Identification of HIV-reservoir cells with reduced susceptibility to antibody-dependent immune response
Figures
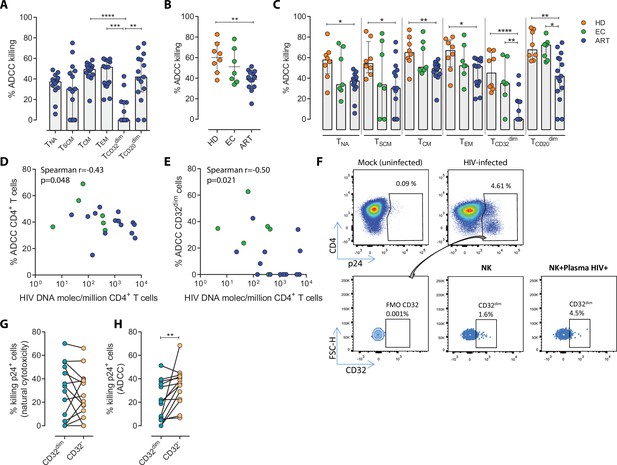
Susceptibility of CD4+ T cell subsets to the Natural Killer (NK) immune response.
The susceptibility of different cell subpopulations that compose the HIV-reservoir to Natural Cytotoxicity (NC) and Antibody-Dependent Cell Cytotoxicity (ADCC) mediated by NK cells was measured by performing functional assays. (A) Percentage of gp120-coated cells killed by ADCC after being exposed to HIV-specific immunoglobulins (Igs) in the presence of NK cells. The intrinsic susceptibility to ADCC was measured in Naïve (TNA), Stem Cell Memory (TSCM), Central Memory (TCM), Effector Memory (TEM), TCD32dim, and TCD20dim subsets. Statistical comparisons were performed using the ANOVA Friedman with Dunn’s multiple comparison test. Median with interquartile range is shown. (B) Percentage of total gp120-coated CD4+ T cells from different cohorts of patients killed by ADCC. Healthy donors (HD), Elite Controllers (EC), and antiretroviral-treated (ART) PLWH. Statistical comparisons were performed using one-way ANOVA with Tukey’s multiple comparison test. Median with interquartile range are shown. (C) Percentage of cell subsets killed by ADCC in cells from HD, EC, and ART. Statistical comparisons were performed using the one-way ANOVA with Tukey’s multiple comparison test. Median with interquartile range is shown. (D–E) Spearman correlations between the size of the HIV-reservoir measured as total HIV-DNA in samples from ART-suppressed PLWH, and the potency of autologous NK cells to kill (D) total CD4+ T cells or (E) TCD32dim cells by ADCC. (F) Representative flow cytometry gating strategy used to quantify HIV infection after ex vivo infection with BaL or NL4.3. Fluorescence minus one (FMO) control was used to determine CD32 expression. Cells were infected for 5 days and the frequency of expression of CD32 on HIV-infected cells was measured for each condition. (G) Percentage of killing by NC of ex vivo HIV-infected TCD32dim and TCD32− cells mediated by autologous NK cells from ART-treated PLWH (n=14). Killing was calculated by normalizing the proportion of each subset within the p24+ fraction in the co-culture condition to the basal condition. (H) Percentage of ADCC killing of ex vivo HIV-infected TCD32dim and TCD32− cells mediated by autologous NK cells from ART-treated PLWH (n=14). Killing was calculated by normalizing the proportion of each subset within the p24+ fraction in the co-culture condition with plasma to the co-culture without plasma. Statistical comparisons were performed using the Wilcoxon matched-pairs signed-rank test. *p<0.05; **p<0.01.
-
Figure 1—source data 1
This file contains the source data used to generate Figure 1.
- https://cdn.elifesciences.org/articles/78294/elife-78294-fig1-data1-v2.xlsx
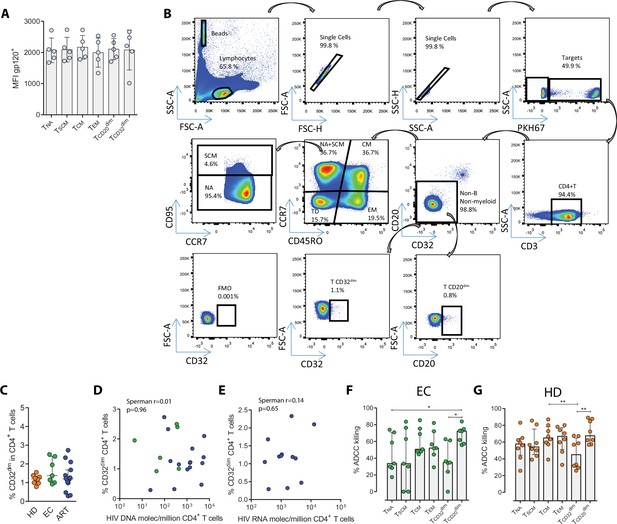
Gp120 cell coating efficiency, gating strategy for the NK-killing assays, and percentages of antibody-dependent cell cytotoxicity (ADCC) killing in elite controllers (EC), and healthy donors (HD).
(A) Cell coating with recombinant gp120 HIV protein. Detection was performed by flow cytometry using the anti-gp120 antibody A32, and a FITC-labeled anti-human secondary antibody. Mean Fluorescence Intensity (MFI) values are shown. (B) Gating strategy used to identify cell killing after the ADCC assay in the different cell subsets. Cell doublets were excluded by forward and side scatter signals and B and myeloid cells discarded based on their high expression of CD20 and CD32. Beads for absolute cell counting were included to calculate ADCC killing by measuring the disappearance of cells in each cell subset. (C) Frequency of expression measured by flow cytometry of CD32 on CD4+ T cells from healthy donors (HD, n=8), Elite controllers (EC, n=7) and ART-treated and virologically-suppressed PLWH (ART, n=15). Median with interquartile range are shown. Statistical comparisons were performed using the Mann-Whitney test. ***p<0.001; ****p<0.0001. (D–E) Correlations of the HIV-reservoir size and CD32 expression. Spearman correlations are shown in samples from HIV-infected individuals. ART-treated and EC are shown in blue and green dots, respectively. (D) Spearman correlation between the total HIV DNA reservoir size and the frequency of expression of CD32 in CD4+ T cells. ART-treated and EC are shown in blue and green dots, respectively. (E) Spearman correlation between HIV RNA levels in CD4+ T cells and the frequency of expression of CD32 in CD4+ T cells. (F–G) Intrinsic susceptibility to NK-mediated ADCC of Naïve (TNA), Stem Cell Memory (TSCM), Central Memory (TCM), Effector Memory (TEM), TCD32dim, and TCD20dim subsets in (F) EC, and (G) HD. Median with interquartile range is shown. Statistical comparisons were performed using the ANOVA Friedman test. *p<0.05; **p<0.01.
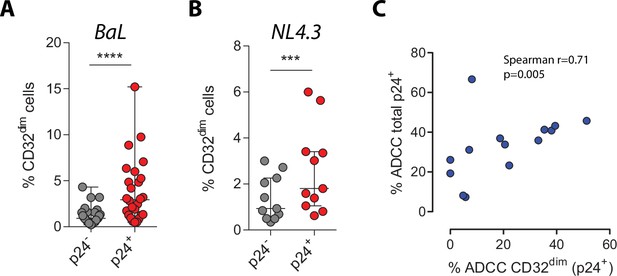
Percentage of CD32 expression on uninfected cells (p24−) and HIV-infected cells (p24+) with BaL (n=26) in (A) or with NL4.3 (n=11) in (B) after 5 days of infection.
Median with interquartile range are shown. Statistical analyses consisted of the Wilcoxon matched-pairs signed-rank test. (C) Spearman correlation between the percentage of ADCC-killing of the total HIV-infected CD4+ T cells and the percentage of ADCC-killing of the TCD32dim subset is shown.
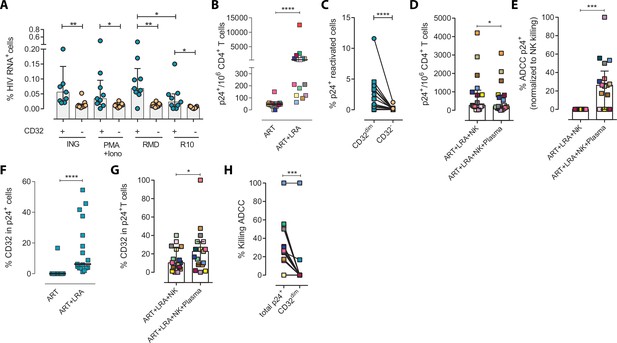
Expression of HIV in TCD32dim reservoir cells after latency disruption and susceptibility to natural killer (NK) immune responses.
Data from the direct ex vivo reactivation of the natural HIV reservoir in ART-suppressed PLWH. (A) Percentage of HIV-RNA expressing cells, measured by the RNA FISH-flow assay, within the TCD32dim and TCD32− subsets after viral reactivation with Ingenol, PMA/ionomycin, or romidepsin (n=9). Statistical comparisons were performed using the Wilcoxon matched-pairs signed-rank test (comparison between CD32dim and CD32− within each drug condition), and the ANOVA Friedman with Dunn’s multiple comparison test (comparison between different drug conditions of the cell subset). Median with interquartile range is represented. (B) Percentage of p24+ cells after 18 hr viral reactivation with PMA/ionomycin (n=17). Each participant is represented by a different color. Statistical comparisons were performed using the Wilcoxon matched-pairs signed-rank test. (C) Frequency of viral reactivation within the total pool of TCD32dim and TCD32− cells (n=17). Statistical comparisons were performed using the Wilcoxon matched-pairs signed-rank test. (D) NK killing assays against viral reactivated cells. Number of p24+ cells per million CD4+ T cells after the addition of NK cells only or together with the autologous plasma is shown (n=17). Statistical comparisons were performed using the Wilcoxon matched-pairs signed-rank test. (E) Percentage of ADCC in p24+ cells normalized to the NC control (ART + LRA + NK). Statistical comparisons were performed using one-sample t-test. (F) Percentage of CD32 expression within the total p24+ pool before and after HIV reactivation (n=17). Statistical comparisons were performed using the Wilcoxon matched-pairs signed-rank test. (G) Percentage of TCD32dim within p24+ cells after HIV reactivation and functional NK-mediated assays (n=17). Statistical comparisons were performed using the Wilcoxon matched-pairs signed-rank test. (H) Percentage of NK-mediated killing by ADCC of the reactivated TCD32dim or total p24+ cells (n=17). ADCC was calculated as the reduction of p24+ cells after the co-culture with NK and plasma and normalized to the condition with NK cells alone. Statistical comparisons were performed using the Wilcoxon matched-pairs signed-rank test. *p<0.05; **p<0.01; ***p<0.001.
-
Figure 2—source data 1
This file contains the source data used to generate Figure 2.
- https://cdn.elifesciences.org/articles/78294/elife-78294-fig2-data1-v2.xlsx
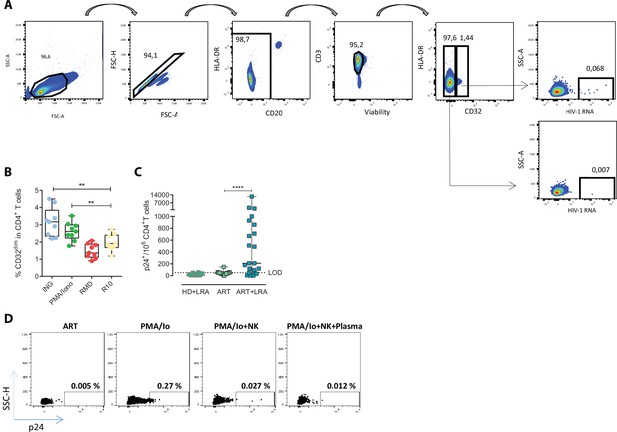
Expression of HIV-RNA and viral protein p24 in TCD32dim reservoir cells after viral reactivation.
(A) Flow cytometry gating strategy used for the identification of TCD32dim cells expressing HIV-RNA after the RNA FISH-flow protocol. (B) Percentage of CD32 expression within the total CD4+ T cells before and after LRA treatment, using samples from ART-suppressed PLWH subjected to RNA FISH-flow assay. Statistical comparisons consisted of the Wilcoxon matched-pairs signed-rank test. **p<0.01. (C) p24+ cells in samples from ART-suppressed individuals at baseline and after viral reactivation with PMA/ionomycin (LRA). The limit of detection is set up at 53 copies/million cells calculated by the formula 3*SD of the mean percentage of p24+ cells detected in healthy donor (HD) samples. (D) Representative flow cytometry plots of viral reactivation levels (measured as p24+ cells) after LRA treatment and natural killer (NK) functional assays.
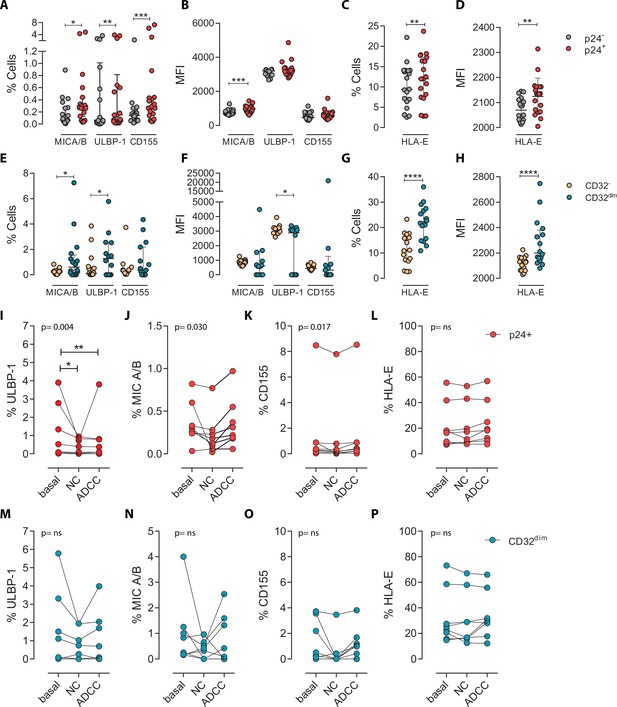
Expression of NK-ligands in cells resistant to NK-mediated killing.
HIV-infected cells from healthy donors were subjected to NK-killing assays and the percentage of expression of different NK-ligands was measured by flow cytometry in different fractions (CD32− and CD32dim) of infected (p24+) or uninfected (p24−) cells. (A–H) Expression of NK-ligands before performing the killing assays (n=19). (A) Percentage of CD4+ T cells expressing MIC A/B, ULBP-1, and CD155. (B) Mean Fluorescence Intensity (MFI) values for the expression of MIC A/B, ULBP-1, and CD155 on CD4+ T cells. (C) Percentage of CD4+ T cells expressing the MHC molecule HLA-E. (D) MFI values for HLA-E expression on CD4+ T cells. All graphs show median with interquartile range and the statistical comparisons were performed using the Wilcoxon matched-pairs signed-rank test (comparison between p24+ and p24−). (E–H) Same analyses as A–D but showing HIV-infected CD32dim and CD32- cells. (I–P) Expression of NK-ligands on HIV-infected cells not killed by the different NK-killing mechanisms. Natural Cytotoxicity (NC) and Antibody-Dependent Cell Cytotoxicity (ADCC). (I–L) Expression of NK-ligands on total infected CD4+ T cells before and after NK killing. (M–P) Expression of NK-ligands on infected TCD32dim cells before and after NK killing. All I-P graphs show median with interquartile range. p values shown in the graphs represent ANOVA Friedman test, and asterisks denote the multiple comparison Dunn’s test.*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
-
Figure 3—source data 1
This file contains the source data used to generate Figure 3.
- https://cdn.elifesciences.org/articles/78294/elife-78294-fig3-data1-v2.xlsx
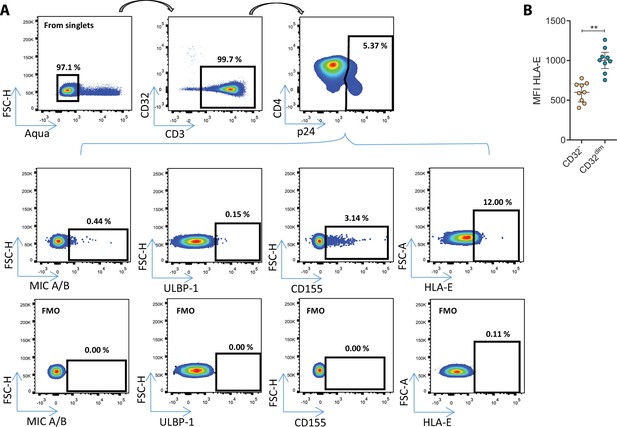
NK-ligands on HIV-infected cells.
(A) Representative flow cytometry gating strategy used to quantify the expression of MIC A/B, ULBP-1, CD155, and HLA-E ligands on HIV-infected cells after ex vivo infection. FMO controls are also shown. (B) Mean Fluorescence Intensity (MFI) values of HLA-E expression on CD4+ T cells in the absence of Human immunodeficiency virus (HIV) infection. Median with interquartile range is shown. Statistical comparison consisted of the Wilcoxon matched-pairs signed-rank test. **p<0.01.
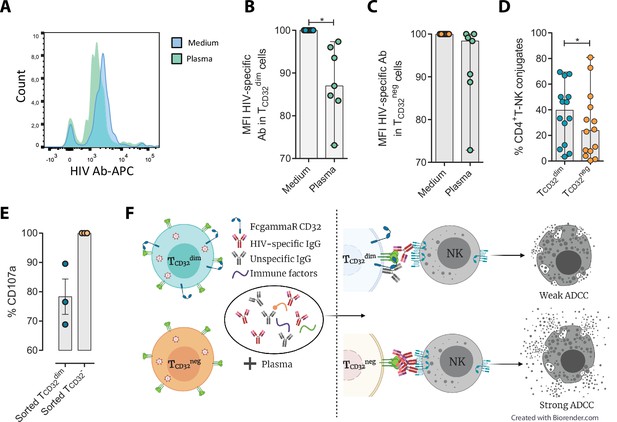
Reduced binding of HIV-specific antibodies to TCD32dim cells and the effect on natural killer (NK) degranulation.
The binding capability of the HIVgp120-specific IgG A32, an antibody (Ab) labeled with allophycocyanin (APC), to gp120-coated TCD32dim and TCD32− cells from healthy donors, before and after incubation with plasma containing non-HIV specific IgGs, was measured by flow cytometry (n=7). Percentage of the Mean Fluorescence Intensity (MFI) signal, normalized to the medium, for A32+ cells after plasma addition is shown in (A) Representative histogram of A32+ TCD32dim cells, (B) TCD32dim and (C) TCD32− cells. (D) Percentage of cell conjugates between ex vivo HIV-infected TCD32dim or TCD32− and NK cells after performing antibody-dependent cell cytotoxicity (ADCC) assays (n=14). (E) Percentage of NK degranulation (CD107a marker) in cell conjugates with sorted TCD32dim or TCD32−-coated with gp120 HIV protein and incubated with plasma HIV+, after a 4 hr antibody-dependent cell cytotoxicity (ADCC) activation assay (n=3). Values are normalized to the CD32 population (F) Schematic illustration of the impaired ADCC response against TCD32dim cells. Graphs show median with range and statistical comparisons were performed using Wilcoxon matched-pairs signed-rank test. *p<0.05.
-
Figure 4—source data 1
This file contains the source data used to generate Figure 4.
- https://cdn.elifesciences.org/articles/78294/elife-78294-fig4-data1-v2.xlsx
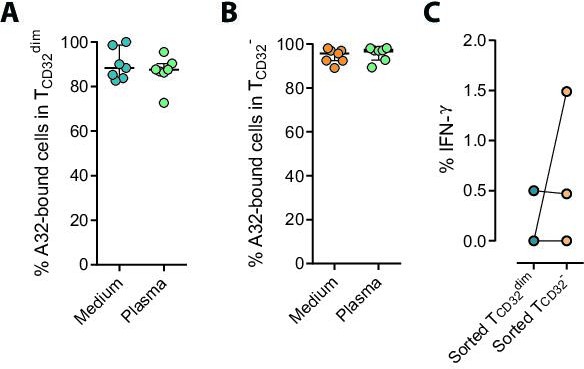
Binding of A32 to T cells and percentage of IFN-γ in cell conjugates.
The binding capability of the A32 HIV-specific antibody (APC-labeled) to gp120-coated TCD32dim and TCD32− before and after incubation with plasma containing non-HIV-specific IgGs, was measured by flow cytometry. (A) Percentage of A32+ TCD32dim cells. (B) Percentage of A32+ TCD32− cells. (C) Percentage of IFN-γ+ NK cells in cell conjugates with sorted TCD32dim and TCD32− cells after ADCC assays. Statistical comparisons were performed using the Wilcoxon matched-pairs signed-rank test.supplementary information.
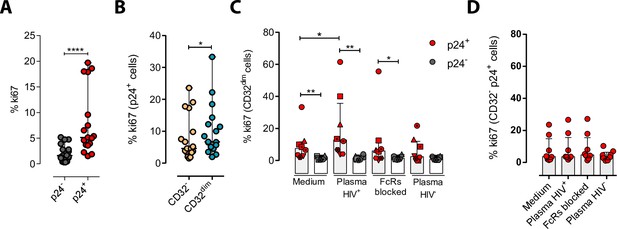
Immune complexes engagement with CD32 induces proliferation in HIV-i-nfected CD32dim cells.
Cells from healthy donors were infected with the viral strain HIVBaL and 5 days post-infection Ki67 expression was measured by flow cytometry. (A) Expression of the proliferation marker Ki67 in uninfected or ex vivo HIV-infected CD4+ T cells. Median and ranges are shown (n=17). Statistical comparisons consisted of the Wilcoxon matched-pairs signed-rank test. (B) Percentage of Ki67+ cells in HIV-infected TCD32dim and TCD32− subsets are shown (n=17). Median and ranges are shown. Statistical comparisons consisted of the Wilcoxon matched-pairs signed-rank test. (C) Percentage of Ki67 expression on TCD32dim cells after immune complexes engagement (Plasma HIV+). Medium alone, FcRs blockers and plasma from an HIV-negative individual were included as controls. Median with interquartile range is shown (n=8). Statistical comparisons were performed using the Wilcoxon matched-pairs signed-rank test (comparison between p24+ and p24− within each condition), and the ANOVA Friedman with Dunn’s multiple comparison test (comparison between different conditions of the p24+ or p24− cells). (D) Percentage of Ki67 expression on HIV-infected TCD32− cells incubated under the same experimental conditions as shown in (C). Median with interquartile range is represented (n=8). Statistical comparisons consisted of the Wilcoxon matched-pairs signed-rank test.*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
-
Figure 5—source data 1
This file contains the source data used to generate Figure 5.
- https://cdn.elifesciences.org/articles/78294/elife-78294-fig5-data1-v2.xlsx
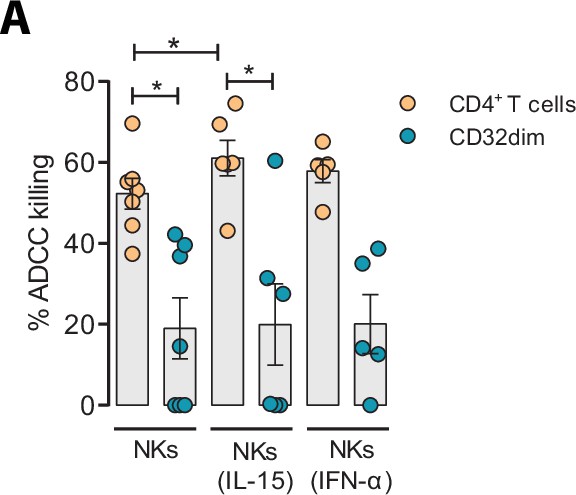
Effect of IL-15 and IFN-α in the natural killer (NK) function.
CD4+ T cells from ART-suppressed participants coated with a gp120 recombinant protein were subjected to antibody-dependent cell cytotoxicity (ADCC) assays in the presence of cytokines. The graph shows the percentage of ADCC killing of HIVgp120-coated CD4+ T cells by autologous NK cells after treatment with IL-15 or IFN-α. Statistical comparisons were performed using the Wilcoxon matched-pairs signed-rank test (comparison between CD4+ T and CD32dim cells within each condition), and the ANOVA Friedman with Dunn’s multiple comparison test (comparison between different conditions of the CD4+ T or CD32dim cells). Mean with SEM is represented. *p<0.05.
-
Figure 6—source data 1
This file contains the source data used to generate Figure 6.
- https://cdn.elifesciences.org/articles/78294/elife-78294-fig6-data1-v2.xlsx
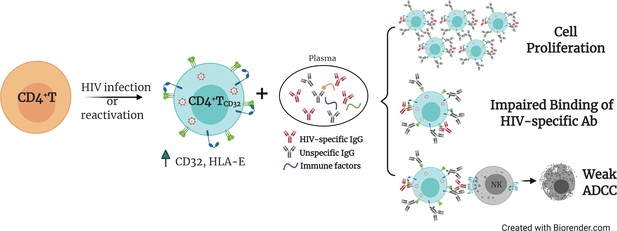
Proposed model for the persistence of CD4+ T CD32dim cells.
A proportion of CD4+T cells will express CD32 molecules upon HIV infection or viral reactivation (spontaneous reactivation or after the use of latency reversal agents). CD4+T CD32dim cells express higher levels of HLA-E, a molecule that inhibits NK cells expressing the receptor NKG2A, and are more susceptible to immune factors present in the plasma, which will (i) induce cell proliferation, (ii) block the recognition of HIV-specific antibodies, and (iii) induce a weak antibody-dependent cell cytotoxicity (ADCC) response.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| antibody | Human monoclonal A32 antibody | AIDS Research and Reference Program | Cat#11,438 | (1/200) |
| antibody | Mouse monoclonal anti-human CD32-PE-Cy7 (FUN-2) | Biolegend | 303,214 | (1/50) |
| antibody | Mouse monoclonal anti-human CD32-FITC (FUN-2) | Biolegend | 303,204 | (1/40) |
| antibody | Mouse monoclonal anti-human CD45RO-BV605 (UCHL1) | Biolegend | 562,790 | (1/40) |
| antibody | Mouse monoclonal anti-human CD20-BV786 (2H7) | Biolegend | 302,355 | (1/40) |
| antibody | Mouse monoclonal anti-human CD95-PE-Cy5 (DX2) | Becton Dickinson | 559,773 | (1/10) |
| antibody | Mouse monoclonal anti-human CD107a-PE-Cy5 (H4A3) | Becton Dickinson | 555,802 | (1/10) |
| antibody | Mouse monoclonal anti-human IFN-ɤ-AF700 (B27) | Life technologies | MHCIFG29 | (1/40) |
| antibody | Mouse monoclonal anti-human ULBP1-PerCP (170818) | R&D System | FAB1380C | (1/20) |
| antibody | Mouse monoclonal anti-human HLA-E-APC (3D12) | Biolegend | 342,606 | (1/20) |
| antibody | Mouse monoclonal anti-human CD155-BV786 (TX24) | Becton Dickinson | 744,720 | (1/100) |
| antibody | Mouse monoclonal anti-human MIC A/B–BV605 (6D4) | Becton Dickinson | 742,324 | (1/166) |
| antibody | Mouse anti-p24-PE (KC57) | Beckman Coulter | 6604667 | (1/200) |
| antibody | Mouse monoclonal anti-human CCR7-PE-CF594 (150503) | Becton Dickinson | 562,381 | (1/100) |
| antibody | Mouse monoclonal anti-human Ki67-BV510 (B56) | Becton Dickinson | 563,462 | (1/83) |
| recombinant DNA reagent | Plasmid encoding HIV-1 strain NL4.3 | NIH AIDS Reagent Program | NA | Malcom Martin |
| peptide, recombinant protein | BaL gp120 | NIH AIDS Reagent Program | NA | 1 µg |
| commercial assay or kit | Human PrimerFlow RNA Assay | EBioscience | NA | NA |
| commercial assay or kit | Human Fc block | Becton Dickinson | 564,219 | (1/20) |
| commercial assay or kit | mirVana miRNA isolation kit | Ambion | AM1560 | NA |
| commercial assay or kit | Zenon Human IgG labeling kit | Invitrogen | Z25451 | NA |
| chemical compound, drug | Ingenol | Sigma Aldrich | SML1318-1MG | 100 nM |
| chemical compound, drug | Romidepsin | Selleckchem | NA | 40 nM |
| chemical compound, drug | Ionomycine | Abcam, Inc. | ab120370 | 1 µM |
| chemical compound, drug | PMA | Abcam, Inc. | ab120297 | 81 nM |
| chemical compound, drug | Raltegravir | AIDS reagent program | NA | 1 µM |
| chemical compound, drug | Darunavir | AIDS reagent program | NA | 1 µM |
| chemical compound, drug | Nevirapine | Sigma Aldrich | SML0097-10MG | 1 µM |
| chemical compound, drug | Q-VD-OPh quinolyl-valyl-O-methylaspartyl-[–2,6-difluorophenoxy]-methyl ketone | Selleckchem | S7311 | 10 µM |
| chemical compound, drug | LIVE/DEAD Fixable Violet Dead Cell Stain Kit | Invitrogen | L34966 | (1/250) |
| software, algorithm | FlowJo software | TreeStar | NA | NA |
Additional files
-
Supplementary file 1
Clinical data of PLWH included in the study.
- https://cdn.elifesciences.org/articles/78294/elife-78294-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/78294/elife-78294-mdarchecklist1-v2.docx





