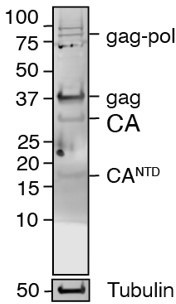
UBQLN2 restrains the domesticated retrotransposon PEG10 to maintain neuronal health in ALS
Figures
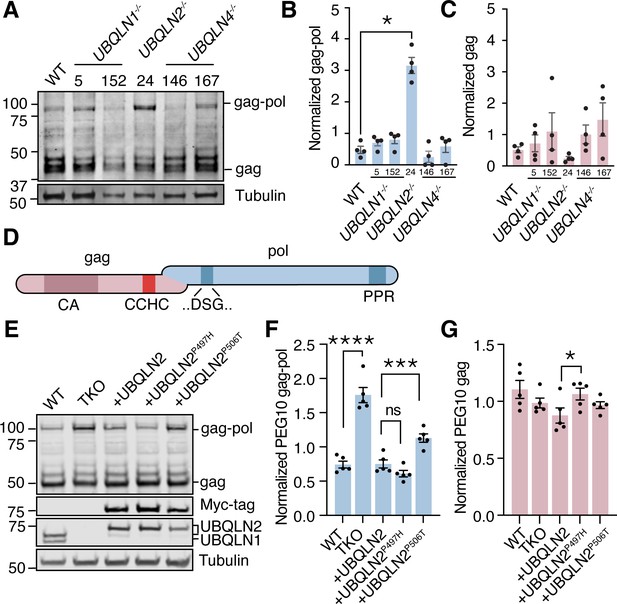
Ubiquilin 2 (UBQLN2) regulates paternally expressed gene 10 (PEG10) gag-pol abundance.
(A) Human embryonic stem cells (ESCs) had individual Ubiquilin (UBQLN) genes deleted by CRISPR gene editing and clones were probed by western blot for endogenous PEG10 protein. Full-length gag-pol protein accumulates only upon UBQLN2 loss. n=4 independent experiments. Two independently generated knockout lines are shown for UBQLN1 and UBQLN4; clone identification numbers are shown at top of the blot. (B–C) Quantification of gag-pol (B) and gag (C) abundance in hESC cell lines of (A). PEG10 protein was normalized to Tubulin, then normalized to the average intensity for each individual experiment. n=4 independent experiments and significance was determined by multiple comparisons test. Mean ± SEM is shown. No differences in gag (C) were detected with an ordinary one-way ANOVA. (D) Schematic of PEG10 protein. The first reading frame (gag) contains a capsid-like (CA) region, as well as a retroviral zinc finger (‘CCHC’). The pol-like sequence contains a retroviral-type aspartic protease with one active site ‘DSG’ motif, as well as a C-terminal polyproline repeat domain (PPR). (E) WT or TKO cells stably transfected with doxycycline- (dox-) inducible constructs expressing Myc-UBQLN2, Myc-UBQLN2P497H, or Myc-UBQLN2P506T were probed for endogenous PEG10. (F) Quantitation of gag-pol abundance in mutant UBQLN2-expressing cells. Gag-pol was normalized to Tubulin and to the average intensity of each experiment. Mean ± SEM is shown for each condition. n=5 wells per condition collected from two different passages. (G) Quantitation of gag abundance. Shown is the mean ± SEM of n=5 wells. Multiple comparison tests were run with Bonferroni correction to compare WT and triple knockout (TKO) cells as well as WT OE with the two mutant lines. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 1—source data 1
Uncropped blot for Figure 1A.
- https://cdn.elifesciences.org/articles/79452/elife-79452-fig1-data1-v2.pdf
-
Figure 1—source data 2
Uncropped blot for Figure 1E.
- https://cdn.elifesciences.org/articles/79452/elife-79452-fig1-data2-v2.pdf
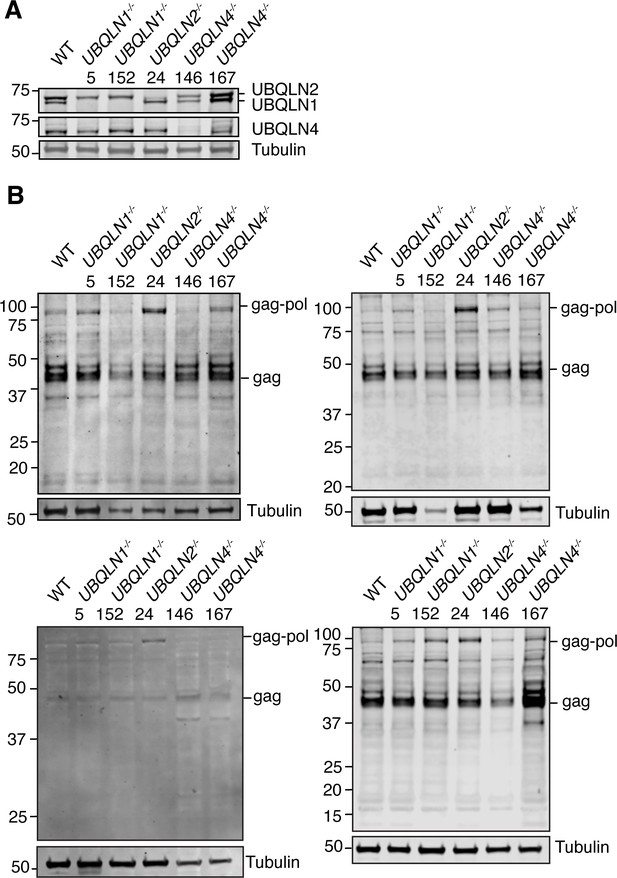
Validation of human embryonic stem cells (hESC) Ubiquilin (UBQLN) loss.
(A) Human ESCs had individual UBQLN genes deleted by CRISPR gene editing and clones were probed by western blot for each Individual UBQLN. n=3 representative blots. (B) Uncropped independent replicates of blots from Figure 1A were used for quantification in B-C. Numbers at top of the blot are clone identification numbers.
-
Figure 1—figure supplement 1—source data 1
Uncropped blot for Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/79452/elife-79452-fig1-figsupp1-data1-v2.pdf
-
Figure 1—figure supplement 1—source data 2
Uncropped blot for Figure 1—figure supplement 1B.
- https://cdn.elifesciences.org/articles/79452/elife-79452-fig1-figsupp1-data2-v2.pdf
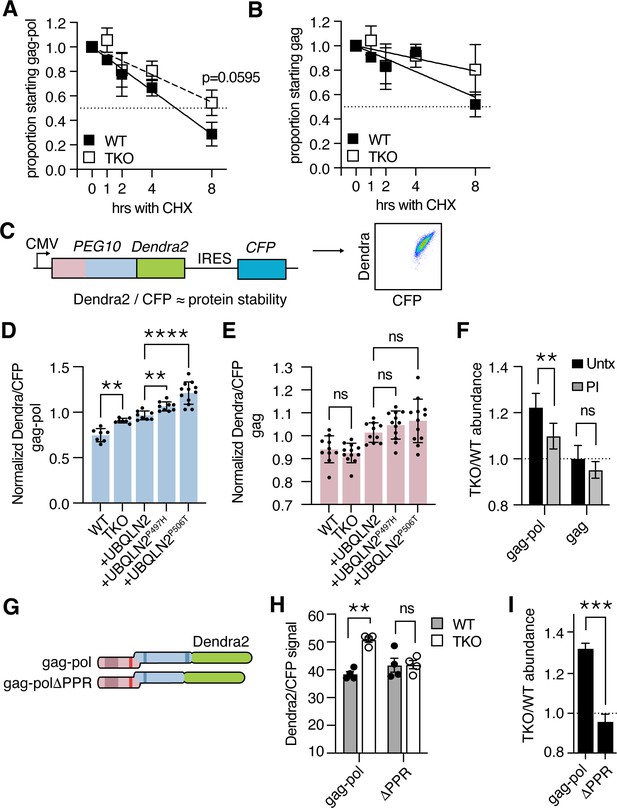
Ubiquilin 2 (UBQLN2) facilitates proteasome-dependent degradation of paternally expressed gene 10 (PEG10) gag-pol through the C-terminus of PEG10.
(A–B) WT (filled squares) and UBQLN TKO (open squares) HEK cells were transfected with an HA-tagged form of PEG10 expressed under the control of a CMV promoter, treated with cycloheximide, and chased for 8 hr followed by western blot. (A) The half-life of PEG10 gag-pol is between 4–8 hr for WT cells, and more than 8 hr for UBQLN-deficient cells. (B) The half-life of the PEG10 gag is longer than 8 hr for WT and TKO cells. For (A–B) n=3 independent experiments and mean ± SEM is shown. No timepoint was deemed significantly different between WT and triple knockout (TKO) cells by paired t-test; p-value of 8 hr gag-pol timepoint (A) is shown. (C) Schematic of PEG10 protein abundance reporter. PEG10 is fused at the 3´ end to Dendra2, followed by an IRES-CFP. Right: example dot plot showing Dendra2 and CFP signal in transfected cells. (D–E) Dendra2 over CFP MFI ratio for PEG10 gag-pol (D) and gag (E) in WT and UBQLN1, 2, and 4 ‘TKO’ HEK293 cells. TKO cells were rescued with the expression of WT or mutant UBQLN2 alleles as in Figure 1E–G. Significance was determined by multiple comparisons test and mean ± SEM is shown of n=7 independent experiments. (F) WT and TKO cells were transfected with either gag-pol or gag and incubated in the presence or absence of proteasome inhibitor (PI) for 12 hr. The ratio of Dendra2/CFP was determined for each cell population and the normalized ratio of PEG10 for TKO/WT cells is shown. Values over 1.0 indicate dependence on UBQLNs for restriction. Significance was determined by Student’s t-test. n=3 independent experiments. (G) WT and truncation mutant of PEG10 gag-pol fused to the fluorophore Dendra2. ΔPPR is missing the last 27 amino acids containing the polyproline repeat. (H) Protein abundance of PEG10-Dendra2 fusions was determined for WT (filled circles) and TKO (open circles) cells by flow cytometry n=4 independent experiments. Significance was determined by multiple comparisons test and mean ± SEM is shown. (I) TKO/WT abundance values for data in (H). Values over 1.0 indicate dependence on UBQLNs for restriction. Mean ± SEM is shown. Significance was determined by Student’s t-test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
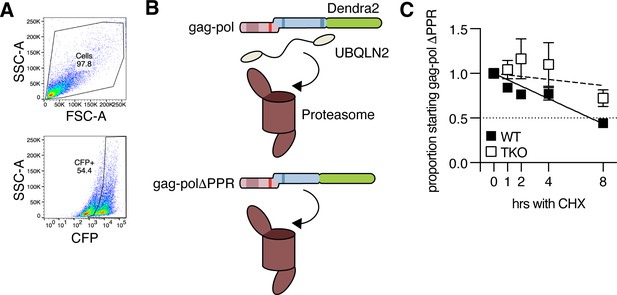
Paternally expressed gene 10 (PEG10) and Ubiquilin 2 (UBQLN2) specificity.
(A) Example gating strategy of abundance reporter test. ‘Cells’ are gated (top), then gated on the CFP +population (bottom, with Dendra2-green fluorescence of CFP +population shown at right). A custom parameter of Dendra2/CFP is then generated on a per-cell basis and the geometric mean fluorescence intensity (MFI) of this custom parameter is averaged per sample. (B) Model of degradation pathways of gag-pol with and without the PPR. Gag-pol containing the polyproline region (PPR) requires UBQLN2 for delivery to the proteasome. Without the PPR, gag-pol can be degraded by typical proteasomal degradation pathways. (C) Western blot data of cycloheximide (CHX) chase of ΔPPR PEG10 in WT (black square) and triple knockout (TKO) (outline square) HEK cells over 8 hr. Shown is the mean ± SEM from three independent experiments.
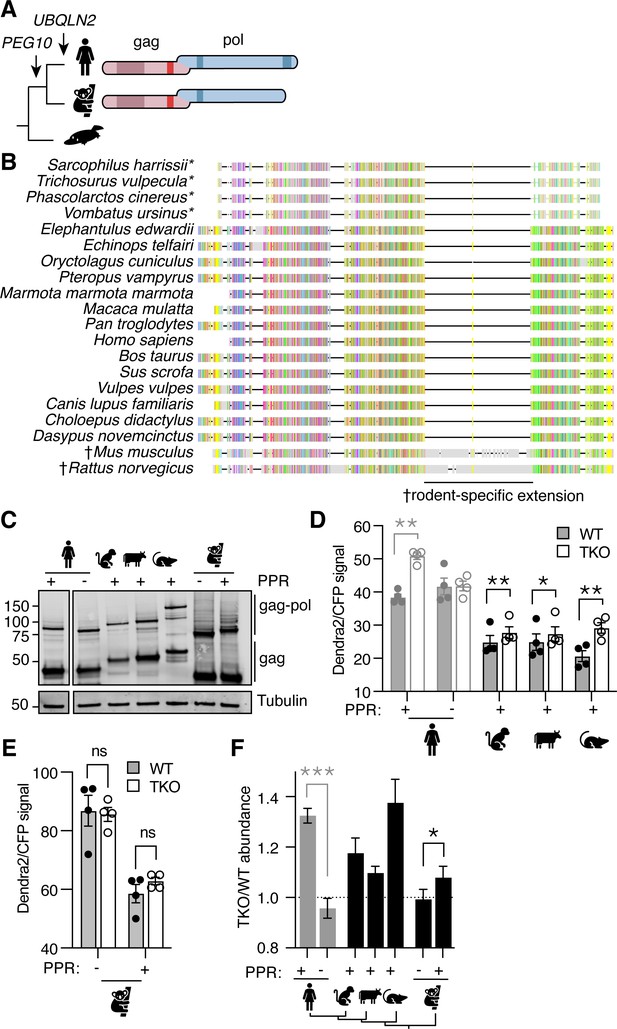
Phylogenetic investigation of the Ubiquilin 2 (UBQLN2)-paternally expressed gene 10 (PEG10) relationship.
(A) Evolutionary schematic of PEG10 protein in eutherian and marsupial mammals. Monotremes (bottom) do not contain PEG10 or UBQLN2 genes. PEG10 and UBQLN2 appearances are highlighted with arrows. PEG10 schematic highlights the lack of C-terminal polyproline repeat region in marsupials. (B) Amino acid alignment of mammalian PEG10 from a diversity of mammalian species showing general conservation and lack of C-terminal polyproline domain in marsupials (starred). Colors represent the conservation of aligned amino acids; the proline is yellow. † highlights the rodent-specific extension within pol. (C) Western blot demonstrating expression of mammalian PEG10 gag and gag-pol in WT cells. PEG10 was detected by N-terminal HA-tag. (D–E) Dendra2-green over CFP MFI ratios for PEG10 from various placental mammals (D) and the marsupial Koala (E) in WT (filled circles) and triple knockout (TKO) (open circles) cells. Shown is the mean ± SEM from four independent experiments, with triplicate transfection wells for each. Human is shown in gray and is duplicated from Figure 2 for ease of visualization. Statistics were determined by multiple comparisons tests. (F) The ratio of Dendra2/CFP was determined for each cell population in (D–E) and the normalized ratio of PEG10 for TKO/WT cells is shown. Values over 1.0 indicate dependence on UBQLNs for restriction. Significance was determined by unpaired (human) or paired (koala) Student’s t-test. n=4 independent experiments. For all experiments, the mean ± SEM is shown. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 3—source data 1
Uncropped blot for Figure 3C.
- https://cdn.elifesciences.org/articles/79452/elife-79452-fig3-data1-v2.pdf
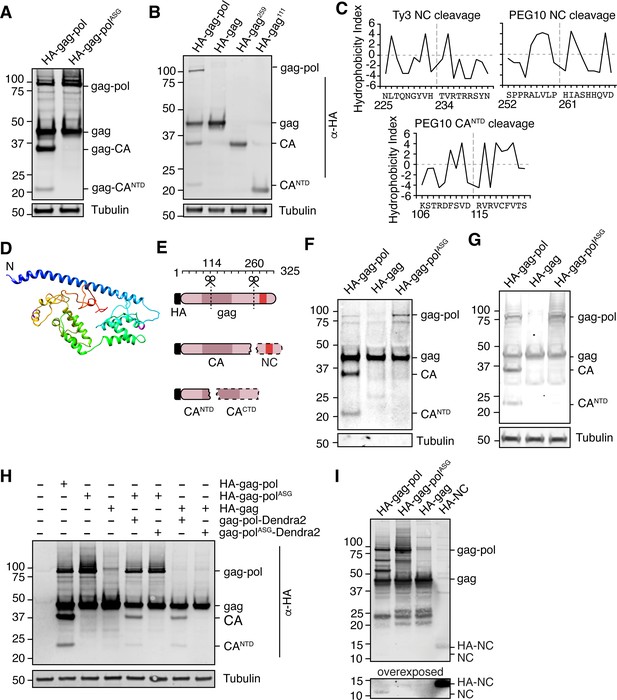
Paternally expressed gene 10 (PEG10) self-cleaves to generate a classical ‘NC’ fragment.
(A) Mutation of the active site aspartic acid in the protease domain results in the disappearance of cleaved PEG10 products. N-terminally HA-tagged PEG10 was expressed either as WT or ‘ASG’ protease mutant in cells and probed by western blot for HA. Based on estimated molecular weight, the fragments are estimated to encompass gag-capsid (CA) and gag-capsid(NTD) (CANTD) fragments. n=4 independent experiments. (B) HA-tagged, C-terminally truncated forms of PEG10 were expressed in cells to approximate self-cleavage sites via Molecular weight. n=3 independent experiments. (C) Kyte-Doolittle (K-D) hydropathy analysis of Ty3 (left, similar to that in Kirchner and Sandmeyer, 1993) and PEG10 nucleocapsid (right) and CANTD (bottom) cleavages. Hydropathy measurements for each amino acid were plotted on a scale from P9 to P9’. Amino acid locations are shown below the amino acid alignment. Estimated cleavage locations by molecular weight in Figure 3D are close in proximity to those estimated by K-D analysis. (D) Structure prediction of PEG10 gag (blue N-terminus) shows that estimated cleavage sites appear accessible by protease. AA114-115 (purple) and AA260-261 (magenta) are highlighted to show estimated cleavages. (E) Model of PEG10 self-cleavage. PEG10 cleaves gag to generate a liberated nucleocapsid (NC) fragment. PEG10 also cleaves the gag-CA domain into CANTD and CACTD. Dotted lines indicate that the fragments are not visible by western blot due to the absence of the N-terminal HA tag. (F) PEG10 gag is capable of being cleaved by PEG10 gag-pol in trans. HA-tagged and PEG10-Dendra2 fusion constructs were co-transfected into cells and the presence of HA-tagged cleavage products was assessed by western blot. n=2 independent experiments. (G) Presence of cleaved PEG10 products in virus-like particles (VLPs). VLPs were isolated from PEG10-transfected HEK cells by ultracentrifugation and probed for cleavage products by western blot. Tubulin was used as a control for contamination of the conditioned medium with cell fragments. n=3 independent experiments. (H) Western blot of accompanying cell lysate from VLP preparations. n=3 independent experiments. (I) Visualization of the endogenously-cleaved NC fragment by western blot. HEK cells were transfected with the listed constructs and prepared for western blot using a custom-generated antibody against PEG10 AA259-325. HA-tagged NC can be seen at ~14 kDa, and endogenously cleaved NC is at ~11 kDa and is only visible upon expression of cleavage-competent gag-pol.
-
Figure 4—source data 1
Uncropped blot for Figure 4A.
- https://cdn.elifesciences.org/articles/79452/elife-79452-fig4-data1-v2.pdf
-
Figure 4—source data 2
Uncropped blot for Figure 4B.
- https://cdn.elifesciences.org/articles/79452/elife-79452-fig4-data2-v2.pdf
-
Figure 4—source data 3
Uncropped blot for Figure 4F.
- https://cdn.elifesciences.org/articles/79452/elife-79452-fig4-data3-v2.pdf
-
Figure 4—source data 4
Uncropped blot for Figure 4G.
- https://cdn.elifesciences.org/articles/79452/elife-79452-fig4-data4-v2.tif
-
Figure 4—source data 5
Uncropped blot for Figure 4H.
- https://cdn.elifesciences.org/articles/79452/elife-79452-fig4-data5-v2.pdf
-
Figure 4—source data 6
Uncropped blot for Figure 4I.
- https://cdn.elifesciences.org/articles/79452/elife-79452-fig4-data6-v2.pdf
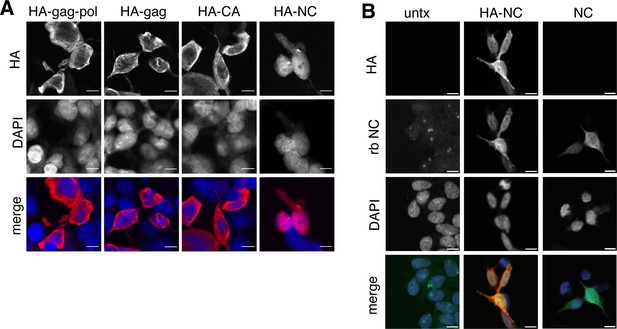
Isolated NC is uniquely found in the nucleus.
(A) Localization of paternally expressed gene 10 (PEG10) fragments using tagged antibodies. Cells were transfected with HA-tagged PEG10 constructs, stained, and imaged by confocal microscopy. Scale bar 10 μm. HA has been colored red and DAPI blue in merged images. Shown are representative cells from 10 fields of view of each construct. n=3 independent experiments. (B) Localization of PEG10 NC fragment using custom NC antibody. Cells were transfected either with HA-tagged NC or untagged NC construct and stained with the listed antibodies. Scale bar 10 μm. HA has been colored red, NC green, and DAPI blue in merged images. Shown are representative cells from 10 fields of view of each construct. n=1 experiment.
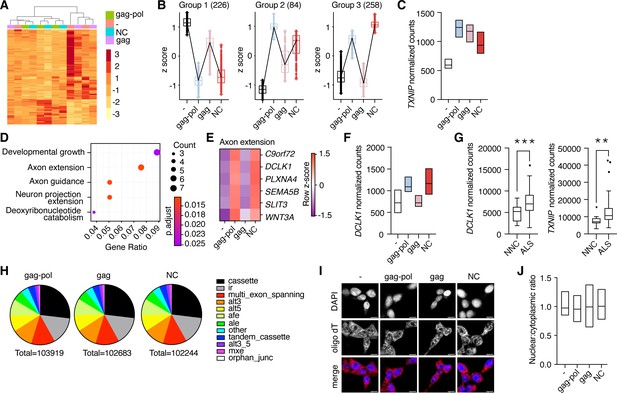
Liberated nucleocapsid alters transcription of axon extension genes.
(A) Heatmap showing Euclidean clustering of expression profiles of HEK cells transfected with PEG10 constructs with top 200 altered genes between gag-pol and negative control plasmid. (B) Cluster profiling of gene expression effects as measured by RNA-seq analysis upon paternally expressed gene 10 (PEG10) construct overexpression. The number of genes in each group is listed in parentheses. Data are shown as box and whiskers min to max with the line at the median. (C) Normalized counts of TXNIP transcript from RNA-seq analysis of three biological replicates from PEG10 transfected or control transfected cells. (D) Top gene expression changes in NC-transfected cells by GO-term enrichment analysis. The top five GO-terms ranked by adjusted p-value are shown. Adjusted p-value is shown by color, and the size of the datapoint reflects the number of genes enriched in the pathway. (E) Heatmap of genes from the Axon extension GO-term showing Row z-score for each gene in the pathway. (F) Normalized counts of DCLK1 from RNA-seq analysis of PEG10-transfected cells. (G) RNA-seq data from the Target Amyotrophic Lateral Sclerosis (ALS) dataset of post-mortem lumbar spinal cords were analyzed for DCLK1 (left) and TXNIP (right) counts. NNC = non-neurological control. n=17 NNC and 127 ALS samples. For (C,F), data are shown as min-max floating bars with the line at mean and significance were determined by DESeq2. For (G), data are shown as a 5–95% box and whisker plot, and significance was determined by DESeq2. (H) Splice pattern changes upon PEG10 overexpression compared to control. Nucleocapsid expression does not alter patterns of splice alteration compared to gag-pol or gag expression. Splice changes were quantified and classified by MAJIQ analysis (Vaquero-Garcia et al., 2016). Total splice alteration counts are shown below in pie charts. ir = intron retention; afe/ale = alternative first/last exon; mxe = mutually exclusive exons. (I) Oligo-dT FISH showed no changes to bulk mRNA trafficking upon PEG10 overexpression. Scale bar 10 μm. Shown are representative images from 10 recorded fields of view. n=3 independent experiments. (J) Quantification of oligo-dT signal in the nucleus versus cytosol for each condition imaged in (I) showing no changes to mRNA trafficking upon PEG10 overexpression. Quantification was performed on a minimum of 60 images from each condition and is representative of 3 independent experiments. *p<0.05, **p<0.01, ***p<0.001.
-
Figure 6—source data 1
RNA-seq results.
- https://cdn.elifesciences.org/articles/79452/elife-79452-fig6-data1-v2.xlsx
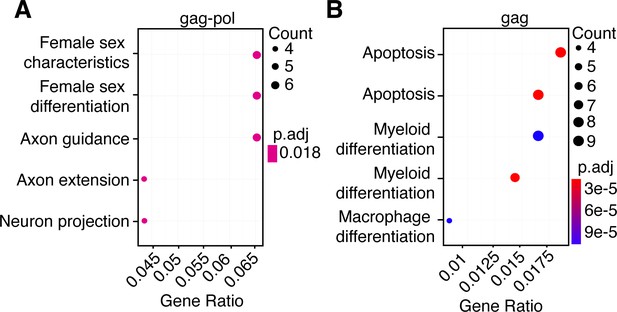
Transcriptional effects of paternally expressed gene 10 (PEG10) nucleocapsid transfection resemble gag-pol, but not gag.
(A–B) Top gene expression changes by GO-term enrichment analysis. The top five GO-terms enriched upon the comparison of gag-pol (A) and gag (B) to control cells are shown with enriched GO-term on the left. Adjusted p-value is shown by color, and the size of the datapoint reflects the number of genes enriched in the pathway.
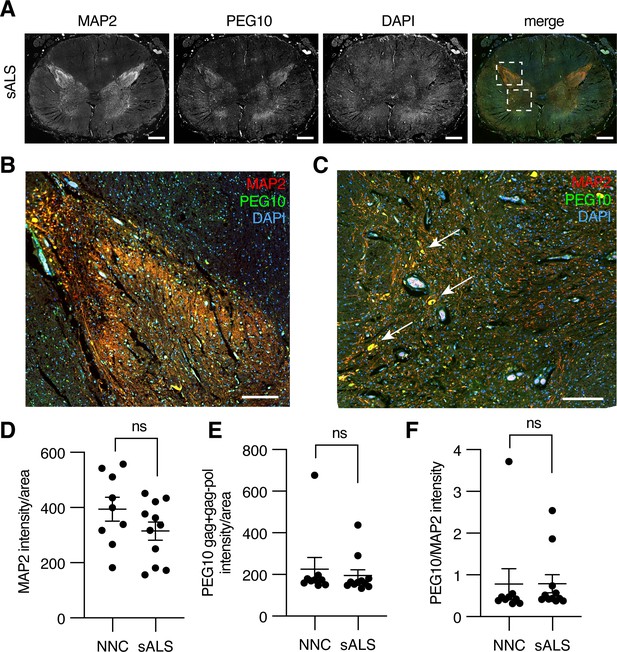
Paternally expressed gene 10 (PEG10) is enriched in horns of the lumbosacral spinal cord.
(A) Tiled image of full-thickness lumbosacral spinal cord section demonstrating Microtubule-associated protein 2 (MAP2), PEG10, and DAPI staining. Merged image shows outlines of boxes for closer examination in (B) and (C). Scale bar 1 mm. (B) Merged image of the posterior horn of the spinal cord showing enrichment for both MAP2 and PEG10 signals. Scale bar 200 μm. (C) Merged image of the anterior horn of the spinal cord showing colocalization of MAP2 and PEG10 signal in cell bodies (arrows). Scale bar 200 μm. (D–E) Quantitation of (D) MAP2 and (E) PEG10 signal in spinal cord sections. Signal intensity of MAP2 and PEG10 were determined per mm2 of spinal cord section for a cohort of non-neurological controls (NNC, n=9) and sporadic Amyotrophic Lateral Sclerosis (ALS) patients (sALS, n=11). (F) Relative PEG10 per MAP2 intensity was quantified for each section, to account for potential loss in neuronal staining due to ALS. No significant differences in intensity or localization were observed in the disease condition. For (D–F) mean ± SEM is shown.
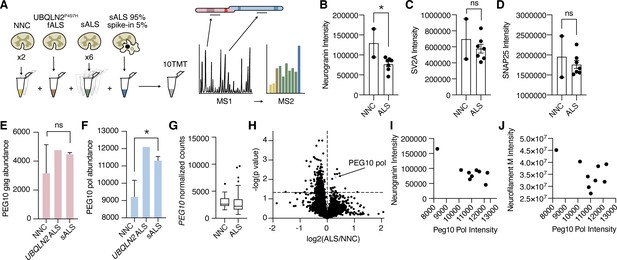
Paternally expressed gene 10 (PEG10) gag-pol protein accumulates in human Amyotrophic Lateral Sclerosis (ALS).
(A) Schematic illustrating multiplexed global proteomic strategy to quantify PEG10 protein from the human lumbar spinal cord. Two non-neurological controls (NNC), one fALS case with a Ubiquilin 2 (UBQLN2) mutation, and six sporadic ALS cases were combined with a ‘spike-in’ PEG10 channel containing 5% lysate from cells transfected with HA-PEG10 gag-pol and 95% spinal cord lysate to normalize proteomic background complexity. All ten samples were labeled with tandem mass tags (TMT) and run as a 10-plex on LC-MS2. (B–D) Abundance of neurogranin (B), SV2A (C), and SNAP25 (D), in the human spinal cord. Mean ± SEM is shown and plotted for each marker. A significant decrease in neurogranin was detected between non-neurological controls NNC (n=2) and all ALS patients (n=7) by unpaired t-test (E–F) Abundance of PEG10 gag (E), and pol (F), in the human spinal cord. Mean ± SEM is shown. Significance was determined by Student’s t-test. (G) RNA-Seq counts of PEG10 from post-mortem lumbar spinal cord tissue of a larger cohort of patients with Classical ALS (n=127) or Non-Neurological Controls (NNC, n=17). Data are shown as 5–95% box and whisker plot and significance was determined by DESeq2. (H) Global proteomic analysis with 7465 individual proteins quantified. All ALS samples were grouped together, and two NNC were grouped to generate log2 ratio of protein abundance and significance calculation by homoscedastic unpaired t-test. PEG10 pol is highlighted in blue, and PEG10 gag (not significant) is highlighted in pink. (I) Quantification of neurogranin intensity (y-axis) and PEG10 pol peptides’ intensity (x-axis) are plotted for all ine spinal cord samples to demonstrate the relationship between the two markers. (J) Quantification of neurofilament medium intensity (y-axis) and PEG10 pol peptides’ intensity (x-axis) are plotted for all nine spinal cord samples to demonstrate the relationship between the two markers. *p<0.05.
-
Figure 8—source data 1
Proteomics results.
- https://cdn.elifesciences.org/articles/79452/elife-79452-fig8-data1-v2.xlsx
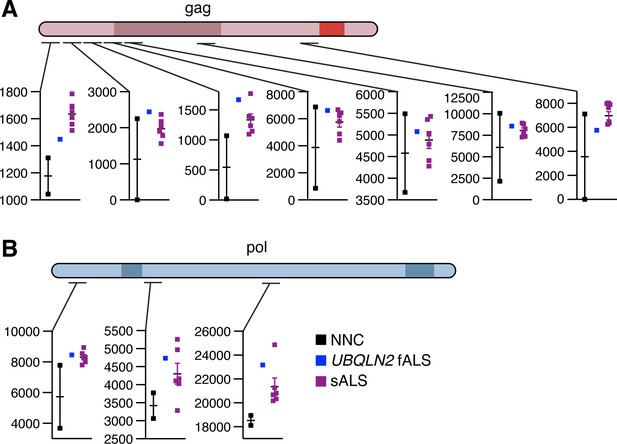
Global proteomic analysis of paternally expressed gene 10 (PEG10) peptides.
(A–B) Schematic of PEG10 protein from gag (A) and pol (B) with regions highlighted where peptides were quantified by global proteomic analysis. More detail on quantified peptides can be found in Figure 8—source data 1. Black: Non-neurological control, blue: UBQLN2-fALS, purple: sporadic Amyotrophic Lateral Sclerosis (ALS). Peptide abundance as measured by the mass spectrometer is shown on the y-axis, with a mean ± SEM of replicates.
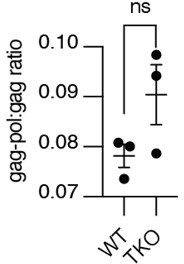
Read-through translation does not appear affected by UBQLN loss, ratio of gag-pol-Dendra2 to gag-Dendra2 to signal in WT or TKO cells treated with bortezomib for 12 hours.
Shown is mean ± SEM from three independent experiments. Significance was determined by unpaired Student's T test.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| strain, strain background (Escherichia coli) | Rosetta | Sigma | 70954–3 | |
| strain, strain background (Escherichia coli) | DH5α | Fisher Scientific | 18265017 | |
| cell line (Homo sapiens) | HEK293 WT and UBQLN1, 2, 4 TKO cells, inducible UBQLN2 expression | Itakura et al., 2016 | ||
| cell line (Homo sapiens) | Human ESCs (H9) WT and UBQLN1, 2, or 4 sinbgle KO cells | Harvard Medical School Cell Biology Initiative for Genome Editing and Neurodegeneration | ||
| antibody | anti-UBQLN1/2 (Mouse Monoclonal M03) | Abnova | H00029978; AB_627374 | WB (1:1000) |
| antibody | anti-UBQLN4 (Rabbit Polyclonal) | GeneTex | GTX85267; AB_10725990 | WB (1:1000) |
| antibody | anti-Myc tab (Mouse Monoclonal GT002) | Sigma | SAB2702192 | WB (1:2000) |
| antibody | anti-PEG10 (Rabbit Polyclonal) | Proteintech | 14412–1-AP; AB_10694427 | WB (1:1000) |
| antibody | anti-HA (Mouse Monoclonal HA-7) | Sigma | H3663; AB_262051 | WB, IF (1:1000) |
| antibody | anti-NC (Rabbit Polyclonal) | Thermo Fisher (custom) | n/a | WB (1:1000) |
| antibody | anti-tubulin (Mouse Monoclonal DM1A) | Novus | NB100-690; AB_521686 | WB (1:20000) |
| antibody | anti-MAP2 (Chicken Polyclonal 28225) | Biolegend | 822501; AB_2564858 | IF (1:2000) |
| antibody | α-mouse IgG 680 (Goat Polyclonal) | Licor | 926–68070; AB_10956588 | WB (1:20000) |
| antibody | α-mouse IgG 800 (Goat Polyclonal) | Licor | 926–32210; AB_621842 | WB (1:20000) |
| antibody | α-rabbit IgG 680 (Goat Polyclonal) | Licor | 926–68071; AB_10956166 | WB (1:20000) |
| antibody | α-rabbit IgG 800 (Goat Polyclonal) | Licor | 926–32211; AB_621843 | WB (1:20000) |
| antibody | Alexa568 α-mouse (Goat Polyclonal) | Invitrogen | A11004; AB_2534072 | IF (1:300) |
| antibody | Alexa568 α-chicken (Goat Polyclonal) | Invitrogen | A11041; AB_2534098 | IF (1:300) |
| antibody | Alexa647 α-rabbit (Goat Polyclonal) | Invitrogen | A32733; AB_AB2633282 | IF (1:300) |
| Transfected construct (Saccharomyces cerevisiae) | LIFEACT (Abp140 AA1-17) | GenBank; Riedl et al., 2008 | n/a | pCDNA3.1 |
| Transfected construct (Homo sapiens) | gag-pol; PEG10 (AA1-708) | GenBank | NP_055883.2 | pCDNA3.1 |
| Transfected construct (Homo sapiens) | gag-polASG;PEG10 (AA1-708)* D370A | GenBank | NP_055883.2 | pCDNA3.1 |
| Transfected construct (Homo sapiens) | Gag; PEG10 (AA1-325) | GenBank | NP_001035242.1 | pCDNA3.1 |
| Transfected construct (Homo sapiens) | CA; PEG10 (AA1-259) | GenBank | NP_001035242.1 | pCDNA3.1 |
| Transfected construct (Homo sapiens) | NC; PEG10 (AA260-325) | GenBank | NP_001035242.1 | pCDNA3.1 |
| Transfected construct (Homo sapiens) | Tag-less NC; PEG10 (AA260-325) | GenBank | NP_001035242.1 | pCDNA3.1 |
| Transfected construct (Homo sapiens) | gag111; PEG10(AA1-111) | GenBank | NP_001035242.1 | pCDNA3.1 |
| Transfected construct (Mus musculus) | gag-pol; PEG10 (AA1-1006) | GenBank | NP_570947.2 | pCDNA3.1 |
| Transfected construct (Mus musculus) | gag-polASG;PEG10 (AA1-1006)* D420A | GenBank | NP_570947.2 | pCDNA3.1 |
| Transfected construct (Homo sapiens) | gag-pol Dendra2; PEG10 (AA1-708)-Dendra2 | GenBank | NP_055883.2 | pDendra2 |
| Transfected construct (Homo sapiens) | gag-polASG Dendra2; PEG10 (AA1-708)* D370A-Dendra2 | GenBank | NP_055883.2 | pDendra2 |
| Transfected construct (Homo sapiens) | ΔPPR Dendra2; PEG10 (AA1-681)-Dendra2 | GenBank | NP_055883.2 | pDendra2 |
| Transfected construct (Macaca Mulatta) | gag-pol Dendra2; PEG10 (AA1-742)-Dendra2 | GenBank | NP_001165893.2 | pDendra2 |
| Transfected construct (Bos taurus) | gag-pol Dendra2; PEG10 (AA1-788)-Dendra2 | GenBank | NP_001120682.1 | pDendra2 |
| Transfected construct (Mus musculus) | gag-pol Dendra2; PEG10 (AA1-1006)-Dendra2 | GenBank | NP_570947.2 | pDendra2 |
| Transfected construct (Phascolarctos cinereus) | gag-pol Dendra2; PEG10 (AA1-624)-Dendra2 | GenBank | XM_021000084.1 (+downstream CDS continuation to pol stop codon) | pDendra2 |
| Transfected construct (Phascolarctos cinereus) | gag-pol Dendra2+HsPPR; Pc PEG10 (AA1-624)+Hs PEG10 (AA682-708) Dendra2 | GenBank | XM_021000084.1 (+downstream CDS continuation to pol stop codon) And NP_055883.2 | pDendra2 |
| Transfected construct (Homo sapiens) | 6XHIS-SUMO2-NC; 6 X His tag + Sumo2 tag+PEG10 (AA260-325) | GenBank | NP_001035242.1 | pETSUMO2 |
| chemical compound, drug | cycloheximide | Sigma | 01810 | |
| chemical compound, drug | Bortezomib | MP Biomedicals | IC0218385905 | |
| other | Prolong Gold DAPI anti-fade mounting media | Invitrogen | P36941 | Mounting medium for coverslips |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/79452/elife-79452-mdarchecklist1-v2.docx
-
Source data 1
All raw western blot images.
- https://cdn.elifesciences.org/articles/79452/elife-79452-data1-v2.zip