Inducible lncRNA transgenic mice reveal continual role of HOTAIR in promoting breast cancer metastasis
Figures
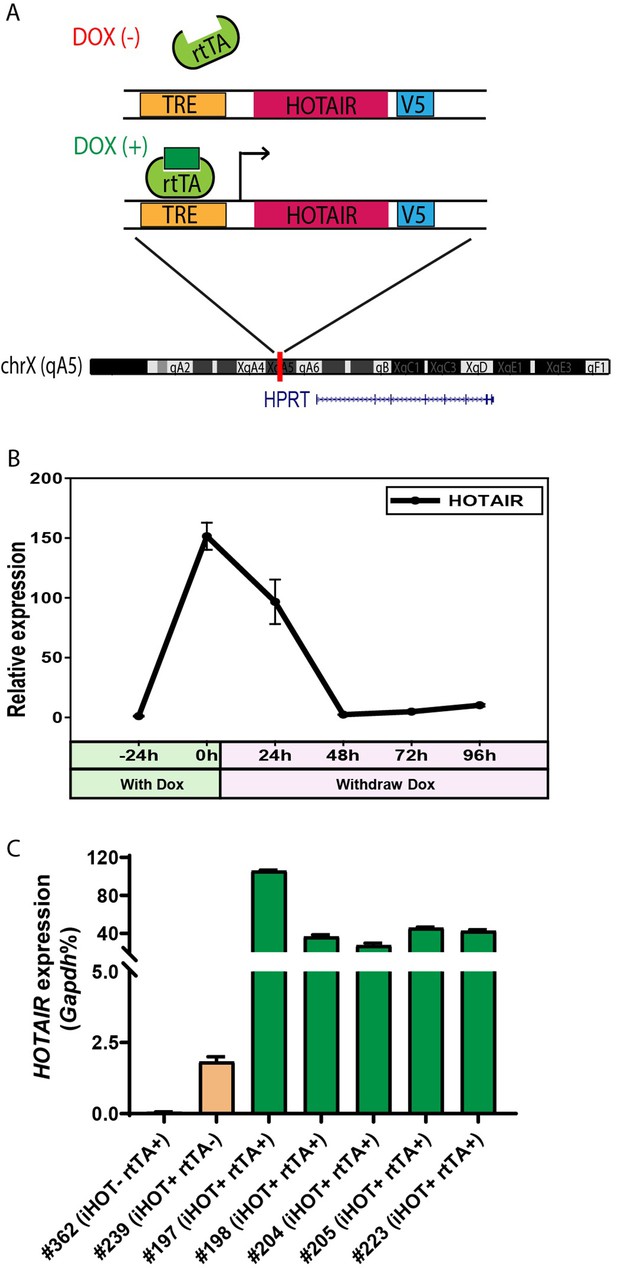
Generation of the inducible HOTAIR murine model.
(A) Schematic description of the Doxycyclin (Dox)-inducible HOTAIR (iHOT) construct and insertion loci. (B) HOTAIR expression decreased to baseline levels after Dox withdrawal in inducible HOTAIR embryonic stem (ES) cells. Inducible HOTAIR ES cells were treated with 1 μg/ml Dox for 24 hr and then withdrew Dox treatment for 96 hr. Human HOTAIR expression was detected by qRT-PCR, normalized to Gapdh and showed as fold change relative to −24 hr. Values are means ± standard deviation (SD). (C) High levels of HOTAIR are induced in HOTAIR-rtTA mice (iHOT+ rtTA+) compare to the control group (iHOT− rtTA− or iHOT+ rtTA−) after Dox administration. All the mice were treated with Dox for about 10 days. RNA was extracted from mice tails. Human HOTAIR levels were quantified by qRT-PCR and calculated as percentages of the mouse Gapdh transcript. Values are means ± SD.
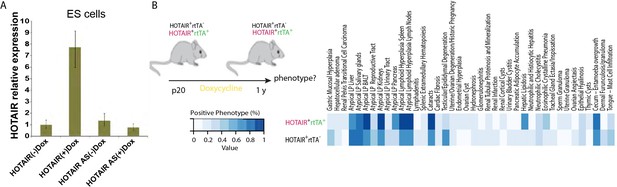
HOTAIR expression can be induced and inducible HOTAIR mice display no obvious phenotypes in different tissues.
(A) HOTAIR expression in inducible HOTAIR embryonic stem (ES) cells (AS: HOTAIR antisense controls). (B) Inducible HOTAIR mice have no obvious phenotypes in different tissues. Eight-week-old mice were fed with Dox water for 1 year. Phenotypes about hyperplasia and neoplasia, lymphoid system, age-related lesions, urinary system, liver and pancreas, respiratory, reproductive, and so on are tested and compared between HOTAIR+ rtTA+ and HOTAIR+ rtTA+ mouse. Each genotype has six mice.
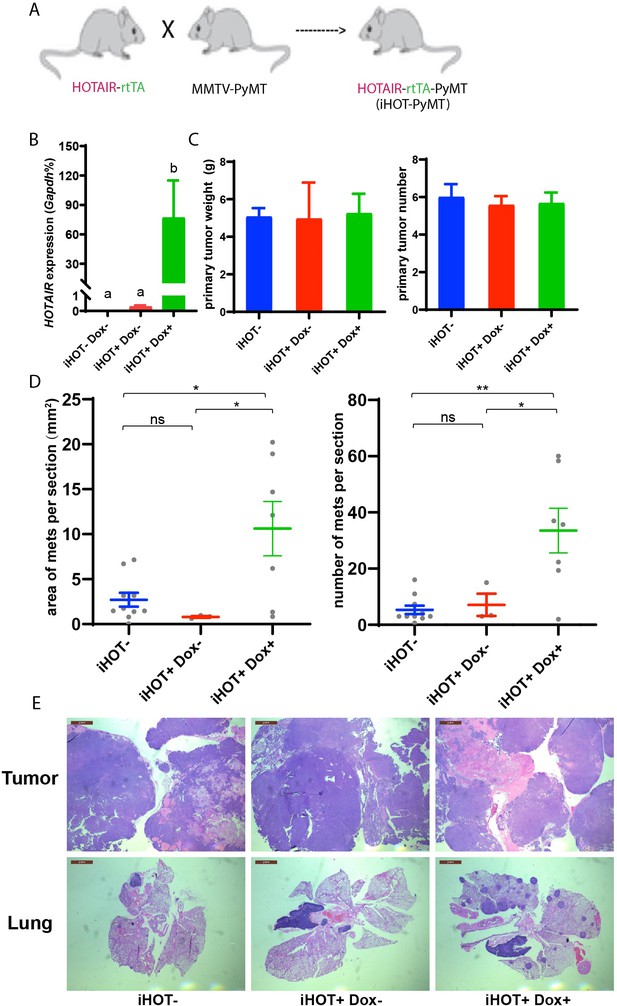
Induced HOTAIR promotes breast cancer metastasis in vivo.
(A) Cross schema for generating the inducible HOTAIR construct in MMTV-PyMT genetic background. (B) qRT-PCR measurements demonstrate that Dox-treated iHOT-PyMT mice display significantly higher levels of HOTAIR expression compared to untreated control groups or controls lacking the iHOT construct. Values are means ± standard error of the mean (SEM), n = 2–3. One-way analysis of variance (ANOVA) was performed to compare the HOTAIR expression levels between each group (p < 0.05). (C) There are no statistically significant differences between primary breast tumor mass in grams or number of tumors between Dox+-treated HOTAIR overexpressing mice compared to the untreated control group or controls lacking the inducible HOTAIR construct. Values are means ± SEM, n = 7–9, mice are 3–4 months old. One-way ANOVA was performed between each group and showed no significant differences. (D, E) iHOT+ mice treated with Dox display an increased number of lung metastases with greater volume. Quantification of lung metastases based on hematoxylin and eosin (H&E) staining of lung and primary tumor sections of iHOT+ Dox-treated mice, untreated controls, and controls lacking the iHOT construct. Tumor area in sections was calculated by ImageJ. Dashes represent means ± SEM, n = 3–10. Spots represent every single value. One-way ANOVA was performed (*p < 0.05, **p < 0.01, ns: not significant). Scale bar = 2 mm.
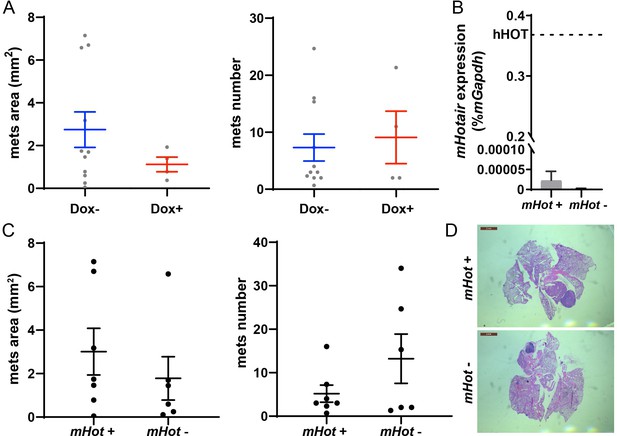
Dox treatment or endogenous mouse Hotair, expressed at a very low level, have no significant effects on breast tumor metastasis.
(A) Dox has no significant influence in tumor progression of MMTV-PyMT mice. MMTV-PyMT mice were treated with or without Dox for about 100–120 days. Quantification of lung metastases was based on hematoxylin and eosin (H&E) staining. Metastases area and numbers in sections were calculated by ImageJ. Dashes represent means ± standard error of the mean (SEM), n = 11 and 4. Spots represent every single value. Student’s t-test was performed and showed no significant difference between Dox− and Dox+ group. (B) Endogenous mouse Hotair expression is very low compared to transformed human HOTAIR. qRT-PCR was performed to measure endogenous mouse Hotair expression in mHotair+ and mHotair− MMTV-PyMT mice. Dashed line demonstrated human HOTAIR level in iHOT-PyMT mice without Dox treated (same as Figure 2B). Values are means ± SEM, n = 3 and 2. mHot is short for mouse Hotair, and hHOT is short for human HOTAIR. (C, D) Endogenous mouse Hotair knockout has no significant influence in breast tumor metastasis in MMTV-PyMT mice. Quantification of lung metastases was based on H&E staining. Metastases area and numbers in sections were calculated by ImageJ. Dashes represent means ± SEM, n = 6 and 7. Spots represent every single value. Student’s t-test was performed and showed no significant difference between mHotair+ and mHotair− group. Scale bar = 2 mm.
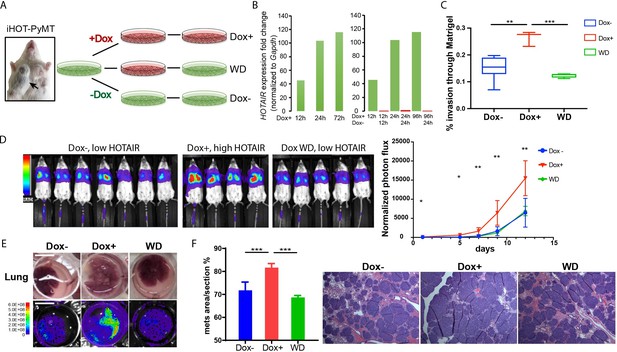
iHOT breast cancer cells display HOTAIR oncogene addiction.
(A) Treatment strategy of iHOT+ cells to assess the physiological properties of HOTAIR overexpressing cells in vitro. iHOT+ cells are isolated from primary mammary tumors of iHOT-PyMT mice. There are three treatment groups: Dox−, Dox+, Dox withdraw after Dox treatment (WD). (B) qRT-PCR measurement of HOTAIR expression in iHOT+ cells after Dox treatment for indicated time and withdrawal at 12 or 24 hr. HOTAIR is effectively induced after Dox treatment and rapidly decreased after Dox withdrawal in iHOT+ breast cancer cells. (C) Dox-induced HOTAIR overexpression promotes increased invasive capacity in iHOT+ cells and the increased invasive capacity of these cells can be rescued when HOTAIR is restored to baseline levels after Dox withdrawal. Matrix invasion assay of iHOT+ breast cancer cells in Dox−, Dox+, Dox withdraw conditions was performed. Invasive capacity was measured by quantifying the displacement of iHOT+ breast cancer cells through Matrigel in media which did or did not contain 2 μg/ml Dox. Values were showed as box plot (n = 3–8). Student’s t-test was performed between each group, **p < 0.01, ***p < 0.001. (D) Tail vein injection assay performed in SCID mice using iHOT+ breast cancer cells in Dox−, Dox+, Dox withdrawal conditions carrying a luciferase reporter. Dox-treated iHOT+ cells overexpressing HOTAIR display higher rates of lung colonization compare to the Dox− group (n = 4–7, another batch in Figure 3—figure supplement 1D). Student’s t-test was performed between Dox+ and Dox− group at each time point, *p < 0.05, **p < 0.05. We show that the potential for lung colonization by iHOT+ cells decreases after Dox withdrawal. (E) Representative photos and bioluminescent imaging of lungs dissected from SCID mice at the 2-week time point after tail vein injection of iHOT+ cells. Scale bar = 1 cm. (F) Quantified percentage of metastatic lung tumor area in Dox−, Dox+, Dox withdrawal cells after tail vein injection in lung sections with HE staining. Values are means ± standard deviation (SD), n = 4–7. One-way analysis of variance (ANOVA) was performed between each group, ***p < 0.001.
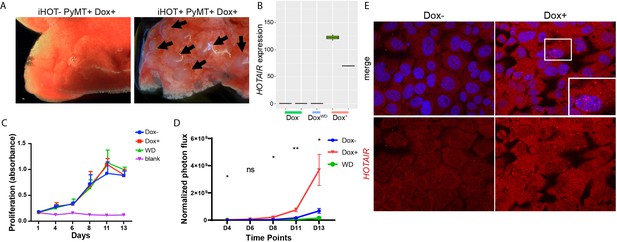
Induced HOTAIR promotes tumor cell metastasis but has no significant effect on cell proliferation.
(A) Images of HOTAIR overexpressing iHOT-PyMT mouse lungs under a dissecting microscope. Arrows indicate metastatic tumors colonizing lung tissue. (B) Expression of HOTAIR level (FPKM value) in RNA-seq data. (C) Dox−, Dox+, Dox withdraw treatments do not have a statistically significant effect on cellular proliferation. Blank controls are cells without substrate. (D) A replicated tail vein injection assay. Dox-treated iHOT+ cells overexpressing HOTAIR display higher rates of lung colonization compare to the Dox− group (n = 5–6). Student’s t-test was performed comparing cellular proliferation in the Dox+ and Dox− groups at each time point, *p < 0.05, **p < 0.05. (E) Human HOTAIR localization in Dox− and Dox+ iHOT+ cells with or without Dox treatment detected by smFISH. Cells were also counterstained with 4',6-diamidino-2-phenylindole(DAPI) to visualize the nuclei.
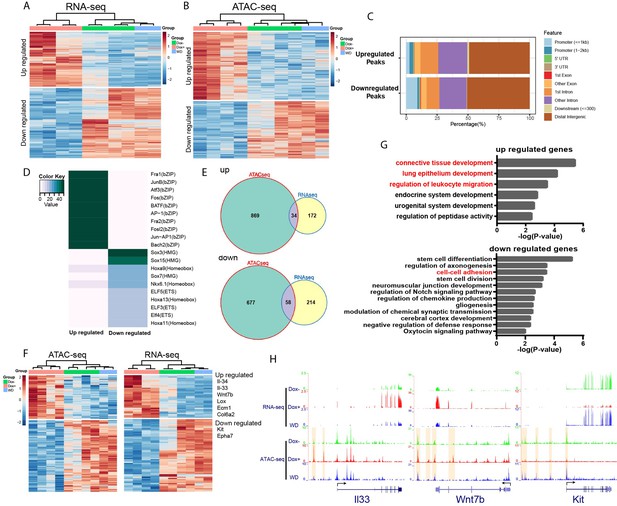
HOTAIR is required for metastasis-associated chromatin accessibility and genes expression.
(A) Hierarchical clustering heatmap of RNA-seq data of iHOT+ cells subject to either Dox−, Dox+, or Dox withdrawal treatment. Dox-induced HOTAIR overexpression results in altered gene expression profiles. There are 499 genes differentially expressed between Dox+ and Dox− cells (FDR <0.05, fold change >2). Transcriptome patterns of Dox withdrawal cells are similar to Dox− cells. (B) Hierarchical clustering heatmap of differential ATAC-seq peaks of iHOT+ cells with Dox−, Dox+, Dox withdraw treatments. Dox-induced HOTAIR overexpression alters chromatin accessibility. There are 1933 differential peaks between Dox+ and Dox− cells. Chromatin accessibility landscape of differential peaks returns to Dox− status after Dox withdrawal. (C) Feature distribution of differentially regulated peaks. We observe significantly more downregulated peaks located in promoter regions (less than 1 kb to transcription start sites) compared to upregulated peaks (p < 0.01). (D) Enriched motifs of differential ATAC-seq peaks by HOMER motif analysis. Color coding indicates −log10 p values. (E) Venn diagram representing overlap between ATAC-seq differential peak-associated genes and differentially expressed genes by RNA-seq (up: upregulated genes; down: downregulated genes). (F) Heatmap of transcriptome and corresponding chromatin status of overlapping genes in (E) in Dox−, Dox+, and Dox withdrawal conditions. Listed on the right are several representative genes related to cancer cell metastasis. (G) Gene ontology of overlapping genes in (E). Terms highlighted in red are examples of terms related to cancer cell metastasis. (H) HOTAIR regulates chromatin accessibility and gene expression. Normalized ATAC-seq and RNA-seq sequencing tracks of metastasis-related genes Il33, Wnt7b, and Kit. Alteration of chromatin accessibility correlates well with transcriptional changes.
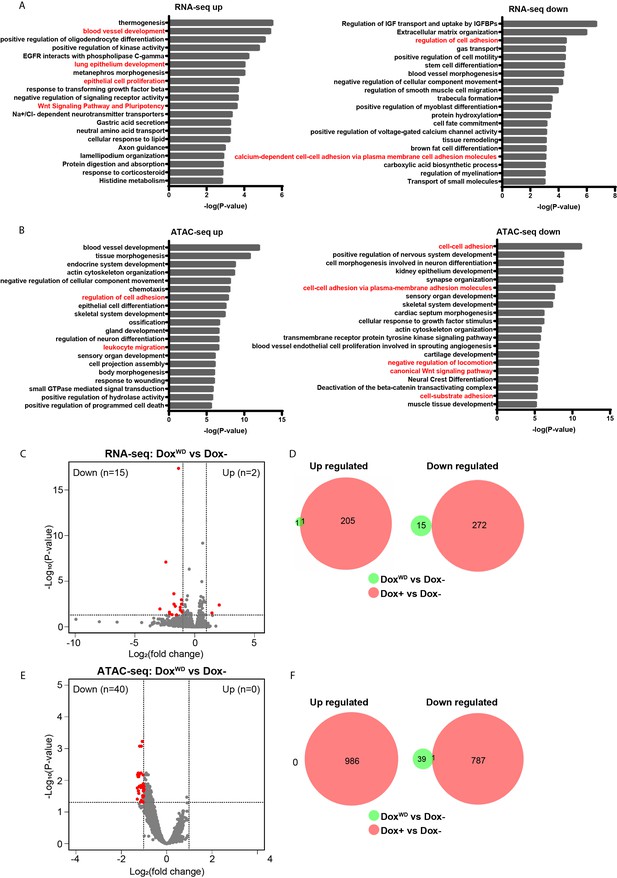
Gene ontology (GO) enrichment analysis of differential genes of Dox− vs. Dox+ in RNA-seq and ATAC-seq data and differential analysis between DoxWD and Dox−.
(A) GO enrichment analysis of differentially expressed genes (DEGs) by RNA-seq in Dox− and Dox+ iHOT cells. (B) GO enrichment analysis of differential peaks associated genes by ATAC-seq in Dox− and Dox+ iHOT cells. Terms highlighted in red are examples of terms related to cancer cell metastasis. (C) Volcano plot showing the DEGs between DoxWD and Dox− cells. Red dots indicate significant DEGs (p < 0.05, fold change >2). (D) Venn diagram representing overlapping DEGs between DoxWD vs. Dox− group and Dox+ vs. Dox− group. (E) Volcano plot illustrating the differential ATAC-seq peaks between DoxWD and Dox− cells. Red dots indicate significant differential peaks (p < 0.05, fold change >2). (F) Venn diagram representing overlapping differential ATAC-seq peaks between DoxWD vs. Dox− group and Dox+ vs. Dox− group.
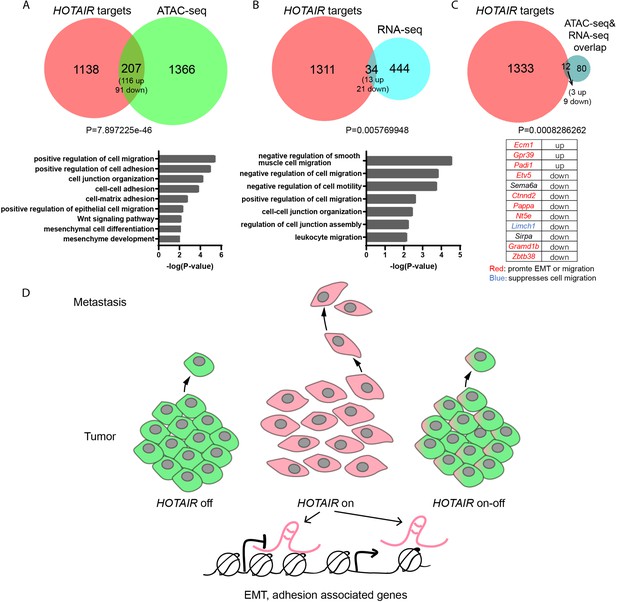
Potential targets of HOTAIR and model of HOTAIR’s function.
Venn diagrams representing overlap between genes associated with HOTAIR occupancy sites by ChIRP-seq (HOTAIR targets) and genes associated with ATAC-seq differential peaks (A) or differentially expressed genes by RNA-seq (B). p values were calculated by hypergeometric test to test for significance. Gene ontology (GO) terms correlated with overlapping genes are listed below the Venn diagrams. The HOTAIR ChIRP-seq data are from Chu et al., 2011. (C) Venn diagram illustrating overlap between HOTAIR targets and overlapping genes within ATAC-seq and RNA-seq data. Hypergeometric test was performed to test the significance. The overlapping genes and their expressing alterations in Dox+ cells were listed below. Genes marked in red were reported to promote migration and genes marked in blue were reported to suppress migration in previous publications. (D) A schematic figure illustrating the effect of HOTAIR in breast cancer cell metastasis. HOTAIR governs the expression of multiple epithelial-to-mesenchymal transition (EMT) and adhesion-associated genes by regulating their chromatin accessibility to promote tumor metastasis. Subsequent withdrawal of HOTAIR overexpression can revert the metastatic phenotype.
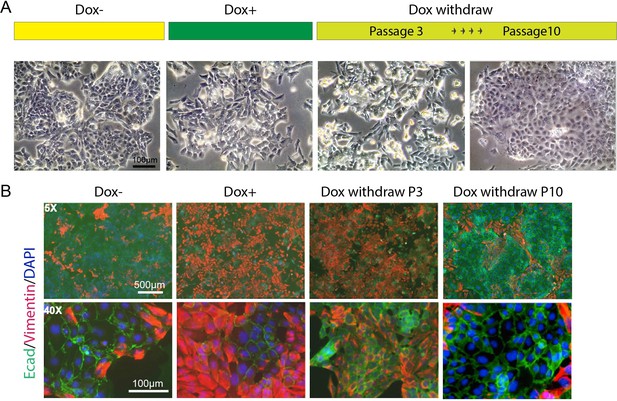
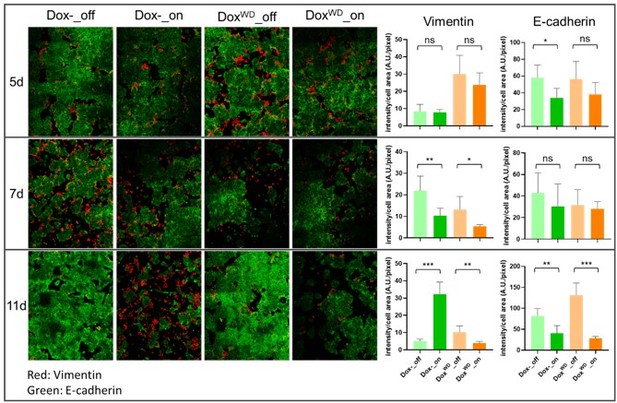
HOTAIR continually enforces epithelial–mesenchymal transition of breast cancer cells.
(A) Scheme demonstrating how iHOT+ cells were treated with Dox and microphotographs of cell morphology in each condition. (B) Immunofluorescence staining of E-cadherin (green), Vimentin (red), and DAPI (blue) of iHOT+ cells in each condition.