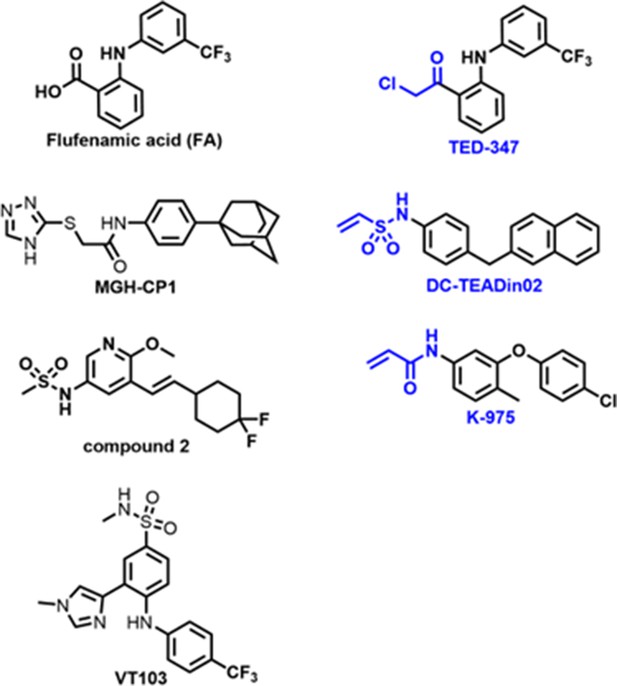
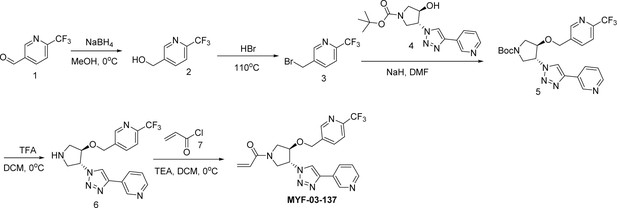
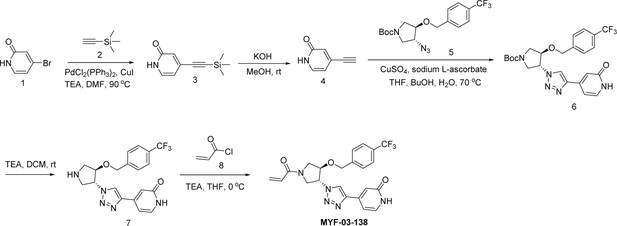
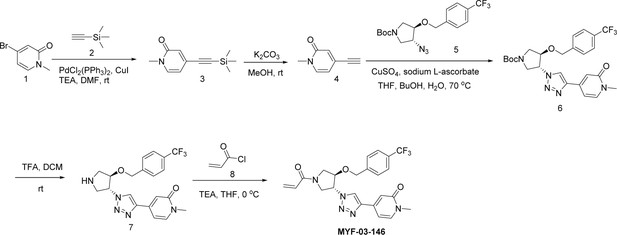
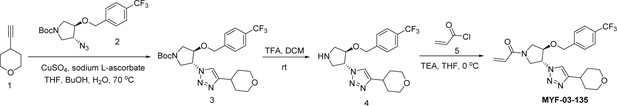
Covalent disruptor of YAP-TEAD association suppresses defective Hippo signaling
Figures
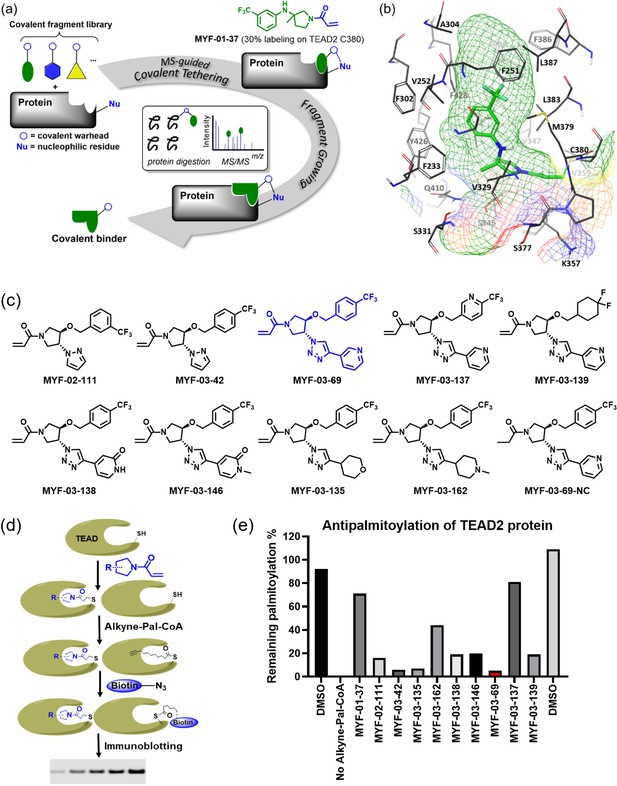
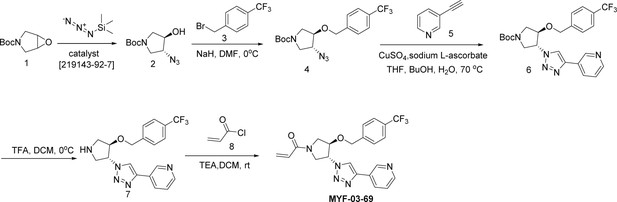
Covalent fragments screening and structure-guided design to identify Y-shaped compound MYF-03–69.
(a) Illustration of covalent fragment library screening and optimization for TEAD inhibitor. (b) Surface of TEAD2 palmitate pocket depicted in mesh with MYF-01–37 (in green) modeled in. Residues forming the pocket were labeled. The color of mesh indicates hydrophobicity of the pocket surface. (c) Chemical structures of representative Y-shaped compounds. (d) The schematic diagram of in vitro palmitoylation assay. (e) Anti-palmitoylation activity of MYF-01–37 and Y-shaped compounds after TEAD2 protein was pre-incubated with 2 μM compound at 37 °C for 2 hr.
-
Figure 1—source data 1
Uncropped gel/blots and raw files related to Figure 1.
- https://cdn.elifesciences.org/articles/78810/elife-78810-fig1-data1-v2.zip
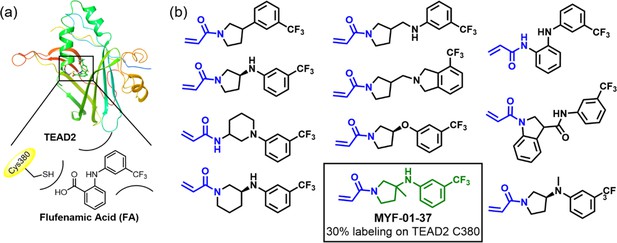
Rationale of covalent fragments design.
(a) Binding model of FA in TEAD2 palmitate pocket. (b) Chemical structures of representative covalent fragments. This figure is related to Figure 1a.
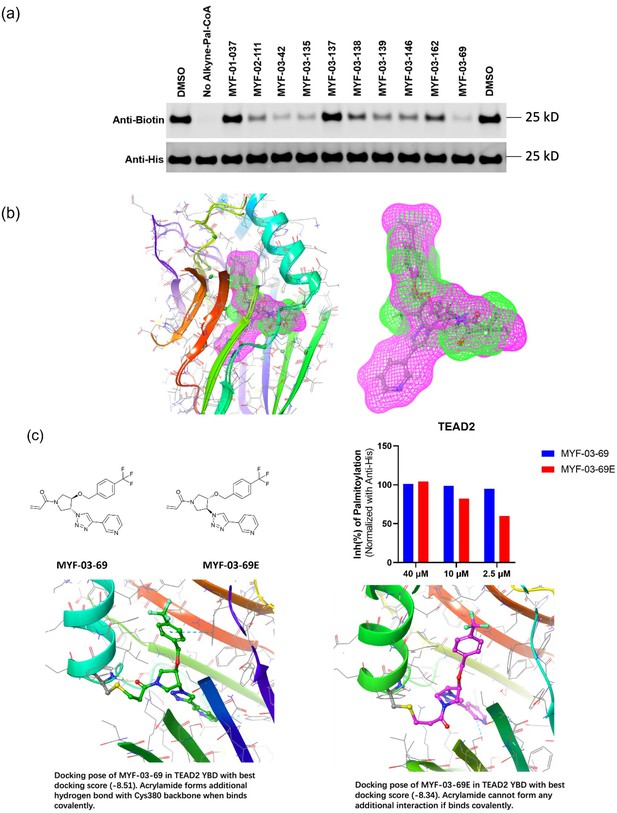
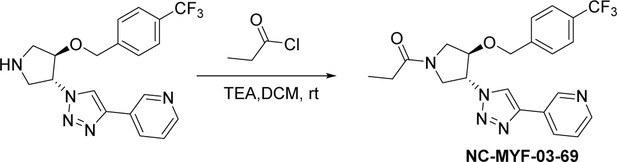
MYF-03–69 is the most potent TEAD inhibitor among derivatives that occupy a Y-shaped pocket.
(a) Original western blot image of palmitoylation assay in YBD protein of TEAD2. This figure is related to Figure 1d-e. (b) The binding poses of MYF-03-69 and K-975 in complex with TEAD1-YBD. (c) The inhibitory activities and docking results of MYF-03-69 and its enantiomer against TEAD2-YBD protein in the gel-based palmitoylation assay.
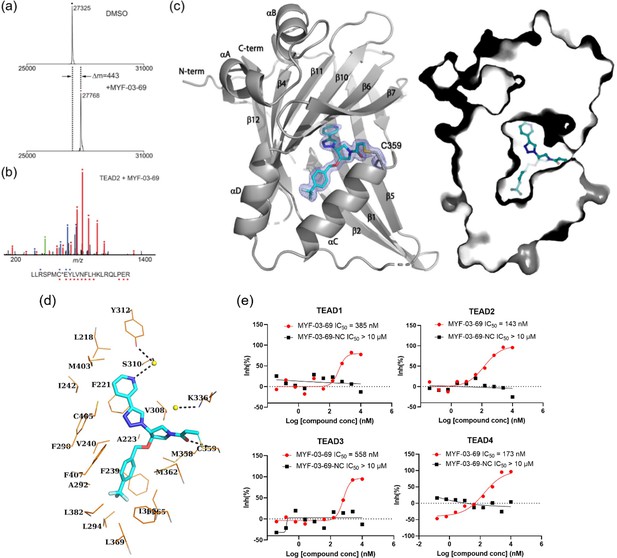
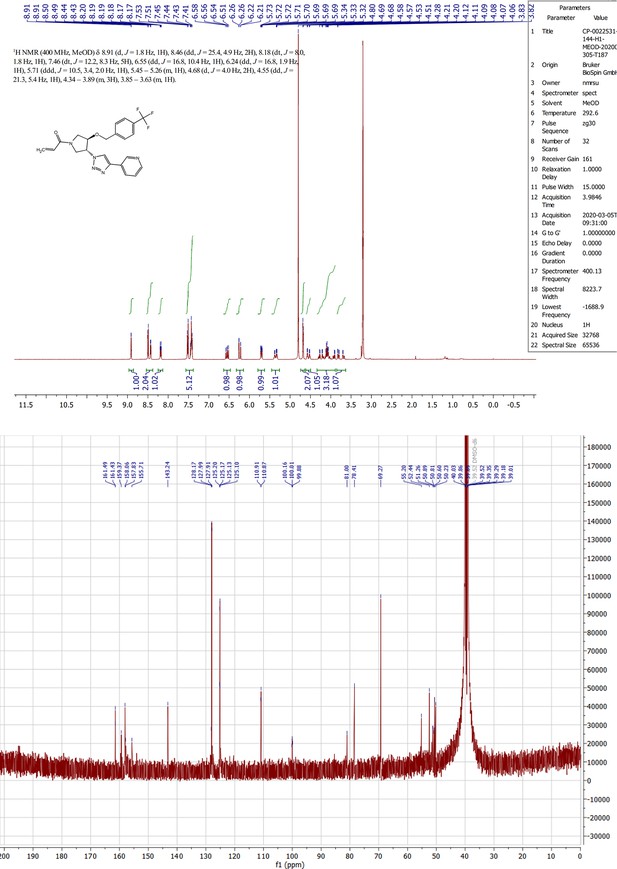
MYF-03–69 binds in TEAD palmitate pocket covalently through the conserved cysteine.
(a) The mass labeling of intact TEAD2 protein by MYF-03–69. (b) Trypsin digestion and tandem mass spectrum (MS/MS) localize labeling site of cysteine 380. (c) Co-crystal structure of MYF-03–69 with TEAD1 indicates covalent bond formation with Cys359. The compound adopts Y-shape and binds in both lipid tunnel and hydrophilic side pocket. (d) Interactions between MYF-03–69 and TEAD1 palmitate pocket. (e) A dose titration of MYF-03–69 and MYF-03–69-NC in anti-palmitoylation assay on TEAD1-4. Recombinant YBD protein of TEADs were preincubated with compounds at 37 °C for 2 hr. Data are representative of n=3 independent experiments.
-
Figure 2—source data 1
Uncropped gel/blots and raw files related to Figure 2.
- https://cdn.elifesciences.org/articles/78810/elife-78810-fig2-data1-v2.zip
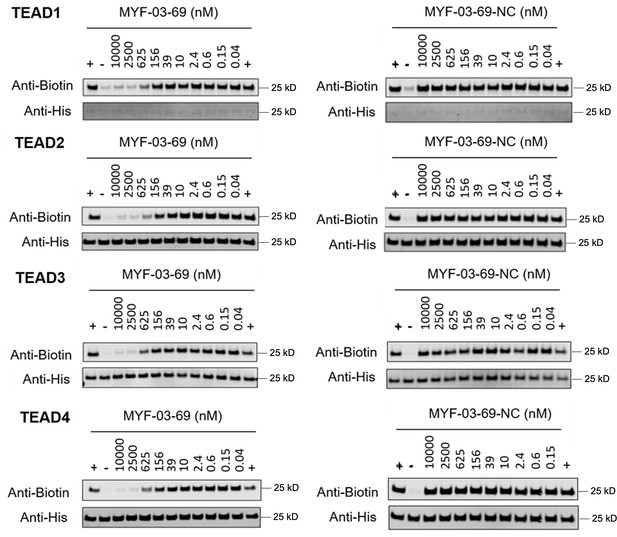
Original western blot images of palmitoylation assays in YBD protein of TEAD1-4.
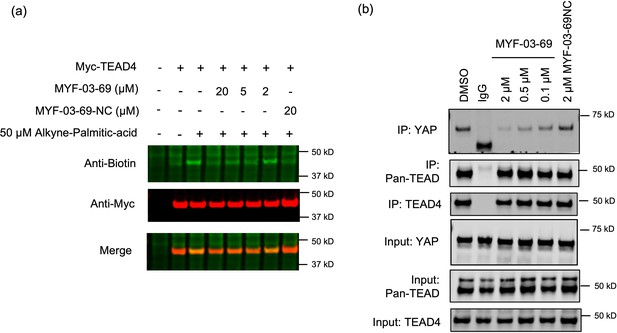
MYF-03–69 inhibits palmitoylation of TEAD protein and disrupts its association with YAP in cells.
(a) Palmitoylation of Myc-TEAD4 in HEK293T cells after treatment with MYF-03–69 and MYF-03–69-NC indicated by an alkyne-palmitic-acid probe and click chemistry. Cells were treated for 24 hr. (b) Co-immunoprecipitation (Co-IP) of endogenous YAP and TEAD in NCI-H226 cells after treatment with MYF-03–69 and MYF-03–69-NC at indicated doses. Cells were treated for 24 hr.
-
Figure 3—source data 1
Uncropped gel/blots and raw files related to Figure 3.
- https://cdn.elifesciences.org/articles/78810/elife-78810-fig3-data1-v2.zip
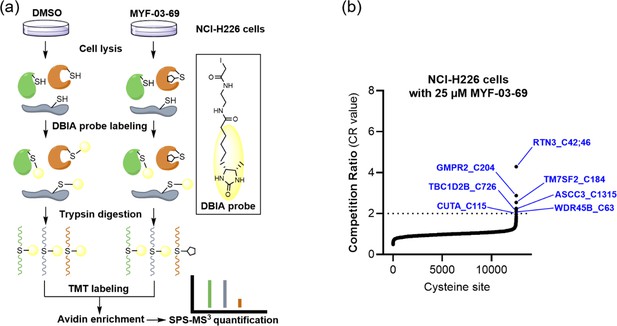
MYF-03–69 exhibited low reactivity towards the cysteine proteome.
(a) Overview of the SLC-ABPP proteome-wide selectivity approach for profiling of MYF-03-69. NCI-H226 cells were treated with 0.5, 2, 10 or 25 μM MYF-03-69 and DMSO over 3 hours in triplicates before cell lysis followed by labeling with DBIA probe. The SLC-ABPP approach was used to profile competition of MYF-03-69 in proteome cysteine labeling. Competition ratio (CR) was calculated in order to quantitatively assess labeling at every cysteine residue. (b) The 7 cysteine sites that were significantly labeled (i.e. exhibited >50% conjugation or CR>2) by 25 µM of MYF-03-69 are in blue, all of which exhibited dose-dependent engagement.
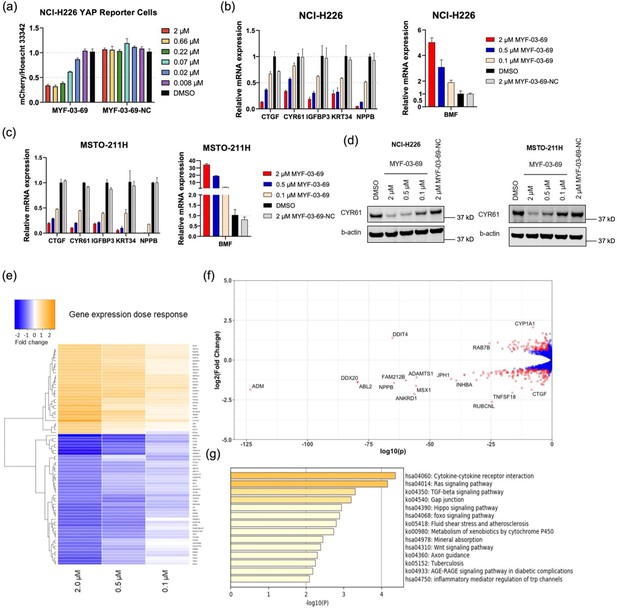
MYF-03–69 inhibits YAP-TEAD transcription.
(a) MYF-03–69, but not MYF-03–69-NC, inhibits YAP-TEAD transcriptional activity in NCI-H226 mCherry reporter cells. Cells were treated for 72 hr. Data were presented as mean ± SD of n=3 biological independent samples.(b–c) MYF-03–69, but not MYF-03–69-NC, downregulates YAP target genes and upregulates a pro-apoptotic gene BMF. Cells were treated for 24 hr. Data were presented as mean ± SD of n=3 biological independent samples. (d) MYF-03–69, but not MYF-03–69-NC, downregulates CYR61 protein level in NCI-H226 and MSTO-211H cells. (e) Heatmap for gene expression change with MYF-03–69 treatment at indicated concentrations. (f) Differential gene expression from RNA sequencing of NCI-H226 cells treated with 2 μM MYF-03–69. The differentially expressed genes with FC ≥1.5 and p ≤ 0.05 were colored in red and labeled. Not all differentially expressed genes were labeled. (g) Pathway enrichment analysis of differentially expressed genes from 2 μM compound treatment samples.
-
Figure 4—source data 1
Uncropped gel/blots and raw files related to Figure 4.
- https://cdn.elifesciences.org/articles/78810/elife-78810-fig4-data1-v2.zip
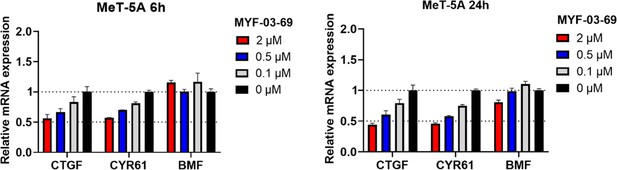
MYF-03–69 showed comparatively weak inhibition on YAP-TEAD downstream gene expression in normal mesothelium cell MeT-5A upon 6 hr and 24 hr treatment.
Data were presented as mean ± SD of n=3 biological independent samples.
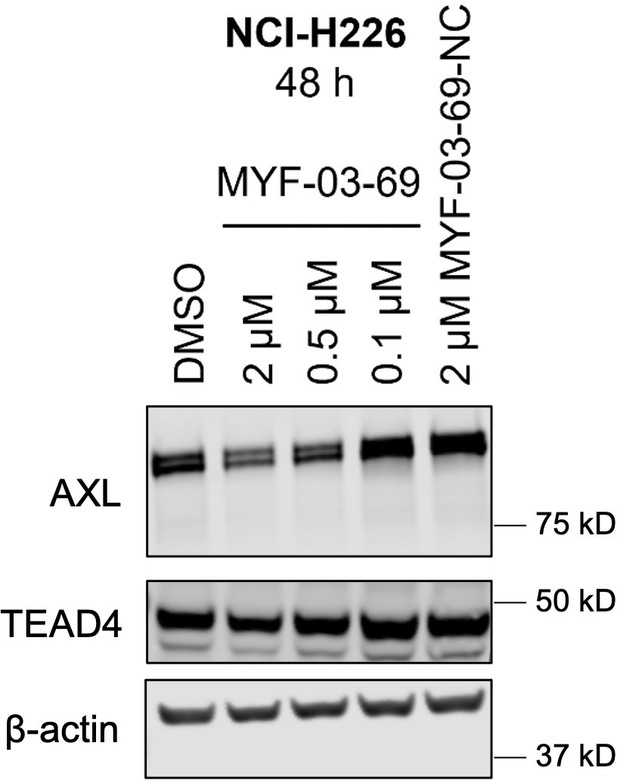
MYF-03–69 downregulated product of canonical YAP downstream gene AXL with minimal effect on TEAD stability.
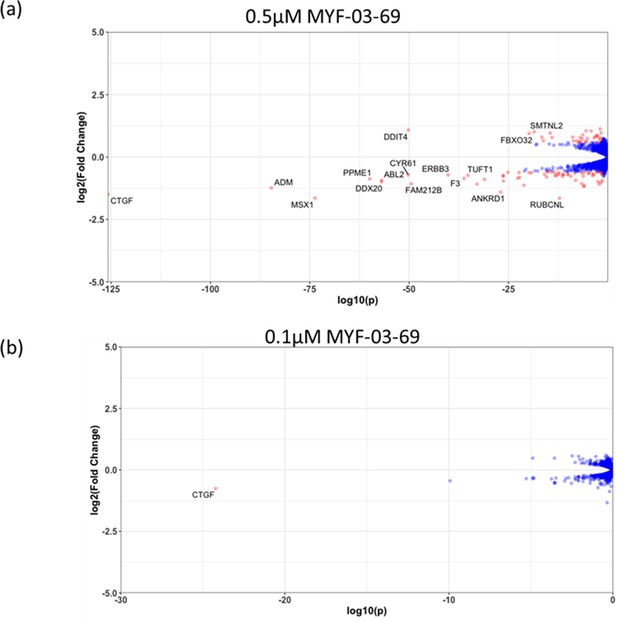
The number of genes with significantly altered expression level decreased with lower concentration treatment in NCI-H226 cells.
(a) 0.5 μM MYF-03-69, (b) 0.1 μM MYF-03-69.
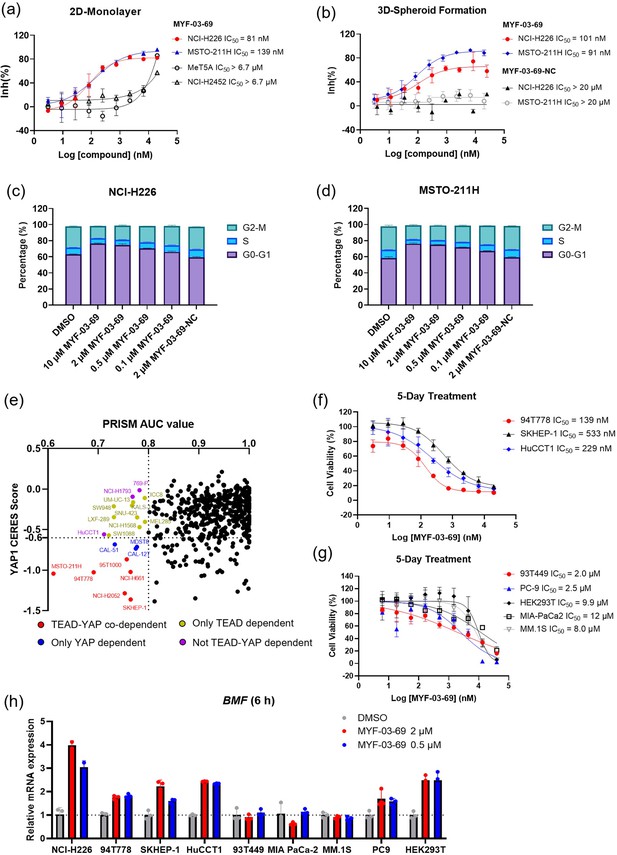
MYF-03–69 selectively inhibits proliferation of cancer cells with YAP or TEAD dependency regardless of lineage.
(a) Antiproliferation IC50 curve of MYF-03–69 on three mesothelioma cell lines (NCI-H226, MSTO-211H and NCI-H2452) and a normal noncancerous mesothelium cell line (Met-5A). Data were presented as mean ± SD of n=3 biological independent samples. (b) Antiproliferation IC50 curve of MYF-03–69 and MYF-03–69-NC in 3D cell culture. Data were presented as mean ± SD of n=3 biological independent samples. (c–d) Cell cycle arrest induced by MYF-03–69, but not MYF-03–69-NC. Cells were treated for 48 hr at indicated doses. Data were presented as mean ± SD of n=3 biological independent samples. (e) PRISM profiling across a broad panel of cell lineages. 903 cancer cells were treated with MYF-03–69 for 5 days. The viability values were measured at 8-point dose manner (3-fold dilution from 10 μM) and fitted a dose-response curve for each cell line. Area under the curve (AUC) was calculated as a measurement of compound effect on cell viability. CERES score of YAP1 or TEADs from CRISPR (Avana) Public 21Q1 dataset (DepMap) were used to estimate gene-dependency. Cell lines without CERES Score of YAP1 were excluded from the figure. The CERES Score of most dependent TEAD isoform was used to represent TEAD dependency. Cell lines with a dependency score less than –0.6 were defined as the dependent cell lines. (f) Antiproliferation curves of cell lines that are sensitive to MYF-03–69 treatment besides mesothelioma. Data were presented as mean ± SD of n=3 biological independent samples. (g) Antiproliferation curves of cell lines that are insensitive to MYF-03–69 treatment. Data were presented as mean ± SD of n=3 biological independent samples. (h) BMF expression level after treatment with 0.5 or 2 μM MYF-03–69 over 6 hr in different cells. Data were presented as mean ± SD of n=3 biological independent samples.
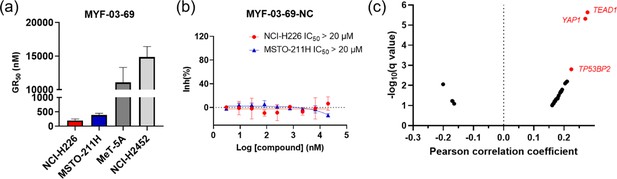
MYF-03–69 inhibited the proliferation of TEAD-dependent cancer cells.
(a) The calculated GR50 values of MYF-03-69 in 5-day proliferation assay on mesothelioma cells. NCI-H226 and MSTO-211H are Hippo signaling defective mesothelioma cells. NCI-H2452 is Hippo signaling intact mesothelioma cell. MeT-5A is non-cancerous mesothelium cell. Data were presented as mean ± SD of n=3 biological independent samples. This figure is related to Figure 5a. (b) MYF-03-69-NC did not inhibit cell proliferation of NCI-H226 and MSTO-211H cells in 5-day treatment. Data were presented as mean ± SD of n=3 biological independent samples. This figure is related to Figure 5a. (c) Correlation analysis between compound PRISM sensitivity (log2.AUC of each cell line) and dependency of certain gene (CRISPR knockout score for each cell line, from DepMap Public 20Q4 Achilles_gene_effect.csv dataset) across the PRISM cell line panel. The Pearson correlation coefficients (X axis) and associated p-values were computed. Positive correlations correspond to dependency correlating with increased sensitivity. The q-values (a corrected significance value accounting for false discovery rate) are computed from p-values using the Benjamini Hochberg algorithm. Associations with q-values above 0.1 are filtered out. Top 3 correlated genes are in red.
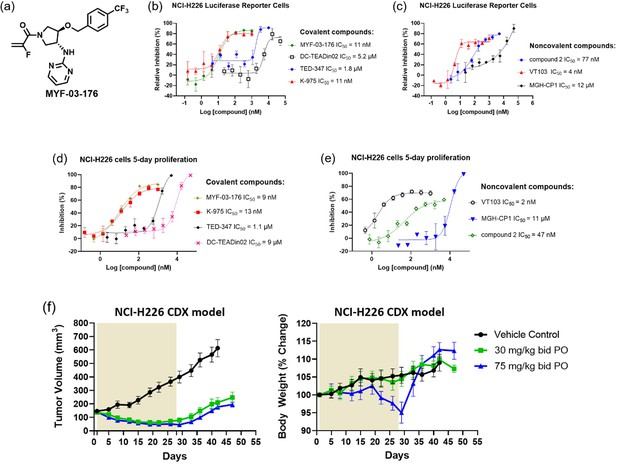
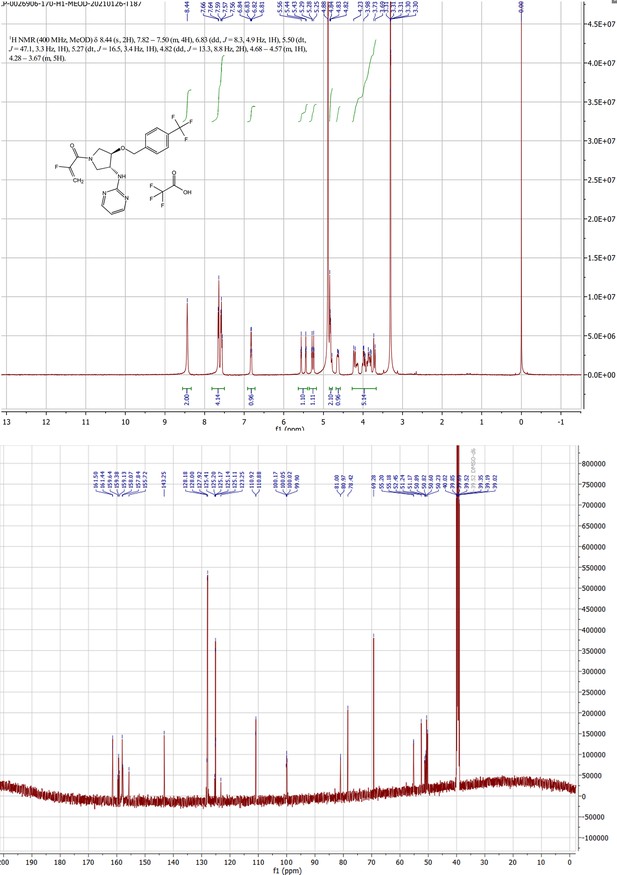
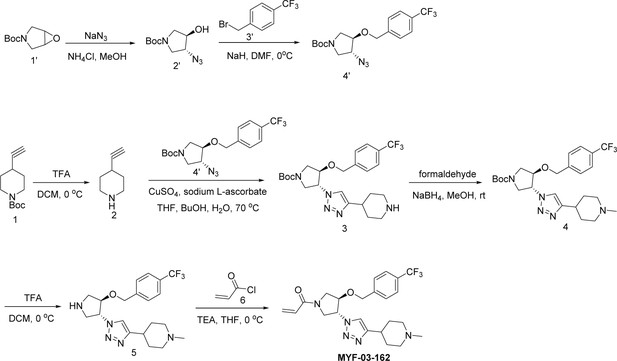
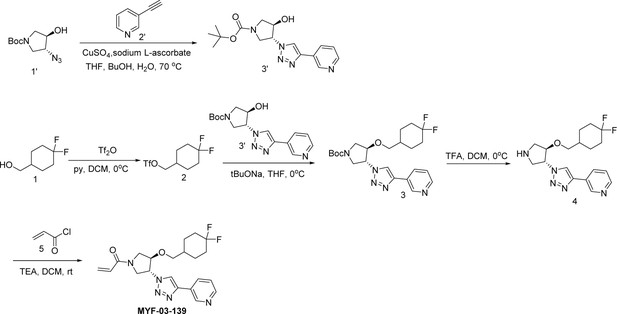
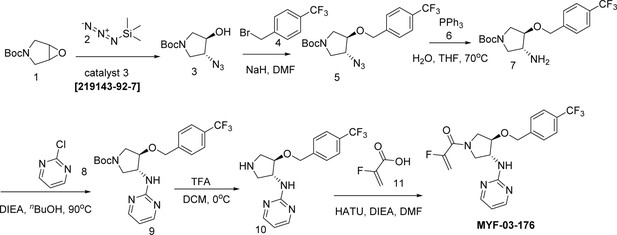
MYF-03–176 is a potent and orally bioavailable YAP-TEAD transcription inhibitor and suppresses tumor growth in mesothelioma xenograft mouse model.
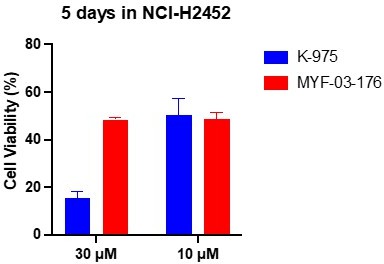
(a) Chemical structure of MYF-03–176. (b–c) Inhibitory effect of MYF-03–176 and other TEAD PBP binders on YAP-TEAD transcription in NCI-H226 luciferase reporter cells. Cells were treated for 72 hr. Data were presented as mean ± SD of n=3 biological independent samples. (d-e) Antiproliferation effect of MYF-03–176 and other TEAD PBP binders in NCI-H226 cells. Cells were treated for 5 days. Data were presented as mean ± SD of n=3 biological independent samples. (f) In vivo efficacy of MYF-03–176 in NCI-H226 CDX mouse model (n=8–9 per group).
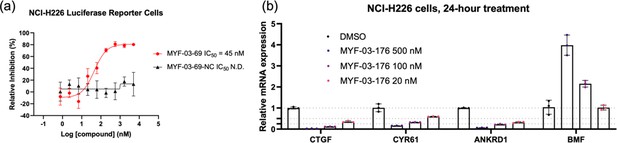
MYF-03–69 and MYF-03–176 inhibited TEAD transcriptional activity.
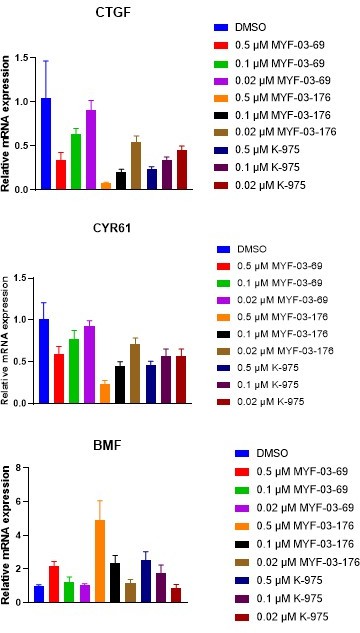
(a) MYF-03-69, but not MYF-03-69-NC, inhibits YAP-TEAD transcriptional activity in NCI-H226 luciferase reporter cells. Data were presented as mean ± SD of n=3 biological independent samples. This figure is related to Figure 6b-c. (b) MYF-03-176 downregulates YAP target genes and upregulates a pro-apoptotic gene BMF. Data were presented as mean ± SD of n=3 biological independent samples.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | NCI-H226 (Mesothelioma, Male) | ATCC | CRL-5826 | |
| Cell line (Homo sapiens) | MSTO-211H (Mesothelioma,Male) | ATCC | CRL-2081 | |
| Cell line (Homo sapiens) | NCI-H2452 (Mesothelioma, Male) | ATCC | CRL-5946 | |
| Cell line (Homo sapiens) | MeT-5A (Mesotheliumcells) | ATCC | CRL-9444 | |
| Antibody | Anti-human YAP (Rabbit monoclonal) | Cell Signaling | Cat#14074 S | WB (1:1000) |
| Antibody | Anti-human pan-TEAD (Rabbit monoclonal) | Cell Signaling | Cat#13295 S | WB (1:1000) |
| Antibody | Anti-human TEAD4 (Mouse monoclonal) | Abcam | Cat#ab58310 | WB (1:1000) |
| Antibody | Anti-human b-actin (Mouse monoclonal) | Cell Signaling | Cat#3700 S | WB (1:2000) |
| Recombinant DNA reagent | Myc-TEAD4 (plasmid) | Addgene | RRID: Addgene_24638 | Overexpression of Myc-TEAD4 |
| Recombinant DNA reagent | TBS-mCherry vector (plasmid) | Cancer Cell. 2020 Jan 13; 37(1): 104–122.e12 | Monitor TEAD transcriptional activity | |
| Commercial assay or kit | Dynabeads Co-Immunoprecipitation Kit | ThermoFisher Scientific | Cat#14321D | |
| Commercial assay or kit | SuperScript III First-Strand Synthesis System Kit | ThermoFisher Scientific | Cat#18080051 | |
| Chemical compound, drug | Palmitoyl alkyne-coenzyme A | Cayman chemical | Cat#15968 | |
| Chemical compound, drug | Alkyne palmitic acid | Click Chemistry Tools | Cat#1165–5 | |
| Software, algorithm | GraphPad9 | GraphPad | ||
| Other | TEAD luciferase reporter lentivirus | BPS Biosciences | Cat#79833 | Used to establish a stable cell line to monitor TEAD transcriptional activity; related to Figure 6—figure supplement 3 (a) |
| Other | IRDye 800CW Streptavidin | LI-COR | Cat# 926–32230 | WB (0.2 μg/mL) |
Additional files
-
Supplementary file 1
Table of diffraction data collection and refinement statistics for TEAD1 and MYF-03–69 co-crystal structure.
- https://cdn.elifesciences.org/articles/78810/elife-78810-supp1-v2.docx
-
Supplementary file 2
Proteome-wide selectivity profile of MYF-03–69 on cysteines labeling using SLC-ABPP approach.
- https://cdn.elifesciences.org/articles/78810/elife-78810-supp2-v2.xlsx
-
Supplementary file 3
List of differentially expressed genes under MYF-03–69 treatments.
- https://cdn.elifesciences.org/articles/78810/elife-78810-supp3-v2.xlsx
-
Supplementary file 4
Area under the curve (AUC) data of PRISM cell viability screen and corresponding CERES scores of YAP1 and TEADs.
- https://cdn.elifesciences.org/articles/78810/elife-78810-supp4-v2.xlsx
-
Supplementary file 5
Correlation analysis of MYF-03–69 PRISM sensitivity profile and gene DepMap dependency scores.
- https://cdn.elifesciences.org/articles/78810/elife-78810-supp5-v2.xlsx
-
Supplementary file 6
Liver microsome stability and hepatocyte stability of MYF-03–176 and K-975.
- https://cdn.elifesciences.org/articles/78810/elife-78810-supp6-v2.docx
-
Supplementary file 7
Liver microsome stability and PK parameters of MYF-01–37, MYF-03–69 and MYF-03–176.
- https://cdn.elifesciences.org/articles/78810/elife-78810-supp7-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/78810/elife-78810-mdarchecklist1-v2.docx