analysis of the dendritic spine density (right) are shown (n = 4 for each group, 8–12 neurons for each mouse were analyzed). (I–K) Effects of chronic restraint stress (CRS) and fluoxetine treatment (FLX). For CRS, C57BL/6 mice received two-hour of daily restraint stress procedures for 15 days. CRS + FLX group was administered CRS while receiving i.p. injections of 10 mg/ kg of FLX 10 min before each CRS session. (I) Effects of CRS and FLX on body weight gain (n = 6 for CTL, 7 for CRS, and 7 for CRS + FLX). (J) Relative Rai14 mRNA level in the prefrontal cortex of the mice prepared in (I). (K) Relative Rai14 protein level in the prefrontal cortex of the mice prepared in (I). Representative western blot image (left) and densitometric analysis of Rai14 band intensity normalized to α-tubulin (right). Error bars indicate the mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 determined by student’s t-test for (E) and (F), one-way ANOVA with Bonferroni’s multiple comparison test for (G), (H), (J), and (K), and two-way ANOVA for (I). See also Figure 5—figure supplement 1, and Figure 5—source data 1.
-
Figure 5—source data 1
Source data for RNA seq and depressive-like behaviors in Rai14+/- mice.
- https://cdn.elifesciences.org/articles/77755/elife-77755-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Uncropped western blot images with relevant bands labelled.
- https://cdn.elifesciences.org/articles/77755/elife-77755-fig5-data2-v2.pdf
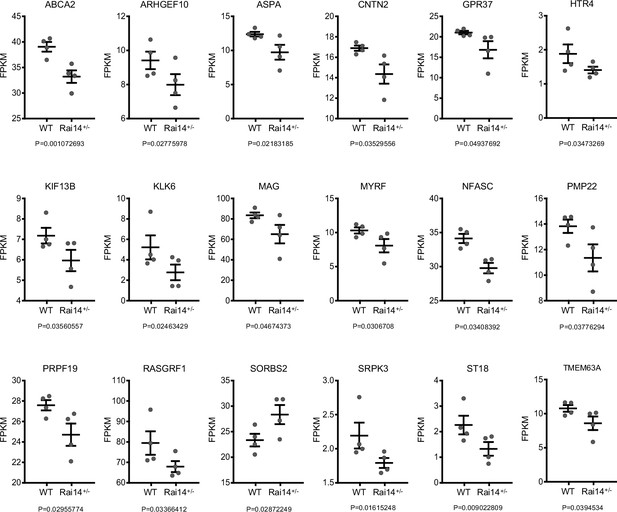
Alteration of gene expression profile in Rai14+/- mice.
FPKM values of individual genes in (Figure 5D). Data are plotted as mean ± SEM. p-Values were obtained from RNA sequencing analysis. FPKM: Fragments Per Kilobase of exon per Million.
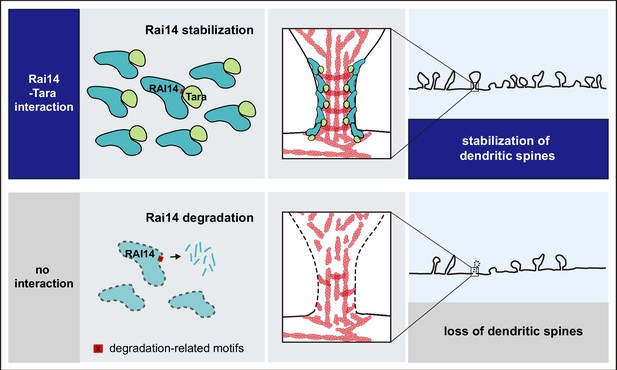
A schematic model; Tara-mediated stabilization of Rai14 for the regulation of dendritic spine dynamics Rai14-Tara interaction stabilizes Rai14 by masking degradation-related motifs within its C-terminal tip. Stabilized Rai14-Tara complex accumulates at the neck of dendritic spines via the ankyrin repeat domain of Rai14. The Rai14 cluster at the spine neck contributes to maintaining spines, thereby upregulating dendritic spine density. Rai14 deficiency leads to reduced dendritic spine density, in association with synaptic impairments relevant to depressive-like behaviors.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background(M. musculus) | IcrTac:ICR | IMSR | Cat# TAC:icr, RRID:IMSR_TAC:icr | |
| Strain, strain background(M. musculus) | C57BL/6NJ | IMSR | Cat# JAX:005304 RRID:IMSR_JAX:005304 | |
| Strain, strain background(M. musculus) | C57BL/6NJ-Rai14em1(IMPC)J | MGI | Cat# MGI:5755416 | |
| Cell line(H. sapiens) | HEK293 | ATCC | Cat# PTA-4488, RRID:CVCL_0045 | |
| Antibody | Anti-Rai14 (rabbit polyclonal) | Proteintech Group | Cat# 17507–1-AP, RRID: AB_2175992 | WB (1:1,000)IP (1:1,000) |
| Antibody | Anti-Tara (rabbit polyclonal) | Thermo Fisher Scientific | Cat# PA5-29092, RRID: AB_2546568 | WB (1:1,000) |
| Antibody | Anti-PSD95, clone 7E31B8 (mouse monoclonal) | Enzo Life Sciences | Cat# ADI-VAM-PS001-E, RRID: AB_2039457 | ICC (1:50) |
| Antibody | Anti-Synaptophysin 1, Rb7.2 (rabbit monoclonal) | Synaptic Systems | Cat# 101 008, RRID: AB_2864779 | ICC (1:200) |
| Antibody | Anti-FLAG (rabbit polyclonal) | Sigma-Aldrich | Cat# F7425, RRID: AB_439687 | WB (1:2,000)ICC (1:200) |
| Antibody | Anti-FLAG, M2 (mouse monoclonal) | Sigma-Aldrich | Cat# F1804, RRID:AB_262044 | WB (1:2,000)IP (1:1,000)ICC (1:200) |
| Antibody | Anti-GFP (rabbit polyclonal) | Molecular Probes | Cat# A-11122, RRID:AB_221569 | WB (1:3,000) |
| Antibody | Anti-GFP, B-2 (mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-9996, RRID:AB_627695 | WB (1:1,000)IP (1:200) |
| Antibody | Anti-α-tubulin, DM1A (mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-32293, RRID:AB_628412 | WB (1:1,000) |
| Antibody | Anti-α-tubulin, 1E4C11 (mouse monoclonal) | Proteintech Group | Cat# 66031–1-Ig, RRID:AB_11042766 | WB (1:2,000) |
| Antibody | Anti-c-Myc, clone 9E10 (mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-40, RRID:AB_627268 | WB (1:1,000) |
| Antibody | Rabbit IgG, polyclonal – Isotype Control (rabbit polyclonal) | Abcam | Cat# ab37415, RRID:AB_2631996 | IP (1:1,000) |
| Antibody | Sheep Anti-Mouse IgG - Horseradish Peroxidase antibody (sheep monoclonal) | GE Healthcare | Cat# NA931, RRID:AB_772210 | WB (1:7,500) |
| Antibody | Donkey Anti-Rabbit IgG, Whole Ab ECL Antibody, HRP Conjugated | GE Healthcare | Cat# NA934, RRID:AB_772206 | WB (1:7,500) |
| Antibody | Goat anti-Rabbit IgG (H + L) cross-Adsorbed antibody, Alexa Fluor 488 (goat polyclonal) | Thermo Fisher Scientific | Cat# A-11008, RRID:AB_143165 | ICC (1:200) |
| Antibody | Goat Anti-Rabbit IgG (H + L) Antibody, Alexa Fluor 568 Conjugated (goat polyclonal) | Molecular Probes | Cat# A-11011, RRID:AB_143157 | ICC (1:200) |
| Antibody | Goat anti-rabbit IgG, Flamma 648 (goat polyclonal) | BioActs | Cat# RSA1261 | ICC (1:200) |
| Antibody | Goat anti-mouse IgG (H + L) cross-Adsobed Secondary antibody, Alexa Fluor 488 (goat polyclonal) | Molecular Probes | Cat#A-11001,RRID: AB_2534069 | ICC (1:200) |
| Antibody | Goat Anti-Mouse IgG (H + L) Antibody, Alexa Fluor 568 Conjugated (goat polyclonal) | Molecular Probes | Cat# A-11004, RRID:AB_141371 | ICC (1:200) |
| Antibody | Goat Anti-Mouse IgG (H + L) Antibody, Alexa Fluor 647 Conjugated (goat polyclonal) | Molecular Probes | Cat# A-21235, RRID:AB_141693 | ICC (1:200) |
| Recombinant DNA reagent | pEGFP-N1 | Clontech | Cat# 6085–1 | |
| Recombinant DNA reagent | pEGFP-C3 | Clontech | Cat# 6082–1 | |
| Recombinant DNA reagent | pFLAG-CMV2 | Sigma-Aldrich | Cat# E7033 | |
| Recombinant DNA reagent | pcDNA3.1/myc-His | Invitrogen | Cat# V80020 | |
| Recombinant DNA reagent | pDsRed2-N1 | Clontech | Cat# 632,406 | |
| Recombinant DNA reagent | hRai14-EGFP | This paper | N/A | Subcloned from EGFP-hRai14 |
| Recombinant DNA reagent | EGFP-hRai14 | This paper | N/A | Insertion of hRai14 CDS into pEGFP-C3 |
| Recombinant DNA reagent | FLAG-hRai14 | This paper | N/A | Insertion of hRai14 CDS into pFLAG-CMV2 |
| Recombinant DNA reagent | hRai14 Δ401-600-EGFP | This paper | N/A | Subcloned from hRai14-EGFP with fusion-PCR method |
| Recombinant DNA reagent | hRai14 Δ601-800-EGFP | This paper | N/A | Subcloned from hRai14-EGFP with fusion-PCR method |
| Recombinant DNA reagent | hRai14 Δ801-980-EGFP | This paper | N/A | Subcloned from hRai14-EGFP with fusion-PCR method |
| Recombinant DNA reagent | hRai14 Δ801-860-EGFP | This paper | N/A | Subcloned from hRai14-EGFP with fusion-PCR method |
| Recombinant DNA reagent | hRai14 Δ861-920-EGFP | This paper | N/A | Subcloned from hRai14-EGFP with fusion-PCR method |
| Recombinant DNA reagent | hRai14 Δ921-980-EGFP | This paper | N/A | Subcloned from hRai14-EGFP with fusion-PCR method |
| Recombinant DNA reagent | hRai14 Δ921-940-EGFP | This paper | N/A | Subcloned from hRai14-EGFP with fusion-PCR method |
| Recombinant DNA reagent | hRai14 Δ941-960-EGFP | This paper | N/A | Subcloned from hRai14-EGFP with fusion-PCR method |
| Recombinant DNA reagent | hRai14 Δ961-980-EGFP | This paper | N/A | Subcloned from hRai14-EGFP with fusion-PCR method |
| Recombinant DNA reagent | hRai14 Δ948-967-EGFP | This paper | N/A | Subcloned from hRai14-EGFP with fusion-PCR method |
| Recombinant DNA reagent | hRai14 ΔANK -EGFP | This paper | N/A | Subcloned from hRai14-EGFP with fusion-PCR method |
| Recombinant DNA reagent | EGFP-hTara | Woo et al., 2019 (PMID:31815665) | N/A | |
| Recombinant DNA reagent | FLAG-hTara | Woo et al., 2019 (PMID:31815665) | N/A | |
| Recombinant DNA reagent | hTara-Myc | Woo et al., 2019 (PMID:31815665) | N/A | |
| Recombinant DNA reagent | EGFP-hTara 1-160 | This paper | N/A | Subcloned from EGFP-hTara |
| Recombinant DNA reagent | EGFP-hTara 161-499 | This paper | N/A | Subcloned from EGFP-hTara |
| Recombinant DNA reagent | EGFP-hTara 500-593 | This paper | N/A | Subcloned from EGFP-hTara |
| Recombinant DNA reagent | EGFP-hTara 241-330 | This paper | N/A | Subcloned from EGFP-hTara |
| Recombinant DNA reagent | EGFP-hTara Δ161-240 | This paper | N/A | Subcloned from EGFP-hTara with fusion-PCR method |
| Recombinant DNA reagent | EGFP-hTara Δ241-330 | This paper | N/A | Subcloned from EGFP-hTara with fusion-PCR method |
| Recombinant DNA reagent | EGFP-hTara Δ331-412 | This paper | N/A | Subcloned from EGFP-hTara with fusion-PCR method |
| Recombinant DNA reagent | EGFP-hTara Δ413-499 | Woo et al., 2019 (PMID:31815665) | N/A | |
| Recombinant DNA reagent | RFP-N1-LifeAct | Woo et al., 2019 (PMID:31815665) | N/A | |
| Recombinant DNA reagent | EGFP-N1-LifeAct | This paper | N/A | Subcloned from RFP-N1-LifeAct |
| Recombinant DNA reagent | pLL3.7-scrambled shRNA-EGFP | Woo et al., 2019 (PMID:31815665) | N/A | |
| Recombinant DNA reagent | pLL3.7-hTara shRNA-EGFP | Woo et al., 2019 (PMID:31815665) | N/A | |
| Recombinant DNA reagent | pLL3.7-mTara shRNA-EGFP | This paper | N/A | Core sequence: GAAGGAGAATGAACTCCAGTA |
| Recombinant DNA reagent | pLL3.7-hRai14 shRNA-EGFP | This paper | N/A | Core sequence: TCGGGAAAGGAATCGGTATTT |
| Recombinant DNA reagent | pLL3.7-mRai14 shRNA-EGFP | This paper | N/A | Core sequence: CGAACACTGTGGACGCCTTAA |
| Commercial assay or kit | EndoFree plasmid maxi kit | Qiagen | Cat# 12,362 | |
| Commercial assay or kit | FD Rapid GolgiStainTM Kit | FD Neurotechnologies | Cat# PK401 | |
| Commercial assay or kit | MAX Efficiency DH5α Competent Cells | Invitrogen | Cat# 18258012 | |
| Chemical compound, drug | Ara-C (Cytosine β-D-arabinofuranoside) | Sigma-Aldrich | C1768 | |
| Chemical compound, drug | B27 supplement | Gibco | Cat# 17504044 | |
| Chemical compound, drug | Clarity Western ECL Substrate | Bio-Rad | Cat# 1705061 | |
| Chemical compound, drug | Complete Protease Inhibitor Cocktail | Roche | Cat# 11697498001 | |
| Chemical compound, drug | DNase I | Sigma-Aldrich | Cat# DN25 | |
| Chemical compound, drug | fetal bovine serum (FBS) | Gibco | Cat# 10082147 | |
| Chemical compound, drug | Fluoxetine hydrochloride | Sigma-Aldrich | Cat# 1279804 | |
| Chemical compound, drug | Ketamine hydrochloride | Yuhan Corporation | N/A | |
| Chemical compound, drug | Laminin | Corning | Cat# 354,239 | |
| Chemical compound, drug | Latrunculin A | Cayman Chemical | Cat# CAY-10010630–2 | |
| Chemical compound, drug | Lipofectamine 2000 | Invitrogen | Cat# 11668019 | |
| Chemical compound, drug | penicillin/streptomycin | Gibco | Cat# 15140122 | |
| Chemical compound, drug | Poly-D-lysine hydrobromide | Sigma-Aldrich | Cat# P6407 | |
| Chemical compound, drug | Polyethylenimine | Polysciences | Cat# 23,966 | |
| Chemical compound, drug | RNAlaterTM Solution | Invitrogen | Cat# AM7020 | |
| Chemical compound, drug | Surgipath FSC22 Clear OCT solution | Leica Biosystems | Cat# FSC22 | |
| Chemical compound, drug | Vivamagic | Vivagen | Cat# VM001 | |
| Chemical compound, drug | Xylazine | Bayer AG | N/A | |
| Software, algorithm | ImageJ (Fiji) | Schindelin et al., 2012 | RRID:SCR_002285 | |
| Software, algorithm | Imaris | Bitplane | RRID:SCR_007370 | |
| Software, algorithm | Olympus cellSens Software | Olympus | RRID:SCR_016238 | |
| Software, algorithm | GraphPad Prism | GraphPad | RRID:SCR_002798 | |
| Other | ECM 830 Square Wave Electroporation System | Harvard Apparatus | Cat# W3 45–0052 | Materials and methods – In utero electroporation |
| Other | Leica VT1000S vibrating blade microtome | Leica Microsystems | N/A | Materials and methods – Golgi-Cox impregnation |
| Other | Olympus Confocal Laser Scanning Microscope Fluoview FV3000 | Olympus | RRID:SCR_017015 | Materials and methods – Microscopy, Time-lapse imaging of live neurons |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/77755/elife-77755-transrepform1-v2.docx
-
Source data 1
- https://cdn.elifesciences.org/articles/77755/elife-77755-data1-v2.zip
-
Source data 2
Unedited raw western blot images in Figure 1—figure supplement 1, Figure 2—figure supplements 1 and 2.
- https://cdn.elifesciences.org/articles/77755/elife-77755-data2-v2.zip
-
Source data 3
Unedited raw western blot images in Figure 2—figure supplement 3, and Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/77755/elife-77755-data3-v2.zip





