CriSNPr, a single interface for the curated and de novo design of gRNAs for CRISPR diagnostics using diverse Cas systems
Figures
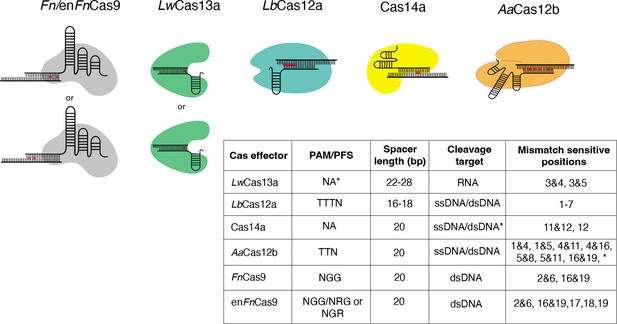
Nucleotide mismatch sensitive crRNA positions have been reported for a variety of Cas systems.
These positions are included in CRISPR-based SNP recognition (CriSNPr) for LwCas13a, LbCas12a, Cas14a, AaCas12b, FnCas9, and enFnCas9, respectively. A table summarizing the protospacer adjacent motif (PAM)/protospacer flanking site (PFS) for each Cas protein is shown, along with mismatched sensitive positions reported in the literature. *LwCas13a does not require PFS when targeting the mammalian genome. *Cas14a cleaves ssDNA without PAM, but dsDNA requires TTTA PAM. *AaCas12b has shown mismatch sensitivity for some other nucleotide positions as well, but they are not included here because the discrimination between wild-type (WT) and mutant is insufficient.
-
Figure 1—source data 1
A table summarizing various Cas proteins previously reported to have mismatched sensitive crRNA nucleotide positions.
- https://cdn.elifesciences.org/articles/77976/elife-77976-fig1-data1-v2.zip
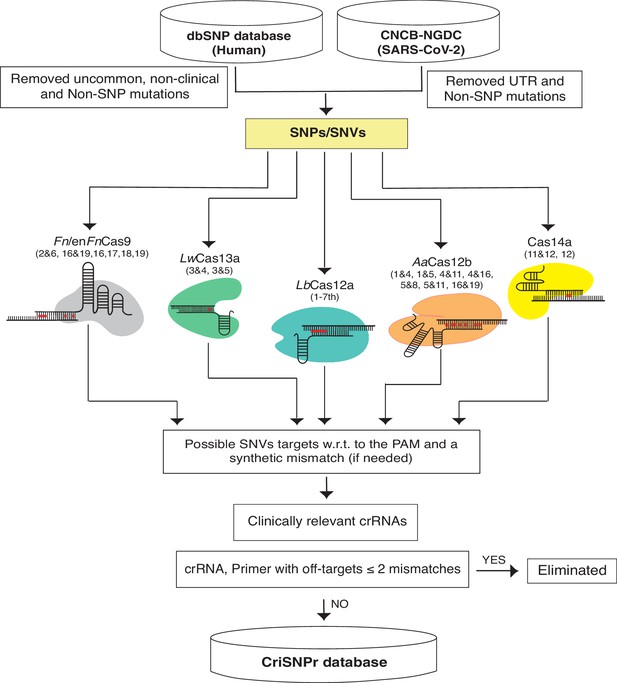
Schematic representation of CRISPR-based SNP recognition (CriSNPr) database curation and the clinically relevant SNP Database (dbSNP) variations were filtered for uncommon and non-SNP mutations and SARS-CoV-2 in non-UTR single nucleotide variants (SNVs).
The filtered SNPs were then checked for targetability by individual Cas systems based on mismatch sensitivity with or without protospacer adjacent motif (PAM). The genome coordinates of target SNPs aided in the acquisition of gene IDs as well as SNP flanking sequences for oligo synthesis, resulting in the creation of an SQLite database. The off-targets were evaluated against representative bacteria, viruses, the human genome, and the transcriptome.
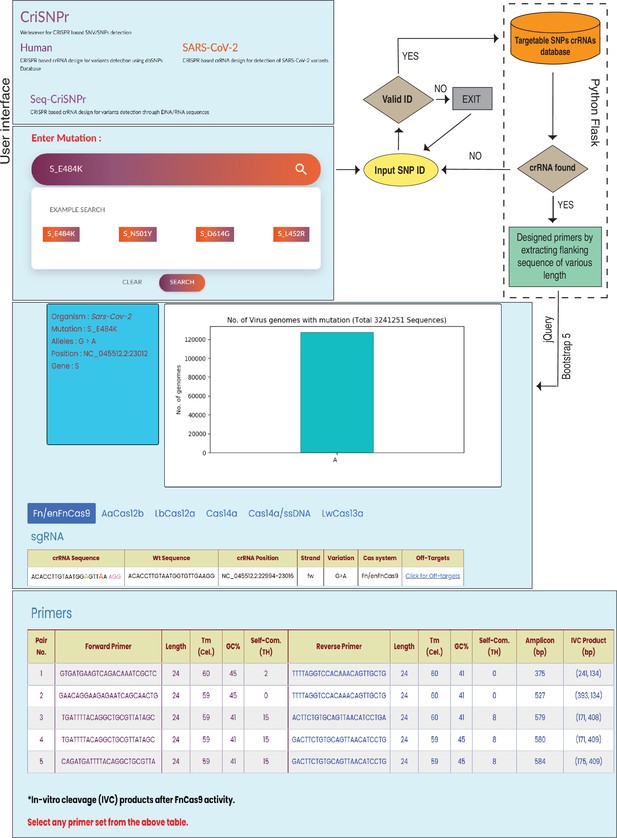
Workflow of the CRISPR-based SNP recognition (CriSNPr) web server.
CriSNPr user interface displays human, SARS-CoV-2, and Seq-CriSNPr subdomains, each of which accepts rsID, mutant amino acid position, and single nucleotide variant (SNV) containing 20–30 nt sequences as inputs. With a valid input, the server will look for matching crRNA sequences in the database created with the Python Flask framework. The results include the sequences of the amplification and SNV-detection primers, the allelic distribution of the SNVs, the crRNAs, and the off-targets.
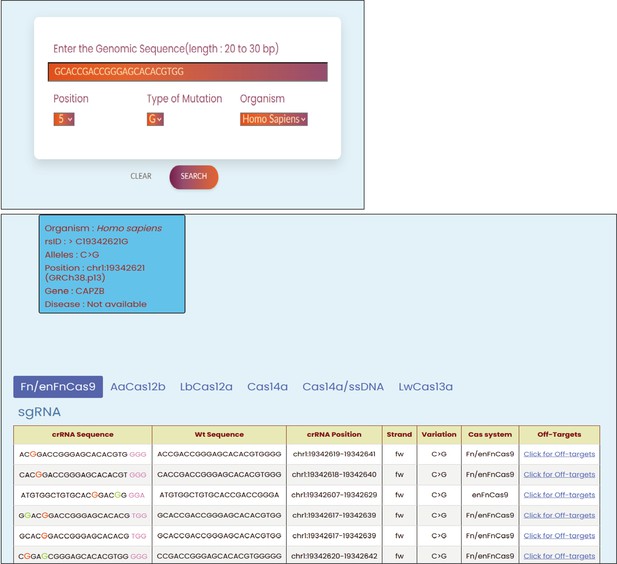
Seq-CRISPR-based SNP recognition (CriSNPr) allows for the entry of any 20–30 nt.
Sequences related to the human or SARS-CoV-2 genomes, as well as single nucleotide variant (SNV) nucleotide position and identity in the query sequence, in order to produce designing sequence results. For user inputs, representative examples are shown.
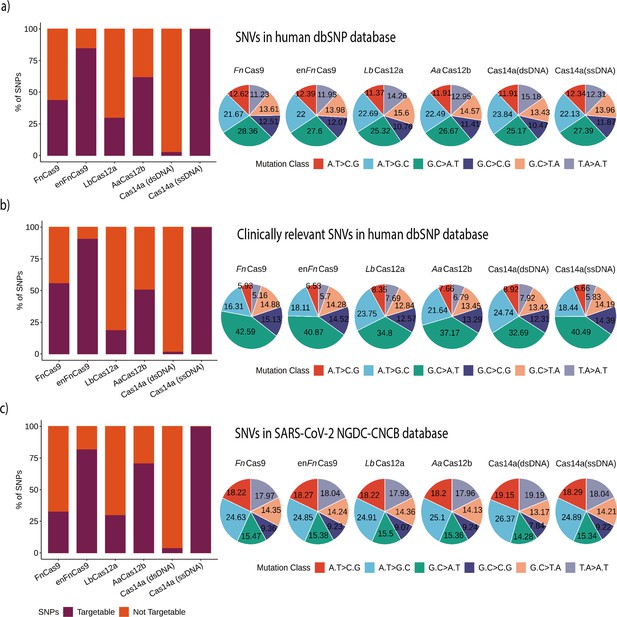
Various Cas systems targeting SNPs/single nucleotide variants (SNVs) in SNP Database (dbSNP) and SARS-CoV-2 genomes.
(a) Shows percent SNP targets for different Cas-systems across the dbSNP, as well as the base distribution of targeted SNPs by individual Cas-system. (b) The percentage of targeted SNPs that have clinical significance or disease relevance in humans, with a percentage base distribution at each SNP position targeted by each Cas-system. (c) The percentage of targeted SNPs in SARS-CoV-2 genomes reported in the GISAID Database, along with the percentage base distribution at each SNP position targeted by each Cas-system. In all bar plots, red depicts the percentage of non-targeting SNVs, while violet indicates the percentage of SNVs that can be targeted.
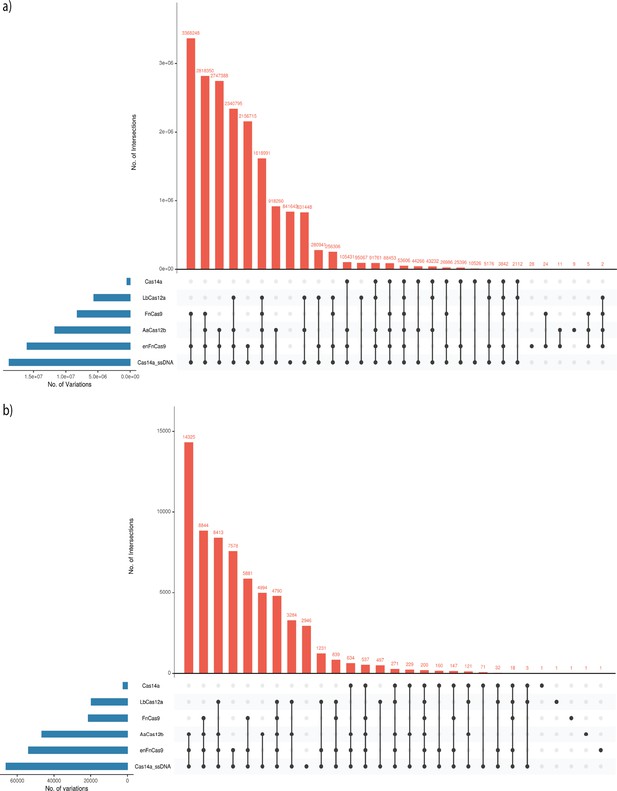
Upset plots of the intersection of the targetable variation of various Cas systems.
(a) Human SNP Databases (dbSNPs) with targetable variations. (b) SARS-CoV-2 CNCB-NGDC variations.
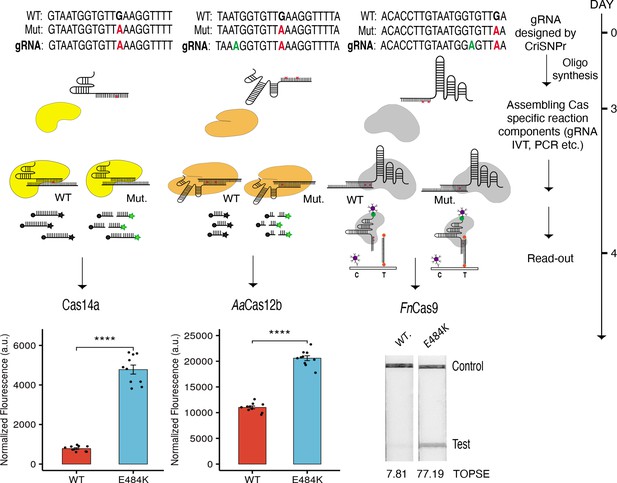
CRISPR-based SNP recognition (CriSNPr) designed guide RNAs (gRNAs) can discriminate SARS-CoV-2 single nucleotide variant (SNV) with multiple Cas proteins.
crRNA sequences designed by CriSNPr for SARS-CoV-2 E484K variant detection by Cas14a, AaCas12b, and FnCas9 can successfully discriminate between wild-type and mutant sequences. A possible implementation schedule for the assays is depicted to the right. SEM, student paired T-test p values **** 0.0001 (dots represent values from independent measurements, n=10).
-
Figure 5—source data 1
The red rectangle denotes the approximate area cropped from the LFA strips for generating Figure 5.
- https://cdn.elifesciences.org/articles/77976/elife-77976-fig5-data1-v2.docx
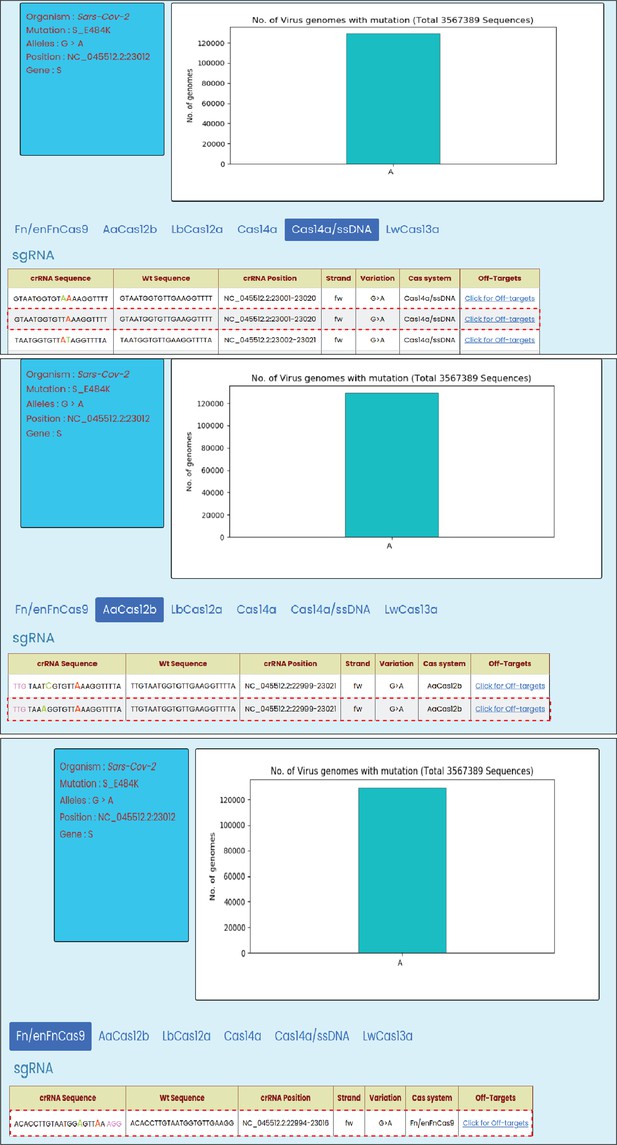
Guide RNA (gRNA) sequences designed by CRISPR-based SNP recognition (CriSNPr) for the detection of the SARS-CoV-2 E484K mutation using Cas14a, AaCas12b, and FnCas9, respectively, as shown in Figure 4 (denoted with a red dotted box).
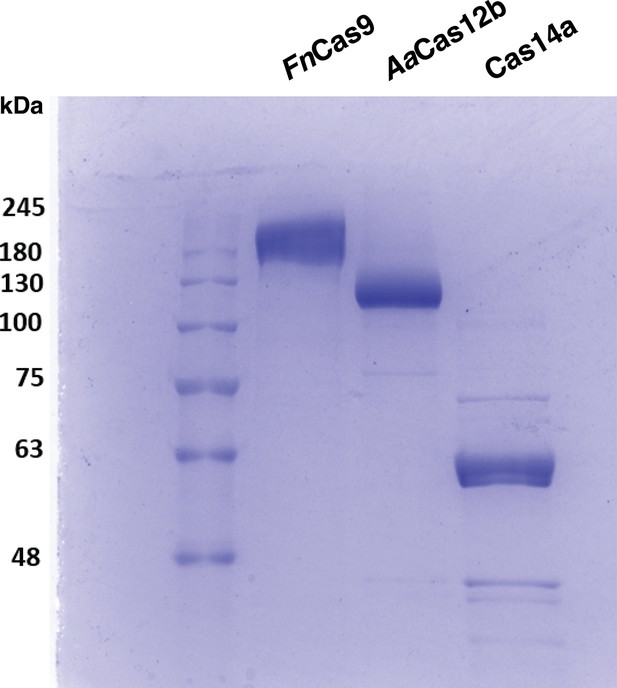
PAGE gel showing purified FnCas9 (~190kDa), AaCas12b (~130kDa), and Cas14a (~61kDa) proteins.
-
Figure 5—figure supplement 2—source data 1
The red rectangle denotes the approximate area cropped from the PAGE gel for generating Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/77976/elife-77976-fig5-figsupp2-data1-v2.zip
-
Figure 5—figure supplement 2—source data 2
Original uncropped PAGE gel of purified FnCas9 (190 kDa), AaCas12b (130 kDa), and Cas14a (61 kDa) proteins.
- https://cdn.elifesciences.org/articles/77976/elife-77976-fig5-figsupp2-data2-v2.zip
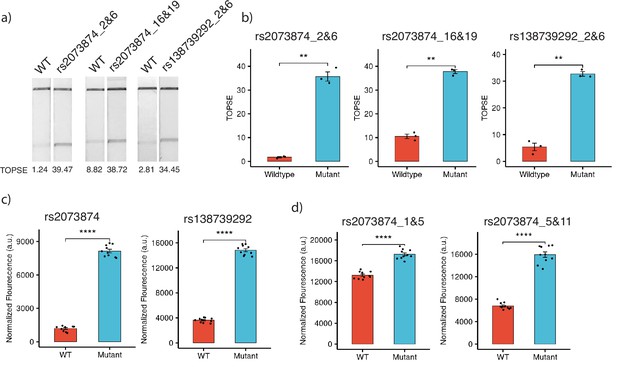
Detection of clinically important human SNPs by CRISPR-based SNP recognition (CriSNPr)-designed guide RNAs (gRNAs) for different Cas proteins.
(a) FnCas9-based detection of WT (wild-type) and mutant sequences containing rs2073874 and rs138739292 using CriSNPr-designed 2 and 6 and 16 and 19 and 2 and 6 position modified crRNAs, respectively. (b) Quantified TOPSE (True Outcome Predicted via Strip Evaluation) intensity values for rs2073874 and rs138739292 detection by FnCas9, SEM, student paired T-test p values ** 0.01 (dots represent independent measurements, n=3). (c) Detection of WT as well as rs2073874 and rs138739292 containing ssDNA sequences using CriSNPr designed gRNAs for Cas14a (dots represent independent measurements, n=10). (d) CriSNPr generated 1 and 5 and 5 and 11 modified crRNAs for use with AaCas12b to distinguish between WT and rs2073874 ssDNA sequences. **** 0.0001 SEM, student paired T-test p values (dots represent independent measurements, n=10).
-
Figure 6—source data 1
The red rectangle denotes the approximate area cropped from the LFA strips for generating Figure 6a.
- https://cdn.elifesciences.org/articles/77976/elife-77976-fig6-data1-v2.docx
Additional files
-
Supplementary file 1
Variation statistics from human SNP Database (dbSNP) for various Cas systems.
- https://cdn.elifesciences.org/articles/77976/elife-77976-supp1-v2.xlsx
-
Supplementary file 2
Variation statistics from SARS-CoV-2 CNCB-NGDC database for various Cas systems.
- https://cdn.elifesciences.org/articles/77976/elife-77976-supp2-v2.xlsx
-
Supplementary file 3
Comparison between different sgRNA designing tools for single nucleotide variants (SNVs) in a sequence.
- https://cdn.elifesciences.org/articles/77976/elife-77976-supp3-v2.xlsx
-
Supplementary file 4
List of oligos used in this study.
- https://cdn.elifesciences.org/articles/77976/elife-77976-supp4-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/77976/elife-77976-transrepform1-v2.docx





