Inducible lncRNA transgenic mice reveal continual role of HOTAIR in promoting breast cancer metastasis
Peer review process
This article was accepted for publication as part of eLife's original publishing model.
History
- Version of Record published
- Accepted Manuscript updated
- Accepted Manuscript published
- Accepted
- Preprint posted
- Received
Decision letter
-
Erica A GolemisSenior and Reviewing Editor; Fox Chase Cancer Center, United States
-
Manish CharanReviewer; The Ohio State University, United States
-
Kenneth Nephew PhDReviewer
Our editorial process produces two outputs: (i) public reviews designed to be posted alongside the preprint for the benefit of readers; (ii) feedback on the manuscript for the authors, including requests for revisions, shown below. We also include an acceptance summary that explains what the editors found interesting or important about the work.
Decision letter after peer review:
Thank you for submitting your article "Inducible lncRNA transgenic mice reveal continual role of HOTAIR in promoting breast cancer metastasis" for consideration by eLife. Your article has been reviewed by 3 peer reviewers, and the evaluation has been overseen by a Reviewing Editor and Erica Golemis as the Senior Editor. The following individuals involved in the review of your submission have agreed to reveal their identity: Manish Charan (Reviewer #1); Kenneth Nephew PhD (Reviewer #3).
The reviewers have discussed their reviews with one another, and the Reviewing Editor has drafted this to help you prepare a revised submission.
Essential revisions:
1. The idea that HOTAIR is controlled by epigenetic loss needs to be documented more robustly. Even though the observed phenotypes could be reverted, it is unclear whether the cancer cells, with HOTAIR switched off, had conserved any molecular memory of HOTAIR overexpression. One criticism is that the study defines epigenetic memory only at the chromatin level or RNA levels. Cells might have a non-genetic memory that would distinguish them from cells that never experienced a high level of HOTAIR, without showing any RNA or ATAC seq differences. One way to tackle this issue would be to turn on HOTAIR again and define whether they develop migration and mesenchymal phenotypes more quickly than the control cells, which would experience HOTAIR overexpression for the first time. In the study, ATAC-seq and RNA-seq do show a return to normal after HOTAIR withdrawal, at least for differentially expressed peaks, but the analysis was done using DE genes and peaks identified and selected using no Dox vs Dox. The authors should run differential analyses between +dox vs WD-dox and control vs WD-dox to identify any RNA or ATAC remaining signals that might pinpoint an HOTAIR-mediated memory.
2. The HOTAIR RNA-mediated effect is not clear. The authors should characterize whether the iHOT+ cells also lose also their phenotypes in absence of the RNA to finalize the characterization of their mice model. For example, iHOT+ ASO-treated cells could be challenged for their migration capacity in comparison to control.
In addition, the reviewers have made a number of recommendations meant to improve the manuscript. Please try to address points on this list, consolidated below from the three reviews.
Recommendations for the authors:
1. Inclusion of H327me ChIP-seq experiments would be very interesting on many levels, including providing greater insight into the human HOTAIR-mouse genome chromatin landscape. While these experiments are strongly suggested for inclusion in the manuscript, these experiments could alternatively be included as a future direction in the Discussion section.
2. It would be useful to characterize HOTAIR expression in primary tumors and iHOT cancer cells. To allow the community to subsequently use the mouse model and iHOT cells, and eventually to design drugs for therapeutic purposes, the authors should comprehensively characterize the subcellular localizations of HOTAIR in situ (in tumors and cells) using smiFISH, and also the chromatin-associated loci of HOTAIR in iHOT+ cells using CHIRP. This set of experiments would further validate the mice model and cells.
https://doi.org/10.7554/eLife.79126.sa1Author response
Essential revisions:
1. The idea that HOTAIR is controlled by epigenetic loss needs to be documented more robustly. Even though the observed phenotypes could be reverted, it is unclear whether the cancer cells, with HOTAIR switched off, had conserved any molecular memory of HOTAIR overexpression. One criticism is that the study defines epigenetic memory only at the chromatin level or RNA levels. Cells might have a non-genetic memory that would distinguish them from cells that never experienced a high level of HOTAIR, without showing any RNA or ATAC seq differences. One way to tackle this issue would be to turn on HOTAIR again and define whether they develop migration and mesenchymal phenotypes more quickly than the control cells, which would experience HOTAIR overexpression for the first time. In the study, ATAC-seq and RNA-seq do show a return to normal after HOTAIR withdrawal, at least for differentially expressed peaks, but the analysis was done using DE genes and peaks identified and selected using no Dox vs Dox. The authors should run differential analyses between +dox vs WD-dox and control vs WD-dox to identify any RNA or ATAC remaining signals that might pinpoint an HOTAIR-mediated memory.
We thank the reviewer for their helpful suggestions. The reviewer was interested in whether the DoxWD cells develop any molecular memory.
The first way to assay molecular memory is to compare RNA-seq and ATAC-seq profiles of Dox- and DoxWD cells as the reviewer suggested. We compared Dox- vs DoxWD groups and identified very few DEGs (2 up-regulated and 15 down-regulated) by RNA-seq and similarly few differential peaks by ATAC-seq (0 up-regulated and 40 down-regulated) (Figure 4—figure supplement 1C&E). This result suggests that Dox- and DoxWD cells are quite similar in gene expression levels and chromatin status. In addition, these DEGs and differential peaks showed little overlap with DEGs or differential peaks within Dox+ and Dox- groups (Figure 4—figure supplement 1D&F), indicating that there were very few remaining signals that might suggest HOTAIR-mediated molecular memory. (We have added these results in our manuscript in line 369-372 & line 403-407.)
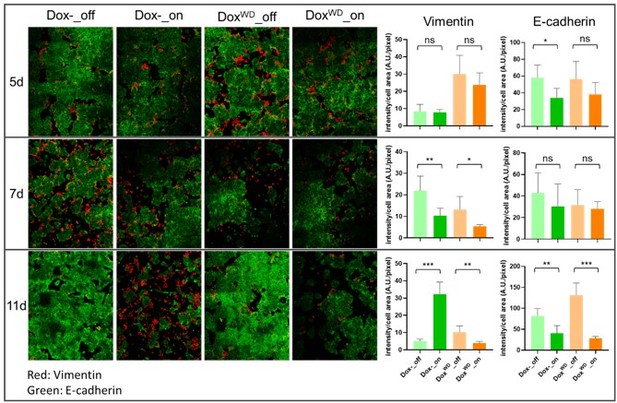
Second, we determined whether DoxWD cells respond differently compared to Dox- control cells when re-inducing HOTAIR expression. We turned on HOTAIR in both the Dox- cells and the DoxWD cells by treating with Dox and compared EMT markers (Vimentin and E-cadherin) between both cell lines. After treating with Dox for either 5 or 7 days, we observed no obvious changes in Vim and E-cad levels in both Dox- and DoxWD groups. After treating with Dox for 11 days, E-cad fluorescence intensity decreased in both the Dox- and DoxWD groups, but Vim fluorescence intensity increased only in the Dox- group. Our results suggest that re-inducing high levels of HOTAIR in DoxWD cells do not cause DoxWD cells to undergo EMT more quickly than Dox- cells which have never been exposed to HOTAIR overexpression, but perhaps even slower. However, upon repeating the experiment several more times, we observed large variations and batch effects between experiments, making it difficult to draw conclusions. Using FISH, we found that HOTAIR induction between cell lines was quite heterogenous, as shown in Figure 3—figure supplement 1E in the manuscript. The data suggests that re-inducing HOTAIR in DoxWD cell groups may result in a mix of cells which have never experienced HOTAIR overexpression or have experienced HOTAIR overexpression once or twice. The observed heterogeneity would make the interpretation of HOTAIR re-induction rather complicated. We will further pursue whether HOTAIR on-off cells show phenotypic memory in our future work.
2. The HOTAIR RNA-mediated effect is not clear. The authors should characterize whether the iHOT+ cells also lose also their phenotypes in absence of the RNA to finalize the characterization of their mice model. For example, iHOT+ ASO-treated cells could be challenged for their migration capacity in comparison to control.
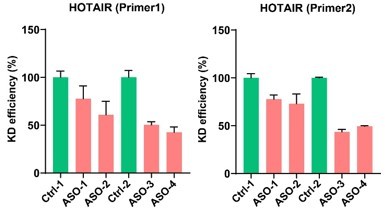
Thank you for the helpful suggestions. We have tried several ASOs, but unfortunately the knockdown efficiency of the tested species is not good. The best efficiency we have observed to date is about 44%. Therefore, it is experimentally challenging to test the migration capacity of ASO-treated iHOT+ cells. As we know that most commercially available synthetic modified ASOs are designed to target intronic sequences, which are ideal targets of RNase H in the nucleus and cause splicing defects or RNA degradation more efficiently. However, in our transgenic model, the human HOTAIR cDNA sequence inserted into the mouse genome does not contain introns. Therefore, the ASO-mediated knockdown approach may not be the best strategy for this model.
In addition, the reviewers have made a number of recommendations meant to improve the manuscript. Please try to address points on this list, consolidated below from the three reviews.
Recommendations for the authors:
1. Inclusion of H327me ChIP-seq experiments would be very interesting on many levels, including providing greater insight into the human HOTAIR-mouse genome chromatin landscape. While these experiments are strongly suggested for inclusion in the manuscript, these experiments could alternatively be included as a future direction in the Discussion section.
Thanks for the helpful comments. We have included this suggestion as a future direction in the Discussion section (line 624-628).
2. It would be useful to characterize HOTAIR expression in primary tumors and iHOT cancer cells. To allow the community to subsequently use the mouse model and iHOT cells, and eventually to design drugs for therapeutic purposes, the authors should comprehensively characterize the subcellular localizations of HOTAIR in situ (in tumors and cells) using smiFISH, and also the chromatin-associated loci of HOTAIR in iHOT+ cells using CHIRP. This set of experiments would further validate the mice model and cells.
Thank you for your suggestion! We have analyzed the HOTAIR expression levels in primary tumors using qRT-PCR as shown in Figure 2B.
For iHOT cells isolated from the tumors, we analyzed the subcellular localizations of HOTAIR in iHOT cells using smFISH according to the reviewer’s suggestions. As shown in Figure 3—figure supplement 1E, our results reveal that HOTAIR localizes to both the nucleus and cytoplasm. The induction of HOTAIR is quite heterogeneous across cell populations. (We have added this result in the manuscript in line 262-265.)
We also further analyzed the published HOTAIR ChIRP-Seq data to see whether HOTAIR-associated genes are altered with HOTAIR induction in iHOT cells. The previous study identified 832 HOTAIR occupancy sites genome-wide in human breast cancer cells, which are associated with 1345 genes (HOTAIR targets) in total (Chu et al., MolCell, 2011). We compared HOTAIR targets established from previous literature with DEGs and differential peak-associated genes identified from our RNA-seq and ATAC-seq assays on Dox- versus Dox+ iHOT cells. As shown in Figure 4—figure supplement 2A, we observed significant overlaps between HOTAIR targets and ATAC-seq differential peak-associated genes (207 genes, ~13% of ATAC-seq differential peak-associated genes). Among these shared genes, the chromatin accessibility of 116 genes were up-regulated and 91 were down-regulated. Our gene ontology (GO) analysis uncovered terms such as cell adhesion, EMT or migration (Figure 4—figure supplement 2A). We also noted significant overlap between HOTAIR targets and DEGs in our RNA-seq data (34 total, 13 genes were up-regulated and 21 genes were down-regulated in Dox+ cells compared to control, Figure 4—figure supplement 2B). GO analysis of these genes also revealed associations with multiple EMT and migration related terms (Figure 4—figure supplement 2B). Furthermore, we compared the HOTAIR targets to genes regulated by HOTAIR through chromatin accessibility, which were represented as overlapping genes shared between our ATAC-seq and RNA-seq datasets (Figure 4E). We identified 12 genes out of a total of 92 genes (~13%) which may be direct targets regulated by HOTAIR in iHOT cells (Figure 4—figure supplement 2C). Interestingly, almost all of these genes were associated with migration. Genes shown to promote migration such as Ecm1, Gpr39, Padi1 were up-regulated and genes shown to suppress migration such as Limch1 were down-regulated (Figure 4—figure supplement 2C). These results suggest that HOTAIR regulates the expression of multiple cell adhesion and EMT associated genes by regulating their chromatin accessibility to promote tumor metastasis. (We have added these results to the Discussion section in line 596-623.)
https://doi.org/10.7554/eLife.79126.sa2