Munc13 supports fusogenicity of non-docked vesicles at synapses with disrupted active zones
Figures
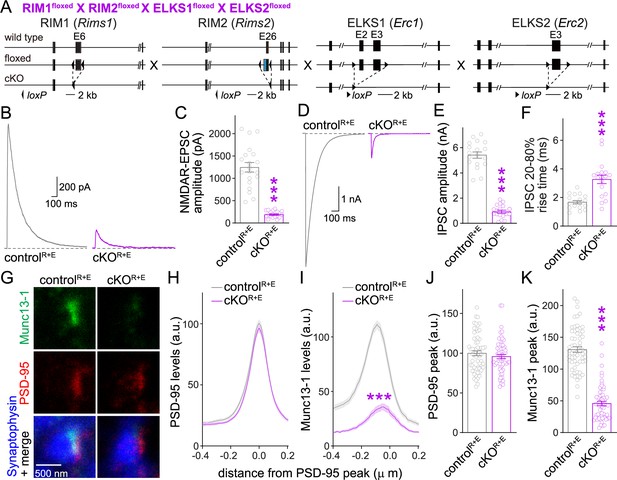
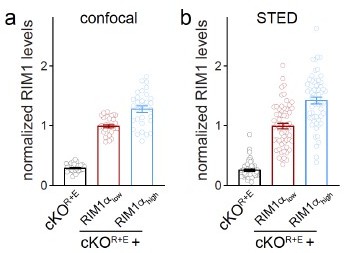
Action potential-evoked neurotransmitter release and Munc13 active zone levels after ablation of RIM + ELKS.
(A) Strategy for deletion of RIM1, RIM2, ELKS1, and ELKS2 in cultured hippocampal neurons. Neurons of mice with floxed alleles for all four genes were infected with Cre-expressing lentiviruses (to generate cKOR+E neurons) or lentiviruses expressing a recombination-deficient version of Cre (to generate controlR+E neurons) as described (Tan et al., 2022; Wang et al., 2016). (B, C) Sample traces (B) and quantification (C) of excitatory postsynaptic currents (EPSCs) evoked by focal electrical stimulation, controlR+E 20 cells/3 cultures, cKOR+E 19/3. (D–F) Sample traces (D) and quantification of amplitudes (E) and 20–80% rise times (F) of inhibitory postsynaptic currents (IPSCs) evoked by focal electrical stimulation, 18/3 each. (G–K) Sample stimulated emission depletion (STED) microscopic images (G) and quantification (H–K) of side-view synapses in cultured hippocampal neurons stained for Munc13-1 (imaged in STED), PSD-95 (imaged in STED), and Synaptophysin (imaged in confocal). In (H, I), fluorescence levels at each position of line profiles (600 nm × 200 nm) positioned perpendicular to the center of the elongated PSD-95 structure and aligned to the PSD-95 peak are shown; in (J, K), peak values for each line profile are shown independent of peak position, 60 synapses/3 cultures each. Data are mean ± SEM; ***p<0.001 as determined by Welch’s t-tests (C, E, F), two-way ANOVA followed by Bonferroni’s multiple-comparisons post-hoc tests (H, I), or unpaired two-tailed Student’s t-tests (J, K). For assessment of Munc13-1 levels after Munc13 knockout using STED microscopy, Synaptophysin levels in cKOR+E synapses, and comparison of Munc13-1 levels by STED microscopy in cKOR+E and cKOM synapses, see Figure 1—figure supplement 1; for assessment of synaptic transmission after Munc13 knockout, see Figure 1—figure supplement 2; for assessment of Munc13-1 expression by confocal microscopy and Western blotting in cKOR+E and cKOM neurons, see Figure 1—figure supplement 3. Source data 1 contains numerical values of all means, errors, and p-values for this and all figures.
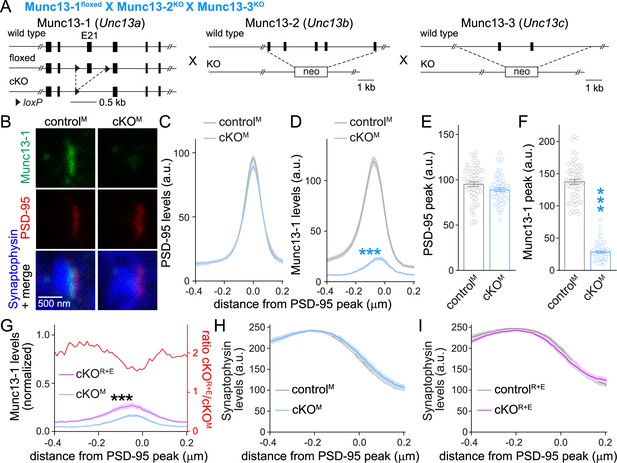
Assessment of Munc13-1 levels with STED microscopy in Munc13 knockout synapses.
(A) Strategy for deletion of Munc13-1, Munc13-2, and Munc13-3 in cultured hippocampal neurons (cKOM). Cultured hippocampal neurons of mice with floxed alleles for Munc13-1 (Banerjee et al., 2022) and constitutive knockout alleles for Munc13-2 (Varoqueaux et al., 2002) and Munc13-3 (Augustin et al., 2001) were infected with Cre-expressing lentiviruses (to generate cKOM neurons) or with lentiviruses expressing a recombination-deficient Cre (to generate controlM neurons). (B–F) Sample images (B) and quantification (C–F) of side-view synapses in cultured hippocampal neurons stained for Munc13-1 (STED), PSD-95 (STED), and Synaptophysin (confocal), 60 synapses/3 cultures each. (G) Comparison of Munc13-1 levels normalized to their own controls in cKOR+E (absolute data from Figure 1I) and cKOM (from D) synapses, 60 synapses/3 cultures each. The red line is the calculated ratio of normalized Munc13-1 levels from cKOR+E to cKOM. (H) Quantification of Synaptophysin levels in side-view synapses of controlM and cKOM neurons for the experiment shown in (B–D), 60 synapses/3 cultures each. (I) Same as (H), but for Synaptophysin levels in controlR+E and cKOR+E neurons for the experiment shown in Figure 1G–I, 60 synapses/3 cultures each. Data are mean ± SEM; ***p<0.001 as determined by two-way ANOVA followed by Bonferroni’s multiple-comparisons post-hoc tests (C, D, G–I; in G, no statistics were performed on ratios), unpaired two-tailed Student’s t-tests (E), or Welch’s t-test (F).
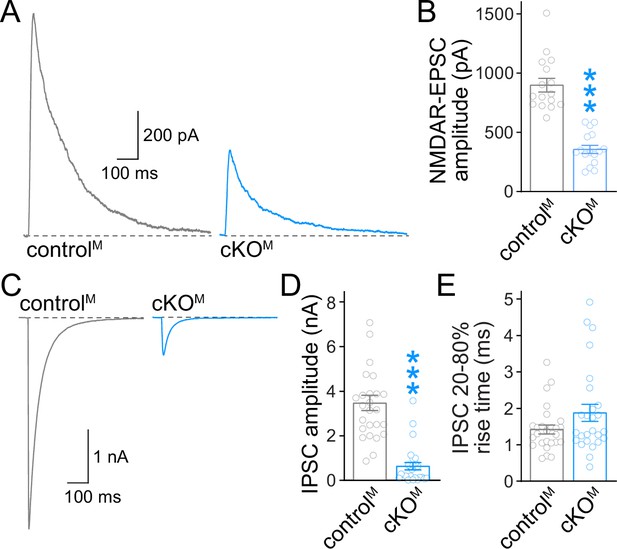
Action potential-evoked neurotransmitter release after ablation of Munc13.
(A, B) Sample traces (A) and quantification (B) of excitatory postsynaptic currents (EPSCs) evoked by focal electrical stimulation in controlM and cKOM neurons, 16 cells/3 cultures each. (C–E) Sample traces (C) and quantification of amplitudes (D) and 20–80% rise times (E) of inhibitory postsynaptic currents (IPSCs) evoked by focal electrical stimulation, 26/3 each. Data are mean ± SEM; ***p<0.001 as determined by Mann–Whitney tests (B, D, E).
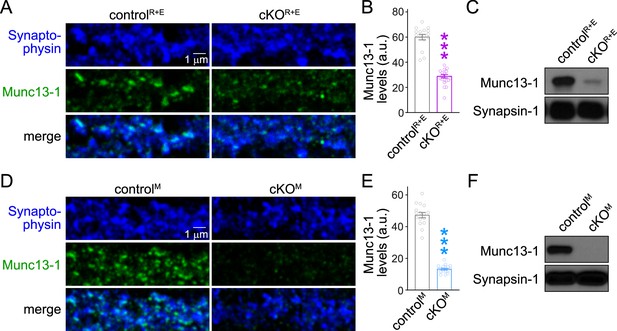
Assessment of Munc13-1 levels with confocal microscopy and Western blot in RIM + ELKS or Munc13 knockout neurons.
(A, B) Sample confocal images (A) and quantification of Munc13-1 levels (B) at synapses of controlR+E and cKOR+E neurons. Confocal scans were taken from the same areas that were analyzed by STED microscopy in Figure 1. Synaptophysin staining was used to define regions of interest (ROIs), and levels of Munc13-1 were quantified within those ROIs, controlR+E 16 images/3 cultures, cKOR+E 19/3. (C) Western blot of homogenates of cultured controlR+E and cKOR+E neurons using anti-Munc13-1 antibodies and anti-Synapsin-1 antibodies as loading controls. Representative examples of six cultures assessed by Western blotting are shown. (D, E) As (A, B), but for controlM and cKOM neurons, controlM 16/3, cKOM 17/3. The Munc13-1 antibody signal measured in confocal microscopy in cKOM neurons was 28% of the control signal, similar to the reduction observed in STED peak analyses (21% of control, Figure 1—figure supplement 1F). (F) As (C), but for controlM and cKOM neurons, the results shown here are from exposures to film that lasted ≤1 min. The very low levels of an alternative variant of Munc13-1 left in Munc13-1 knockout neurons generated from this allele, as described in Fig. S2 of Banerjee et al., 2022, is not detected in these short exposures. Representative examples of six cultures assessed by Western blotting are shown. Data are mean ± SEM; ***p<0.001 as determined by Mann–Whitney tests (B) or Welch’s t-test (E). For source data of (C, F), see Figure 1—figure supplement 3—source data 1.
-
Figure 1—figure supplement 3—source data 1
Western blots for Figure 1—figure supplement 3.
(A) Original scans (1 s and 30 s) of Western blots shown in Figure 1—figure supplement 3C. (B) Raw grayscale scans (left) and brightness- and contrast-adjusted scans (right) of Western blots shown in Figure 1—figure supplement 3C. (C) Original scans (5 s and 1 min) of Western blots shown in Figure 1—figure supplement 3F. (D) Raw grayscale scans (left) and brightness- and contrast-adjusted scans (right) of Western blots shown in Figure 1—figure supplement 3F.
- https://cdn.elifesciences.org/articles/79077/elife-79077-fig1-figsupp3-data1-v2.zip
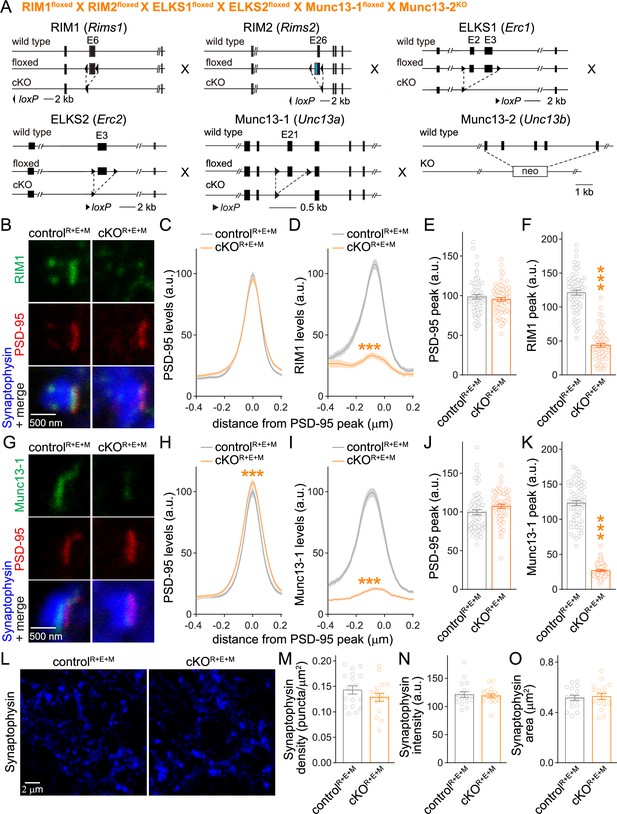
Simultaneous deletion of RIM, ELKS, and Munc13 does not disrupt synapse formation.
(A) Strategy for simultaneous deletion of RIM1, RIM2, ELKS1, ELKS2, Munc13-1, and Munc13-2 in cultured hippocampal neurons (cKOR+E+M). Neurons were infected with Cre-expressing lentiviruses (to generate cKOR+E+M neurons) or viruses expressing a recombination-deficient version of Cre (to generate controlR+E+M neurons). (B–F) Sample images (B) and quantification (C–F) of side-view synapses stained for RIM1 (STED), PSD-95 (STED), and Synaptophysin (confocal), controlR+E+M 65 synapses/3 cultures, cKOR+E+M 66/3. (G–K) Same as (B–F), but for synapses stained for Munc13-1 instead of RIM1, 63/3 each. (L–O) Overview confocal images of anti-Synaptophysin staining (L) and quantification of Synaptophysin puncta density (M), intensity (N), and size (O); Synaptophysin objects were detected using automatic two-dimensional segmentation, confocal images of Synaptophysin staining are from the experiment shown in (B–F), 17 images/3 cultures each. Data are mean ± SEM; ***p<0.001 as determined by two-way ANOVA followed by Bonferroni’s multiple-comparisons post-hoc tests (C, D, H, I), unpaired two-tailed Student’s t-tests (E, J, M–O), or Welch’s t-tests (F, K). For Synaptophysin levels in cKOR+E+M synapses, comparison of Munc13-1 levels by STED microscopy in cKOR+E and cKOR+E+M neurons, and Munc13-1 expression in controlR+E+M and cKOR+E+M neurons assessed by confocal microscopy and Western blotting, see Figure 2—figure supplement 1; for microscopic assessment of synapse formation after lentiviral infection at 2 days in vitro (DIV2) instead of DIV5, see Figure 2—figure supplement 2.
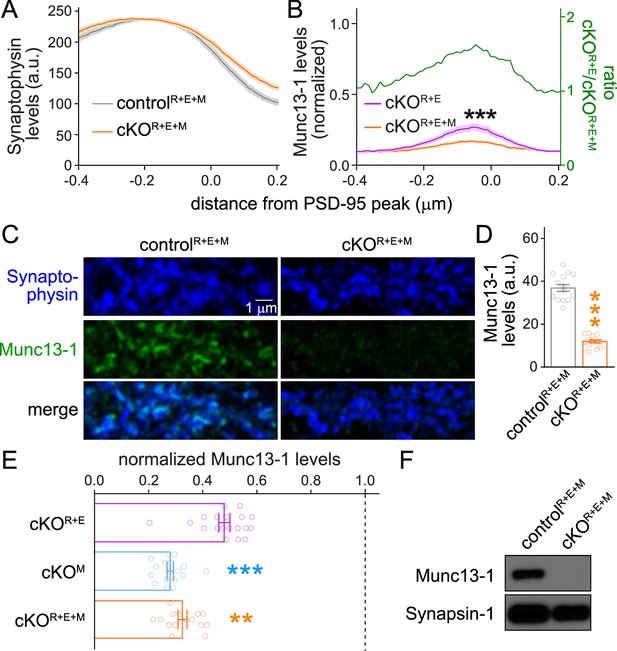
Assessment of Munc13-1 levels in RIM + ELKS + Munc13 knockout neurons.
(A) Quantification of Synaptophysin distribution aligned to the PSD-95 peak in side-view synapses of cultured controlR+E+M and cKOR+E+M neurons from the experiment shown in Figure 2B–D, controlR+E+M 65 synapses/3 cultures, cKOR+E+M 66/3. (B) Comparison of Munc13-1 levels normalized to their own controls in cKOR+E (absolute data from Figure 1I, normalized data replotted from Figure 1—figure supplement 1G) and cKOR+E+M (absolute data from Figure 2I) synapses, cKOR+E 60 synapses/3 cultures, cKOR+E+M 63/3. The green line is the calculated ratio of normalized Munc13-1 levels from cKOR+E to cKOR+E+M. (C, D) Sample confocal images (C) and quantification (D) of Munc13-1 levels at controlR+E+M and cKOR+E+M synapses. Synaptophysin staining was used to define regions of interest (ROIs), and levels of Munc13-1 were quantified within those ROIs, 15 images/3 cultures each. (E) Comparison of Munc13-1 levels normalized to their own controls for cKOR+E (absolute data from Figure 1—figure supplement 3B), cKOM (from Figure 1—figure supplement 3E), and cKOR+E+M (from D), cKOR+E 19/3, cKOM 17/3, cKOR+E+M 15/3. (F) Western blot of homogenates of cultured controlR+E+M and cKOR+E+M neurons using anti-Munc13-1 antibodies and anti-Synapsin-1 antibodies as loading controls. Representative examples of eight cultures assessed by Western blotting are shown. Data are mean ± SEM; **p<0.01, ***p<0.001 as determined by two-way ANOVA followed by Bonferroni’s multiple-comparisons post-hoc tests (A, B; in B, no statistics were performed on ratios), a Welch’s t-test (D), or a Kruskal–Wallis test followed by Dunn’s multiple-comparisons post-hoc tests (E). For source data of (F), see Figure 2—figure supplement 1—source data 1.
-
Figure 2—figure supplement 1—source data 1
Western blots for Figure 2—figure supplement 1.
(A) Original scans (5 s and 1 min) of Western blots shown in Figure 2—figure supplement 1F. (B) Raw grayscale scans (left) and brightness- and contrast-adjusted scans (right) of Western blots shown in Figure 2—figure supplement 1F.
- https://cdn.elifesciences.org/articles/79077/elife-79077-fig2-figsupp1-data1-v2.zip
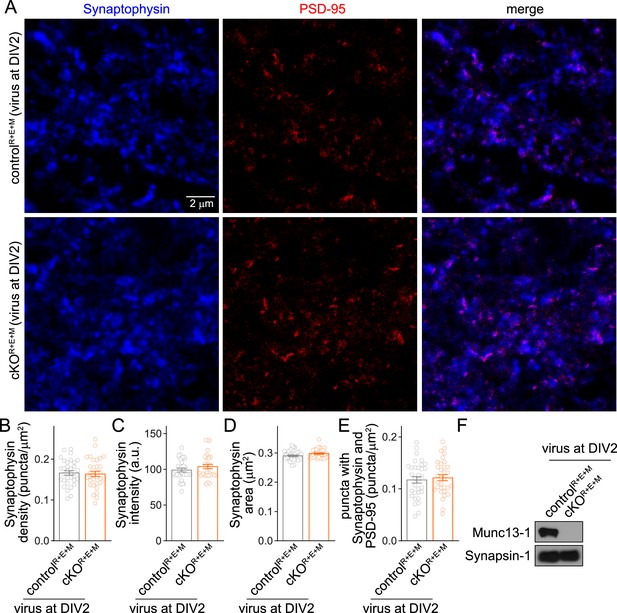
Assessment of Synaptophysin fluorescence in RIM + ELKS + Munc13 knockout neurons after lentiviral infection at DIV2.
(A–E) Overview images of anti-Synaptophysin (confocal) and anti-PSD-95 (STED) staining (A) and quantification of Synaptophysin puncta density (B), intensity (C), size (D), and of the density of puncta that contain both Synaptophysin and PSD-95 (E), from cultured controlR+E+M and cKOR+E+M neurons infected with lentiviruses (expressing Cre recombinase or a recombination-deficient version of Cre) at DIV2 instead of DIV5. Synaptophysin and PSD-95 objects were detected using automatic two-dimensional segmentation, 32 images/3 cultures each. (F) Western blot of homogenates of cultured controlR+E+M and cKOR+E+M neurons (infected with lentiviruses at DIV2) using anti-Munc13-1 antibodies and anti-Synapsin-1 antibodies as loading controls. Representative examples of three cultures assessed by Western blotting are shown. Data are mean ± SEM; unpaired two-tailed Student’s t-tests (B–E) were used. For source data of (F), see Figure 2—figure supplement 2—source data 1.
-
Figure 2—figure supplement 2—source data 1
Western blots for Figure 2—figure supplement 2.
(A) Original scans (1 s and 30 s) of Western blots shown in Figure 2—figure supplement 2F. (B) Raw grayscale scans (left) and brightness- and contrast-adjusted scans (right) of Western blots shown in Figure 2—figure supplement 2F.
- https://cdn.elifesciences.org/articles/79077/elife-79077-fig2-figsupp2-data1-v2.zip
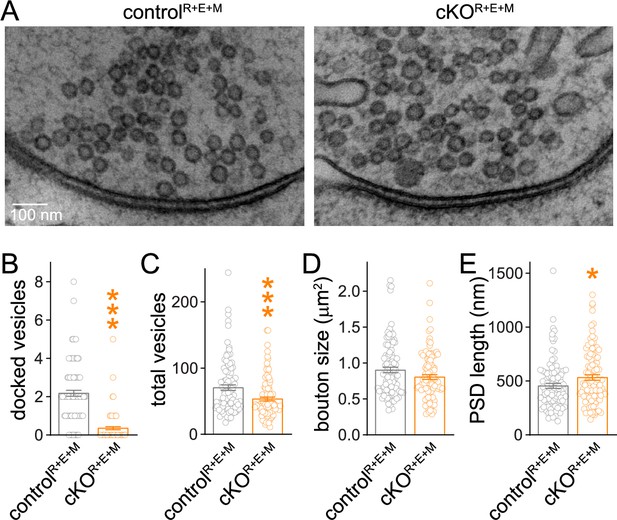
Synaptic ultrastructure after RIM + ELKS + Munc13 knockout.
(A–E) Sample images (A) and analyses (B–E) of synaptic morphology of high-pressure frozen neurons analyzed by electron microscopy; docked vesicles (B), total vesicles (C), bouton size (D), and postsynaptic density (PSD) length (E) per synapse profile are shown. Docked vesicles were defined as vesicles for which the electron density of the vesicular membrane touched that of the target membrane such that the two membranes were not separated by less electron-dense space, controlR+E+M 99 synapses/2 cultures, cKOR+E+M 100/2. Data are mean ± SEM; *p<0.05, ***p<0.001 as determined by Welch’s t-tests (B, C) or unpaired two-tailed Student’s t-test (D, E).
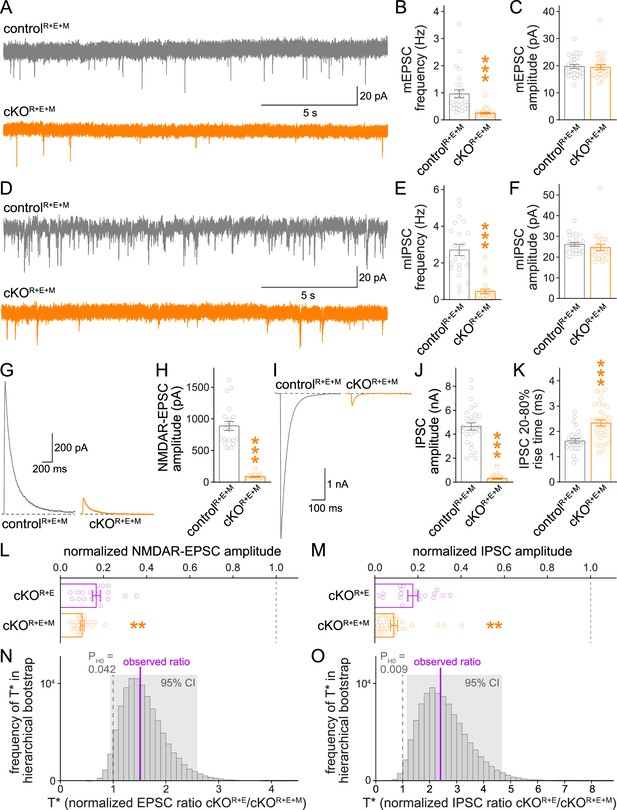
Neurotransmitter release is strongly impaired after RIM + ELKS + Munc13 ablation.
(A–C) Sample traces (A) and quantification of miniature excitatory postsynaptic current (mEPSC) frequencies (B) and amplitudes (C) in controlR+E+M and cKOR+E+M neurons, 27 cells/3 cultures each. (D–F) Sample traces (D) and quantification of miniature inhibitory postsynaptic current (mIPSC) frequencies (E) and amplitudes (F), 22/3 each. (G, H) Sample traces (G) and quantification (H) of EPSCs evoked by focal electrical stimulation, 20/3 each. (I–K) Sample traces (I) and quantification of amplitudes (J) and 20–80% rise times (K) of IPSCs evoked by focal electrical stimulation, controlR+E+M 28/3, cKOR+E+M 31/3. (L) Comparison of EPSCs normalized to their own controls for cKOR+E (absolute data from Figure 1C) and cKOR+E+M (from H) neurons, cKOR+E 19/3, cKOR+E+M 20/3. (M) Comparison of IPSCs normalized to their own controls for cKOR+E (absolute data from Figure 1E) and cKOR+E+M (from J) neurons, cKOR+E 18/3, cKOR+E+M 31/3. (N) Observed ratio and distribution of T* values for NMDAR-EPSCs after 100,000 rounds of hierarchical bootstrap, T* = [mean(cKOR+E)/mean(controlR+E)]/[mean(cKOR+E+M)/mean(controlR+E+M)]; 95% confidence intervals (Cl) and the probability of the null hypothesis PH0 are indicated, n as in (L). (O) As (N), but for IPSCs, n as in (M). Data are mean ± SEM unless noted otherwise; **p<0.01, ***p<0.001 as determined by Welch’s t-tests (K, L) or Mann–Whitney tests (B, C, E, F, H, J, M). For mEPSC and mIPSC kinetics in cKOR+E+M neurons, see Figure 4—figure supplement 1. For a workflow of the hierarchical bootstrap, see Figure 4—figure supplement 2.
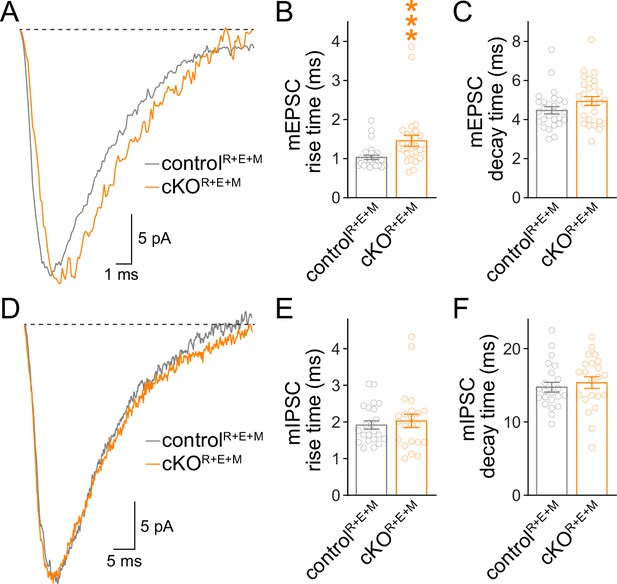
Assessment for the kinetics of spontaneous vesicle release in RIM + ELKS + Munc13 knockout neurons.
(A–C) Sample traces of the average miniature excitatory postsynaptic current (mEPSC) of a single controlR+E+M or cKOR+E+M neuron (A) and quantification of mEPSC rise times (B, 20%–80%) and decay times (C, τ), 27 cells/3 cultures each. (D–F) Same as (A–C), but for miniature inhibitory postsynaptic currents (mIPSCs), 22 cells/3 cultures each. Data are mean ± SEM; ***p<0.001 as determined by Mann–Whitney tests (B, C, E) or unpaired two-tailed Student’s t-tests (F).
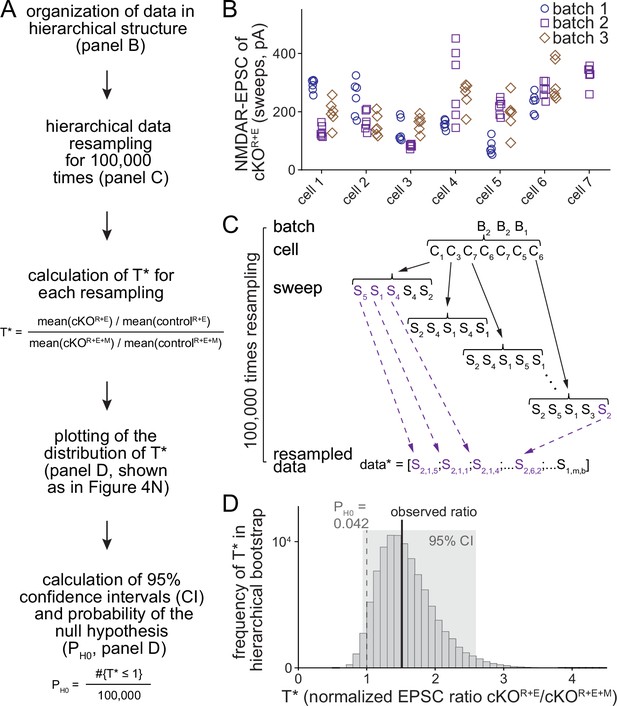
Workflow of the hierarchical bootstrap analyses.
(A) Analysis pipeline for the hierarchical bootstrap on the example of NMDAR-EPSCs in controlR+E, cKOR+E, controlR+E+M, and cKO R+E+M (data from Figure 4). For all cells, we resampled with replacement from the raw data, while preserving its nested, hierarchical structure. By repeating the process 100,000 times, the sampling distribution of the test statistic T was estimated and the distribution of T* values was used to calculate confidence intervals (CI) and the probability of the null hypothesis PH0 given the data. (B) Example of the raw data, shown for cKOR+E NMDAR-EPSC values. Each data point represents an individual measurement (sweep) from a specific cell within a specific culture. The tendency for repeated measurements from one cell to be more similar to each other than to measurements from other cells in this example is supported by two-way ANOVA (p<0.001). (C) Example of how resampling with replacement was performed in a hierarchical, nested manner. Starting with the cKOR+E group, we first resampled with replacement from the batches of cells. In all cases, there were three batches of cultures per experiment, and B2, B2, B1 might be drawn for this iteration. Starting with batch 2, the number of cells sampled in this batch is determined, and randomly resampled from the cells in this batch, replicating the same number but containing a different combination of cells. In the first example, the bootstrap cells C1, C3, C7, C6, C7, C5, C6 were drawn; cells 6 and 7 were included twice and cells 2 and 4 not included at all. Then we proceeded through this list of cells, each time randomly resampling (always with replacement) from the set of technical replicates for that cell and appending these resampled measurements to the bootstrap sample for this group. After doing this for each of the seven cells in the list, the entire process was repeated, first for batch 2 (again), but selecting a different sample of the seven cells, and then for batch 1. At the end, the bootstrap sample for this experimental group was exactly the same size as the original dataset, but containing a different subset of measurements and, importantly, preserving the nested, hierarchical structure of the original data. The above process was then repeated for the remaining three groups (controlR+E, cKOR+E+M, controlR+E+M), and T* was calculated using the formula T* = [mean(cKOR+E)/mean(controlR+E)]/[mean(cKOR+E+M)/mean(controlR+E+M)]; '*' denotes a bootstrap replicate of the test statistic. The entire procedure was repeated 100,000 times, producing an estimate of the sampling distribution of T. (D) Outcome of the hierarchical bootstrap analysis for NMDAR-EPSCs as shown in Figure 4N following the workflow in (A). The histogram shows the distribution of the 100,000 T* values used to calculate confidence intervals and evaluate the probability of the null hypothesis, PH0.
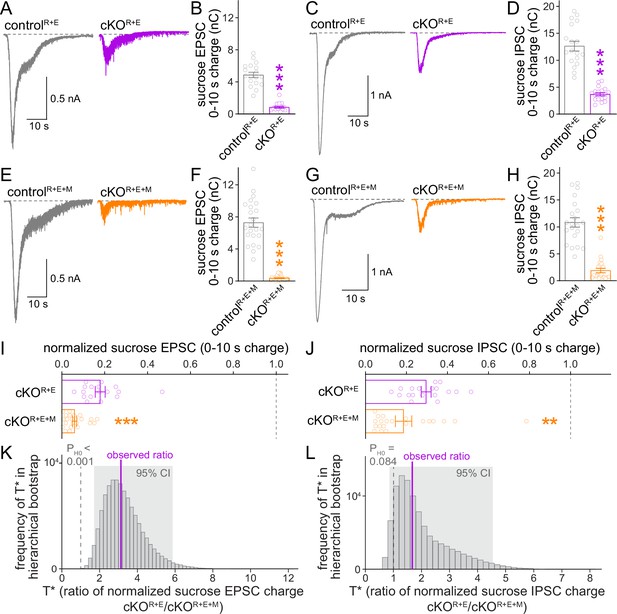
The remaining functional RRP in RIM + ELKS-deficient synapses depends at least in part on Munc13.
(A, B) Sample traces (A) and quantification (B) of excitatory postsynaptic currents (EPSCs) triggered by hypertonic sucrose in controlR+E and cKOR+E neurons, the first 10 s of the EPSC were quantified to estimate the RRP, controlR+E 18 cells/3 cultures, cKOR+E 17/3. (C, D) As (A, B), but for inhibitory postsynaptic currents (IPSCs), 18/3 each. (E–H) As for (A–D), but for controlR+E+M and cKOR+E+M neurons, (F) 23/3 each, (H) 21/3 each. (I) Comparison of EPSCs triggered by hypertonic sucrose normalized to their own controls for cKOR+E (absolute data from B) and cKOR+E+M (from F), cKOR+E 17/3, cKOR+E+M 23/3. (J) Comparison of IPSCs triggered by hypertonic sucrose normalized to their own controls for cKOR+E (absolute data from D) and cKOR+E+M (from H), cKOR+E 18/3, cKOR+E+M 21/3. (K) Observed ratio and distribution of T* values for sucrose EPSC charge after 100,000 rounds of hierarchical bootstrap, n as in (I). (L) As (K), but for sucrose IPSC charge, n as in (J). Data are mean ± SEM unless noted otherwise; **p<0.01, ***p<0.001 as determined by Mann–Whitney tests (B, F, H, I, J) or Welch’s t-tests (D).
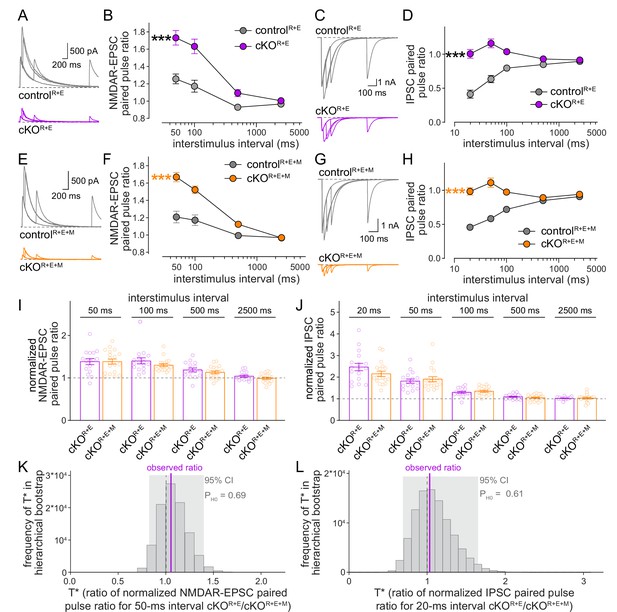
Vesicular release probability is not further impaired by combined RIM + ELKS + Munc13 knockout.
(A, B) Sample traces (A) and quantification (B) of excitatory postsynaptic current (EPSC) paired pulse ratios in controlR+E and cKOR+E neurons, controlR+E 15 cells/3 cultures, cKOR+E, 16/3. (C, D) As (A, B), but for inhibitory postsynaptic currents (IPSCs) (sample traces of 2500 ms intervals are not shown in C for simplicity), 17/3 each. (E–H) As for (A–D), but for controlR+E+M and cKOR+E+M neurons, (F) 19/3 each, (H) 19/3 each. (I) Comparison of EPSC paired pulse ratios across interstimulus intervals normalized to their own controls for cKOR+E (absolute data from B) and cKOR+E+M (from F), cKOR+E 16/3, cKOR+E+M 19/3. (J) Comparison of IPSC paired pulse ratios across interstimulus intervals normalized to their own controls for cKOR+E (absolute data from D) and cKOR+E+M (from H), cKOR+E 17/3, cKOR+E+M 19/3. (K) Observed ratio and distribution of T* values for EPSC paired pulse ratios at 50 ms interstimulus intervals after 100,000 rounds of hierarchical bootstrap, n as in (I). (L) As (K), but for IPSC paired pulse ratios at 20 ms interstimulus intervals, n as in (J). Data are mean ± SEM unless noted otherwise; ***p<0.001 as determined by two-way ANOVA followed by Bonferroni’s multiple-comparisons post-hoc tests (B, D, F, H) or by Mann–Whitney tests (I, J). For miniature EPSCs (mEPSCs) and miniature IPSCs (mIPSCs) in cKOR+E neurons, and for IPSCs evoked by stimulus trains in cKOR+E and cKOR+E+M neurons, see Figure 6—figure supplement 1.
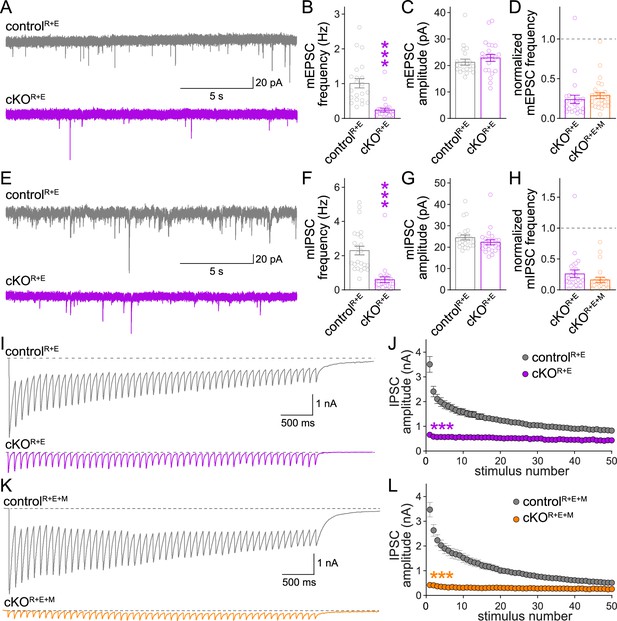
Additional analyses of spontaneous vesicle release and inhibitory postsynaptic currents (IPSCs) evoked by stimulus trains in RIM + ELKS and RIM + ELKS + Munc13 knockout neurons.
(A–C) Sample traces (A) and quantification of miniature excitatory postsynaptic current (mEPSC) frequencies (B) and amplitudes (C) in controlR+E and cKOR+E neurons, controlR+E 21 cells/3 cultures, cKOR+E 23/3. (D) Comparison of mEPSC frequencies normalized to their own controls for cKOR+E (absolute data from B) and cKOR+E+M (from Figure 4B), cKOR+E 23/3, cKOR+E+M 27/3. (E–G) Same as (A–C), but for miniature IPSCs (mIPSCs), 24/3 each. (H) Same as (D), but for comparison of mIPSC frequencies for cKOR+E (absolute data from F) and cKOR+E+M (from Figure 4E), cKOR+E 24/3, cKOR+E+M 22/3. (I, J) Sample traces (I) and quantification (J) of IPSC amplitudes evoked by stimulus trains (50 stimuli at 10 Hz) in controlR+E and cKOR+E neurons, 18/5 each. (K, L) Same as (I, J), but for controlR+E+M and cKOR+E+M neurons, controlR+E+M 21/3, cKOR+E+M 24/3. Data are mean ± SEM; ***p<0.001 as determined by Mann–Whitney tests (B–D, F–H) or two-way ANOVA followed by Bonferroni’s multiple-comparisons post-hoc tests (J, L).
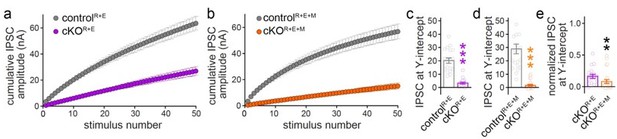
(a, b) Cumulative IPSC amplitude plots in stimulus trains (original data from Figure 6 —figure supplements 1I-1L, n’s as in those figures). (c, d) Back-extrapolation to time zero yields the IPSC-amplitudes at Y-intercept to estimate the recovery-corrected pool size for each cell, back-extrapolation was based on the last ten (4150) responses. (e) Comparison of IPSC at Y-intercept normalized to their own controls in cKOR+E (absolute data from c) and cKOR+E+M (from d) neurons.
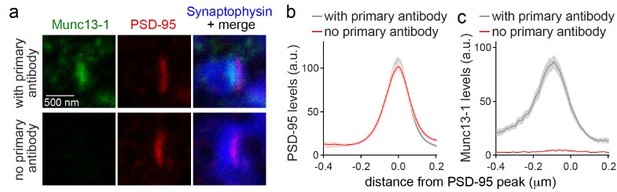
(a-c) Sample STED images (a) and quantification (b, c) of side-view synapses stained for Munc13-1 (imaged in STED), PSD-95 (imaged in STED), and Synaptophysin (imaged in confocal). Munc13-1 primary antibody was not added during staining process as negative control (“no primary antibody”). Peak position and levels were analyzed in line profiles (600 nm x 200 nm) positioned perpendicular to the center of elongated PSD-95 structure and aligned to the PSD-95 peak, 20 synapses/1 cultures each.
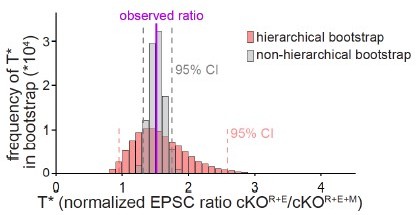
Comparison of hierarchical vs. non-hierarchical bootstrap for the analyses of NMDAR-EPSCs.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | Rims1tm3Sud/J (RIM1αβfl/fl) | Kaeser et al., 2008 | RRID:IMSR_JAX:015832 | |
| Genetic reagent (M. musculus) | Rims2tm1.1Sud/J (RIM2αβγfl/fl) | Kaeser et al., 2011 | RRID:IMSR_JAX:015833 | |
| Genetic reagent (M. musculus) | Erc1tm2.1Sud/J (ELKS1αfl/fl) | Liu et al., 2014 | RRID:IMSR_JAX:015830 | |
| Genetic reagent (M. musculus) | Erc2tm1.2Sud/J (ELKS2αfl/fl) | Kaeser et al., 2009 | RRID:IMSR_JAX:015831 | |
| Genetic reagent (M. musculus) | Unc13atm1.1Bros (Munc13-1fl/fl) | Banerjee et al., 2022 | MGI:7276178 | |
| Genetic reagent (M. musculus) | Unc13btm1Rmnd (Munc13-2-/-) | Varoqueaux et al., 2002 | RRID:MGI:2449706 | |
| Genetic reagent (M. musculus) | Unc13ctm1Bros (Munc13-3-/-) | Augustin et al., 2001 | RRID:MGI:2449467 | |
| Cell line (Homo sapiens) | HEK293T cells | ATCC | Cat# CRL-3216; RRID:CVCL_0063 | |
| Recombinant DNA reagent | pFSW EGFP Cre | Liu et al., 2014 | pHN131014; lab plasmid code (LPC): p009 | |
| Recombinant DNA reagent | pFSW EGFP ΔCre | Liu et al., 2014 | pHN131015; LPC: p010 | |
| Antibody | Anti-RIM (rabbit polyclonal) | SySy | Cat# 140003; RRID:AB_887774; lab antibody code (LAC): A58 | Immunofluorescence (IF) (1:500) |
| Antibody | Anti-PSD-95 (mouse monoclonal) | NeuroMab | Cat# 73-028; RRID:AB_10698024; LAC: A149 | IF (1:500) |
| Antibody | Anti- Synaptophysin (guinea pig polyclonal) | SySy | Cat# 101004; RRID:AB_1210382; LAC: A106 | IF (1:500) |
| Antibody | Anti-Munc13-1 (rabbit polyclonal) | SySy | Cat# 126103; RRID:AB_887733; LAC: A72 | IF (1:500); Western blot (WB) (1:1000) |
| Antibody | Anti-Synapsin-1 (mouse monoclonal) | SySy | Cat# 106001; RRID:AB_2617071; LAC: A57 | WB (1:4000) |
| Antibody | Anti-rabbit Alexa Fluor 488 (goat polyclonal) | Thermo Fisher | Cat# A-11034; RRID:AB_2576217; LAC: S5 | IF (1:200) |
| Antibody | Anti-mouse IgG2a Alexa Fluor 555 (goat polyclonal) | Thermo Fisher | Cat# A-21137; RRID:AB_2535776; LAC: S20 | IF (1:200) |
| Antibody | Anti-guinea pig Alexa Fluor 633 (goat polyclonal) | Thermo Fisher | Cat# A-21105; RRID:AB_2535757; LAC: S34 | IF (1:500) |
| Antibody | Anti-mouse peroxidase-conjugated (goat polyclonal) | MP Biologicals | Cat# 0855550; RRID:AB_2334540; LAC: S52 | WB (1:10000) |
| Antibody | Anti-rabbit peroxidase-conjugated (goat polyclonal) | MP Biologicals | Cat# 0855676; RRID:AB_2334589; LAC: S53 | WB (1:10000) |
| Software, algorithm | MATLAB | MathWorks | RRID:SCR_001622; https://www.mathworks.com/products/matlab.html | |
| Software, algorithm | Prism | GraphPad | RRID:SCR_002798; https://www.graphpad.com/scientific-software/prism | |
| Software, algorithm | Fiji/ImageJ | NIH | RRID:SCR_002285; https://imagej.net/software/fiji/downloads | |
| Software, algorithm | pClamp | Molecular Devices | RRID:SCR_011323; https://www.moleculardevices.com/products/software/pclamp.html | |
| Software, algorithm | MATLAB code for object recognition and analysis of 2D images | This paper | https://doi.org/10.5281/zenodo.6388196 | This code was adapted for 2D images from a previously generated code (Liu et al., 2018; Liu et al., 2022) and is freely accessible at zenodo.org |
| Software, algorithm | MATLAB code for hierarchical bootstrap | This paper | https://github.com/kaeserlab/Hierarchical_Bootstrap_Analysis_RB; Born, 2022 | This code was generated for this paper and is freely accessible at github.com |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/79077/elife-79077-mdarchecklist1-v2.pdf
-
Source data 1
Numerical data for all analyses shown in the figures.
- https://cdn.elifesciences.org/articles/79077/elife-79077-data1-v2.xlsx