Lifelong regeneration of cerebellar Purkinje cells after induced cell ablation in zebrafish
Figures
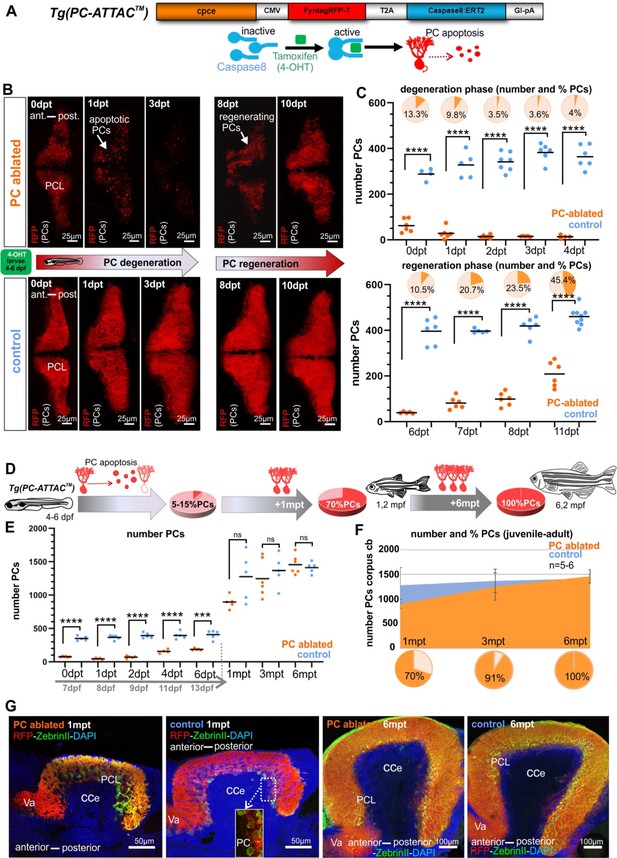
Induced Purkinje cell (PC) ablation in zebrafish larvae: time course of degeneration and regeneration.
(A) Construct used to generate the Tg(PC-ATTAC) transgenic line, modified from Weber et al., 2016. (B) Images of the PC layer (fyn-tagRFP-T fluorescence expressed in mature PCs) after induced PC ablation in larvae at 4–6 dpf monitoring 10 days after 4-hydroxy-tamoxifen (4-OHT) treatment. (C) Number and percentage of PCs in ablated (4-OHT) vs control (EtOH) larvae. (D–G) Quantitative monitoring of PC regeneration after induced PC ablation in larvae. (D–F) Numbers and percentage of PCs until 6 mpt. (G) Images of PCs on sagittal cerebellar sections after immunostaining with anti-tagRFP and anti-ZebrinII antibodies, comparing ablated and control groups at 1 and 6 mpt. Statistical information: sample size n=4-9, statistical method=unpaired t-test two tailed, levels of significance=P <0.0001 (****), P=0.0001 (***). Additional information in Supplementary file 1.
-
Figure 1—source data 1
Purkinje cell (PC) quantification for Figure 1C, D, F.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig1-data1-v2.xlsx
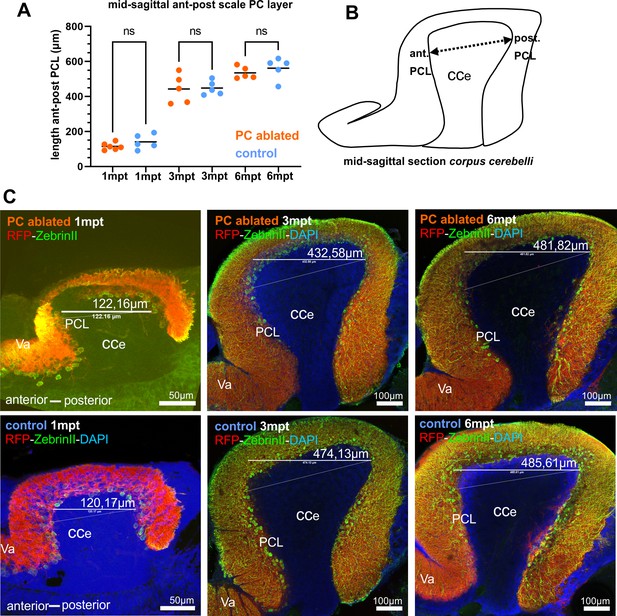
Quantification of the distance between rostral and caudal Purkinje cell (PC) layer in the corpus cerebelli of juveniles and adults after PC ablation in larvae, related to Figure 1.
Quantification (A), schematic drawing (B), and representative images (C) of the distance measured. Significant differences between ablated and control groups were not observed.
-
Figure 1—figure supplement 1—source data 1
Antero-posterior distance measurements of Purkinje cell (PC) layer for Figure 1—figure supplement 1A
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig1-figsupp1-data1-v2.xlsx
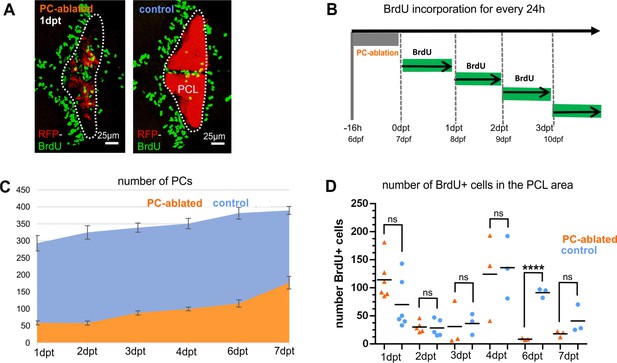
Cell proliferation in the cerebellum after Purkinje cell (PC) ablation: Bromodeoxyuridine (BrdU) analysis after PC ablation in larvae, related to Figure 1.
(A–D) Quantification of cell proliferation by BrdU labeling after PC ablation. (A) Representative images of BrdU-positive cells in the cerebellum of PC-ablated and control larvae. Rostral is to the left. (B) Schematic representation of the BrdU incorporation treatment and analysis carried out for periods of 24 hr. Numbers of PCs (C) and BrdU-positive cells within the PC layer area of the cerebellum (D). Dotted line in A indicates the cell counting area.
-
Figure 1—figure supplement 2—source data 1
Numbers of BrdU+ cells for Figure 1—figure supplement 2D.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig1-figsupp2-data1-v2.xlsx
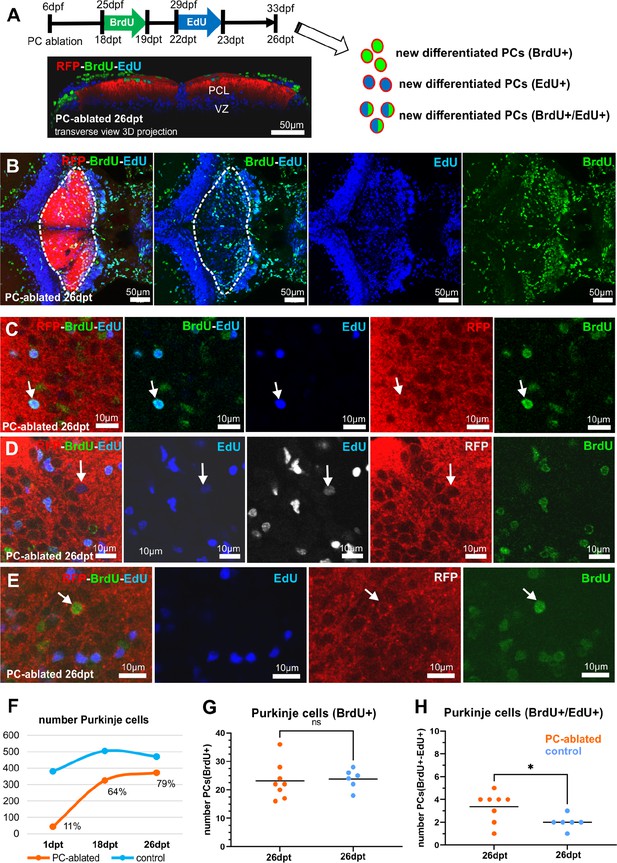
Bromodeoxyuridine (BrdU)/5-Ethynyl-2′-deoxyuridine (EdU) double pulse chase after Purkinje cell (PC) ablation in larvae, related to Figure 1.
Schematic representation of the BrdU and EdU double incorporation assay (A) and representative images (B–E) showing double-positive PCs: RFP(PCs)+/BrdU+/EdU+ (C), FP+/EdU+ (D), and RFP+/BrdU+ (E). Quantitative analysis of the number of PCs (F), number of PCs RFP+/BrdU+ (G), and RFP+/BrdU+/EdU+ (H) double positive.Statistical information: statistical method=unpaired t-test two tailed, levels of significance=P=0.0355 (*). Additional information in Supplementary file 1.
-
Figure 1—figure supplement 3—source data 1
Numbers of Purkinje cells (PCs) for Figure 1—figure supplement 3F and of double-positive BrdU+ and EdU+ PCs for Figure 1—figure supplement 3G, H.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig1-figsupp3-data1-v2.xlsx
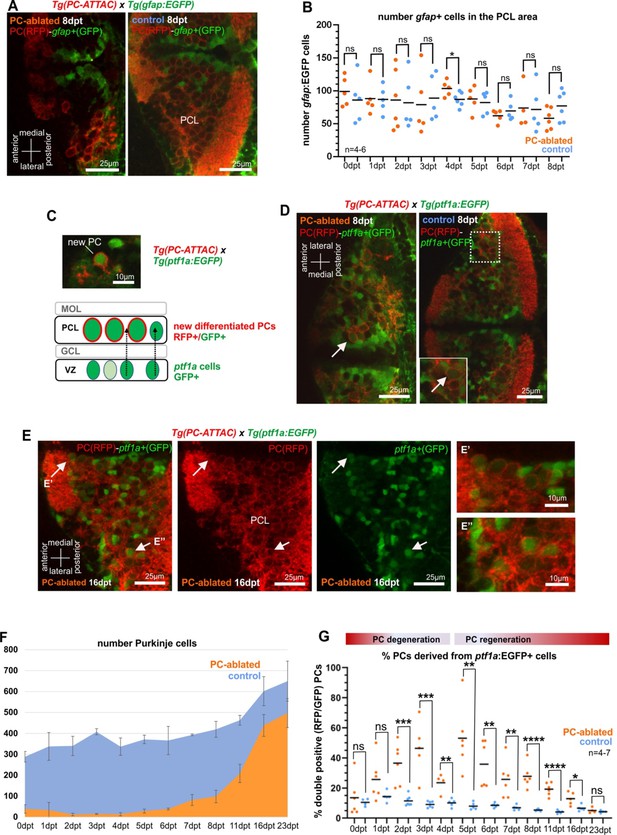
Cellular analysis of potential progenitors of regenerating Purkinje cells (PCs).
(A) Images of larval cerebellum of the double transgenic line Tg(PC-ATTAC)/Tg(gfap:EGFP) after induced PC ablation. (B) Number of gfap+ cells throughout the PC layer area during PC degeneration and beginning of regeneration. (C) Illustration of new PC development from ptf1a+ progenitors. (D, E) Images of larval cerebellum of the double transgenic line Tg(PC-ATTAC)/Tg(ptf1a:EGFP) after induced PC ablation, revealing double-positive cells (arrows). Average of PC numbers (F) and percentage of PCs showing GFP fluorescence double-positive cells, (G) during degeneration and regeneration of PCs. The red fluorescent protein from the PC-ATTAC strain is exclusively expressed in the cell membrane, while EGFP from the ptf1a- and gfap-reporter strains localizes to the cytoplasm. GFP and RFP were enhanced by fluorescence immunohistochemistry in A, C–E. Statistical information: statistical method=unpaired t-test two tailed, levels of significance=P<0.05 (*), P<0.01 (**), P<0.001 (***), P<0.0001 (****). Additional information in Supplementary file 1.
-
Figure 2—source data 1
Quantification of gfap-expressing cells for Figure 2B and quantification of ptf1a:GFP-expressing PC-ATTAC cells for Figure 2G.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig2-data1-v2.xlsx
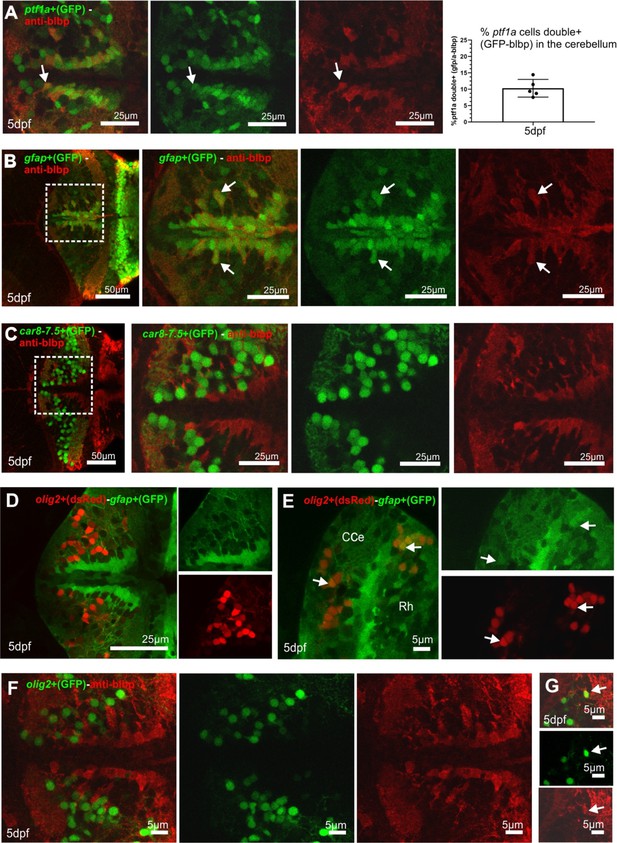
Identity of glial cells in the larvae cerebellum: images of double staining with radial glial markers and cell precursors and specific cell type markers, related to Figure 2.
(A–G) Coexpression analysis of radial glia markers in specific cerebellar cell types. (A) Double staining showing GABAergic cell precursors (ptf1a-GFP-expressing cells) and cells labeled with the anti-brain lipid-binding protein (blbp) antibody, showing double-positive cells (arrows), and percentage of ptf1a-expressing cells colocalizing with the anti-blbp antibody. (B) Double staining of gfap-expressing cells (Tg(gfap:GFP) line) and anti-blbp antibody, showing the majority of cells double positive. (C) Double staining of the line Tg(car8-7,5:GFP) and anti-blbp, not being observed double-positive cells. (D, E) Double transgenic line Tg(gfap:GFP) and Tg(olig2:dsRed) showing most of cells single positive (D), and only scarce double-positive cells at the ventro-lateral cerebellum (left arrow in E) and at the boundary with the caudal rhombencephalon (right arrow in E). (F, G) Double staining of olig2-expressing cells (Tg(olig2:GFP)) and anti-blbp antibody, showing consistent results to that observed with the gfap/olig2 double transgenic line, showing the majority of cells not colocalizing (F), except for isolated cells (G).
-
Figure 2—figure supplement 1—source data 1
Percentage of double-positive ptf1a:GFP/antiBLBP cells.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig2-figsupp1-data1-v2.xlsx
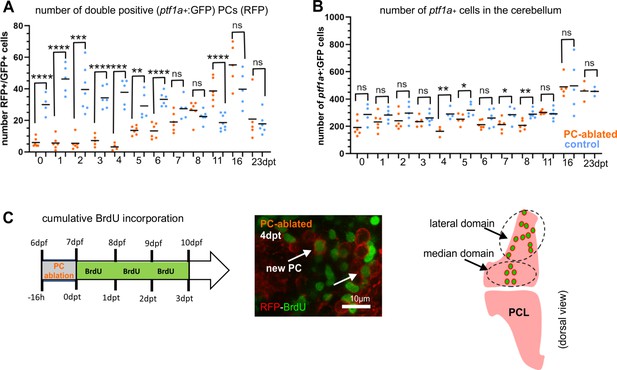
GABAergic cell progenitors in the cerebellum after Purkinje cell (PC) ablation, related to Figure 2.
(A, B) ptf1a:GFP-positive PCs and absolute number of ptf1a:GFP cells in the cerebellum after PC ablations compared to controls. (C) Cumulative Bromodeoxyuridine (BrdU) labeling of regenerating PCs. (A) Number of double-positive PCs showing RFP expression in mature PCs, and green fluorescence from ptf1a-EGFP-expressing cells. (B) Number of ptf1a-expressing cells (GFP positive) in the cerebellum, a general increase of ptf1a-expressing cell progenitors after PC ablation is not detected. (C) Mature PCs with BrdU incorporation (after a cumulative treatment of several days), revealing the location of new PCs during the period of BrdU incorporation, grouped into median and lateral domains of newly generated PCs (this location is similar to newly generated PCs derived from ptf1a-EGFP-expressing progenitors). Statistical information: statistical method=unpaired t-test two tailed, levels of significance=P<0.05 (*), P<0.01 (**), P<0.001 (***), P<0.0001 (****). Additional information in Supplementary file 1.
-
Figure 2—figure supplement 2—source data 1
Quantification of ptf1a:GFP-expressing cells for Figure 2—figure supplement 2A, B.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig2-figsupp2-data1-v2.xlsx
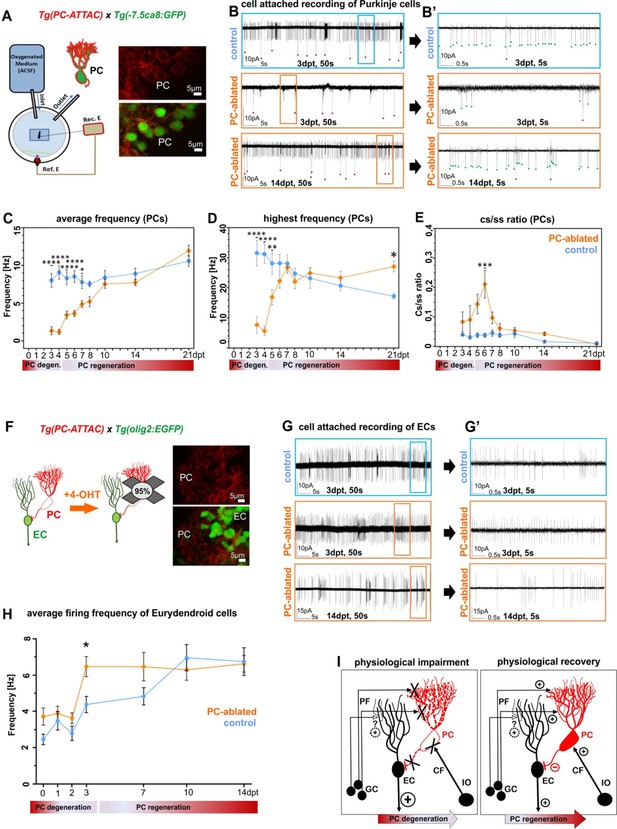
Electrophysiological properties of Purkinje cells (PCs) and eurydendroid cells (ECs) during ablation and regeneration phase.
(A) Patch-clamp recording setup used for all experiments. Fluorescent PCs in larvae of the double transgenic line Tg(PC-ATTAC)/Tg(–7.5ca8:EGFP). (B) 50 s trace of representative recordings of the tonic firing activity in control larvae 3 days after EtOH or in PC-ablated larvae 3 and 14 days after 4-hydroxy-tamoxifen (4-OHT) treatment and (B’) 5 s traces for all three traces shown. Red dots mark complex spikes and green dots mark simple spikes. (C–E) Diagrams representing results of the electrophysiologic investigations in PCs. (C) Average tonic firing frequency plotted vs dpt. (D) Highest spontaneous bursting frequency over an interval of 1 s during a 100 s trace plotted vs dpt. (E) Ratio of the complex spikes to simple spikes vs dpt. (F) Illustration of PC loss after 4-OHT treatment and representative image of the PC layer from a double transgenic Tg(PC-ATTAC)/Tg(olig2:EGFP) larva. (G) 50 s trace of representative recordings of the tonic firing activity of ECs in control larvae 3 days after EtOH or in PC-ablated larvae 3 and 14 days after 4-OHT treatment and (F) 5 s traces for all 3 traces shown. (H) Average tonic firing frequency of ECs after PC ablation plotted vs dpt. (I) Illustration of physiological impairment and recovery model of the input/output of PCs during PC degeneration and regeneration, respectively. Statistical information: sample size n=17-45, statistical method=2-Way ANOVA, Šídák's multiple comparisons test, levels of significance=P<0.05 (*), P<0.01 (**), P<0.001 (***), P<0.0001 (****). Additional information in Supplementary file 1.
-
Figure 3—source data 1
Average firing frequency determination of Purkinje cells (PCs) for Figure 3C, highest burst frequency numbers of PCs for Figure 3D, and dataset of complex spike to simple spike firing ratios of PCs for Figure 3E.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig3-data1-v2.xlsx
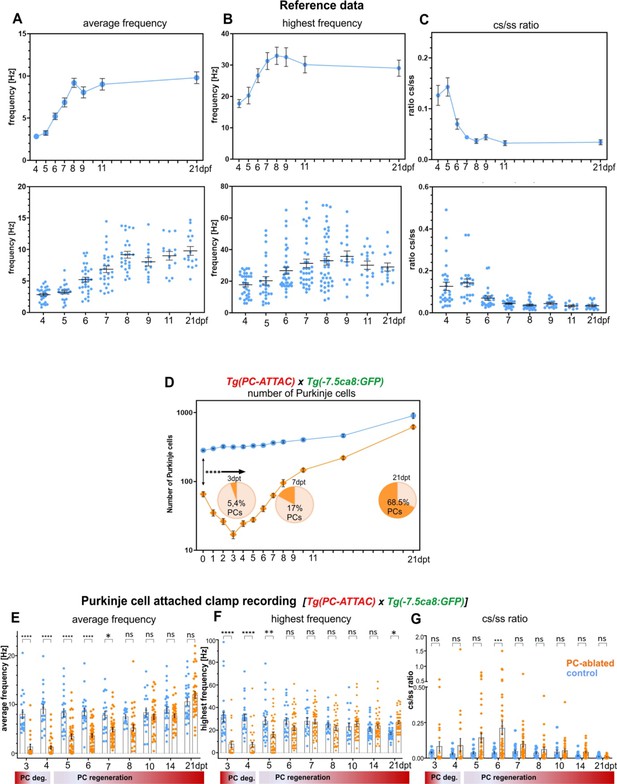
Reference set of physiological properties of Purkinje cells (PCs) and physiological properties of individual PCs after ablation, related to Figure 3.
(A–C) Electrophysiological reference data of differentiating PCs. (D) Quantification of PC ablation efficiency in specimens for electrophysiological recordings. (E–G) Electrophysiological data of regenerating PCs. (A–C) Electrophysiological characterization of PCs in 4- to 21-dpf-old larvae. (A) Average tonic firing frequency plotted vs dpf. (B) Highest spontaneous bursting frequency over an interval of 1 s during a 100 s trace plotted vs dpf. (C) Ratio of the complex spikes to simple spikes vs dpf. (D) Quantification of PC ablation efficiency in specimens for electrophysiological recordings. (E, F) Physiological properties of individual PCs after ablation. (E) Average tonic firing frequency plotted vs dpt. (F) Highest spontaneous bursting frequency over an interval of 1 s during a 100 s trace plotted vs dpt. (G) Ratio of the complex spikes to simple spikes vs dpt. Statistical information: statistical method=2-Way ANOVA, Šídák's multiple comparisons test, levels of significance=P<0.05 (*), P<0.01 (**), P<0.001 (***), P<0.0001 (****). Additional information in Supplementary file 1.
-
Figure 3—figure supplement 1—source data 1
Average firing frequency determination of Purkinje cells (PCs) for Figure 3—figure supplement 1A, highest burst frequency numbers of PCs for Figure 3—figure supplement 1B, and dataset of complex spike to simple spike firing ratios of PCs for Figure 3—figure supplement 1C.
Quantification of PC numbers for Figure 3—figure supplement 1D, E. Average firing frequency determination of PCs for Figure 3—figure supplement 1E, highest burst frequency numbers of PCs for Figure 3—figure supplement 1F, and dataset of complex spike to simple spike firing ratios of PCs for Figure 3—figure supplement 1G.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig3-figsupp1-data1-v2.xlsx
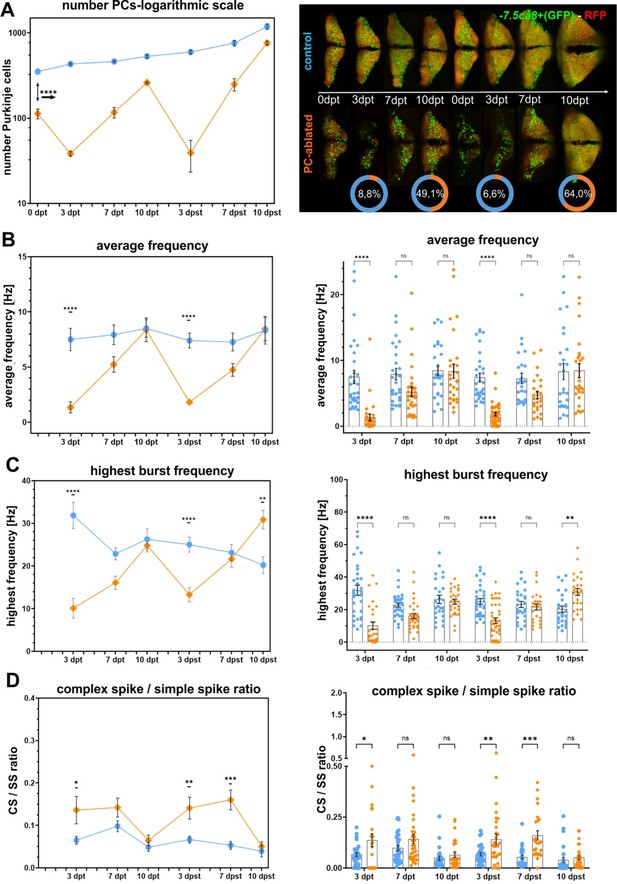
Purkinje cell (PC) activity recovery after repeated cell type-specific ablation, related to Figure 3.
(A) Quantification of PC ablation efficiency in specimens for electrophysiological recordings after repeated 4-hydroxy-tamoxifen (4-OHT) treatment and representative images of the PC layer from double transgenic Tg(PC-ATTAC)/Tg(–7.5ca8-GFP) larvae. (B–D) Electrophysiological characterization of PCs in 0–10 days post (second) treatment old larvae. (B) Average tonic firing frequency plotted vs dpt/dpst. (C) Highest spontaneous bursting frequency over an interval of 1 s during a 100 s trace plotted vs dpt/dpst. (D) Ratio of the complex spikes to simple spikes vs dpt/dpst. The data for individual PCs in plotted on the right. Statistical information: statistical method=2-Way ANOVA, Šídák's multiple comparisons test, levels of significance=P<0.05 (*), P<0.01 (**), P<0.001 (***), P<0.0001 (****). Additional information in Supplementary file 1.
-
Figure 3—figure supplement 2—source data 1
Quantification of Purkinje cells (PCs) for Figure 3—figure supplement 2A, and PC activity for Figure 3—figure supplement 2B-D.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig3-figsupp2-data1-v2.xlsx
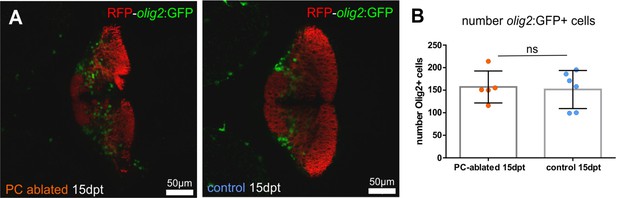
Olig2-positive cells after Purkinje cell (PC) ablation in larvae, related to Figure 3.
(A) Images of the double transgenic line Tg(olig2:GFP) and Tg(PC-ATTAC). (B) Number of olig2-GFP-expressing cells 2 weeks post-ablation, showing not significant differences between ablated and control groups.
-
Figure 3—figure supplement 3—source data 1
Numbers of olig2:GFP-expressing cells at 15 dpt for Figure 3—figure supplement 3B.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig3-figsupp3-data1-v2.xlsx
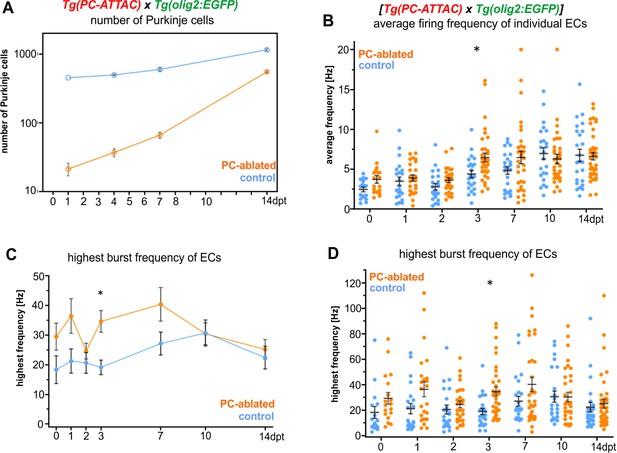
Eurydendroid cell (EC) activity recovery after Purkinje cell (PC) ablation, related to Figure 3.
(A) Quantification of PC ablation efficiency in specimens for electrophysiological recordings. (B) Average tonic firing frequency of individual ECs plotted vs dpt. (C) Highest spontaneous bursting frequency of ECs over an interval of 1 s during a 100 s trace plotted vs dpt. (D) Highest spontaneous bursting frequency of individual ECs over an interval of 1 s during a 100 s trace plotted vs dpt. Statistical information: statistical method=2-Way ANOVA, Šídák's multiple comparisons test, levels of significance=P<0,05 (*). Additional information in Supplementary file 1.
-
Figure 3—figure supplement 4—source data 1
Quantification of Purkinje cells (PCs) for Figure 3—figure supplement 4A, and eurydendroid cell (EC) activity for Figure 3—figure supplement 4B-D.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig3-figsupp4-data1-v2.xlsx
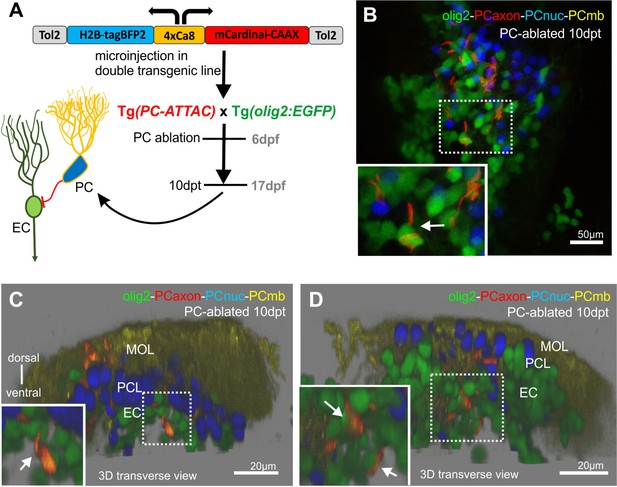
Axon tracing of regenerating Purkinje cells (PCs), related to Figure 3.
(A) Schematic representation of the construct used for injections, the temporal outline of the experiment and the expected fluorescence pattern. (B) Dorsal view of a double transgenic Tg(PC-ATTAC)/Tg(olig2:GFP) larva, which expresses the injected construct. (C, D) 3D reconstructions of double transgenic Tg(PC-ATTAC)/Tg(olig2:GFP) larvae, which express the injected construct.
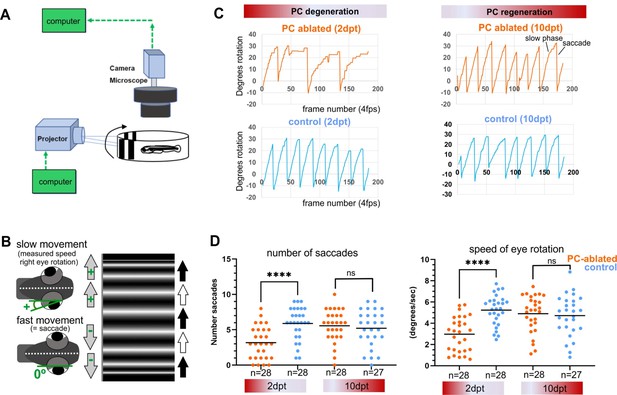
Visuo-motor behavior analysis: optokinetic response (OKR) after induced Purkinje cell (PC) ablation in larvae.
Illustration of OKR setup (A) and eye movements in larvae (B) during performance of the OKR test. (C) Representative graphs showing OKR response performance during acute PC degeneration (2 dpt) and regeneration (10 dpt) phases in PC-ablated (4-hydroxy-tamoxifen [4-OHT] treated) vs control group (EtOH treated). (D) Quantification of OKR response: number of normal saccades (eye rotation >19.5°) and speed of eye rotation during slow phase movements at 2 and 10 dpt. The data correspond to the results of two independent trials that were pooled. Statistical information: statistical method=Mann-Whitney test or unpaired t-test, two tailed, level of significance=P<0.0001 (****). Additional information in Supplementary file 1.
-
Figure 4—source data 1
Datasets of eye deflection measurements during optokinetic response (OKR) behavior for Figure 4C, D.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Datasets of eye deflection measurements during optokinetic response (OKR) behavior for Figure 4C, D.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Eye rotation and saccade quantification for Figure 4C, D.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig4-data3-v2.xlsx
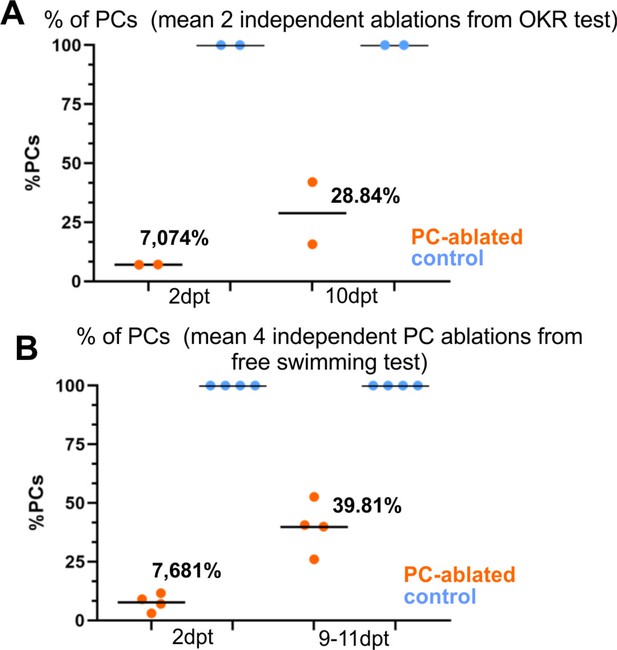
Average percentage of Purkinje cells (PCs) at acute degeneration phase and beginning of regeneration from independent trials of optokinetic response (OKR) (A) and free-swimming tests (B) after PC ablation in larvae, related to Figures 4 and 5.
-
Figure 4—figure supplement 1—source data 1
Quantification of Purkinje cells (PCs) for Figure 4—figure supplement 1A, B.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig4-figsupp1-data1-v2.xlsx
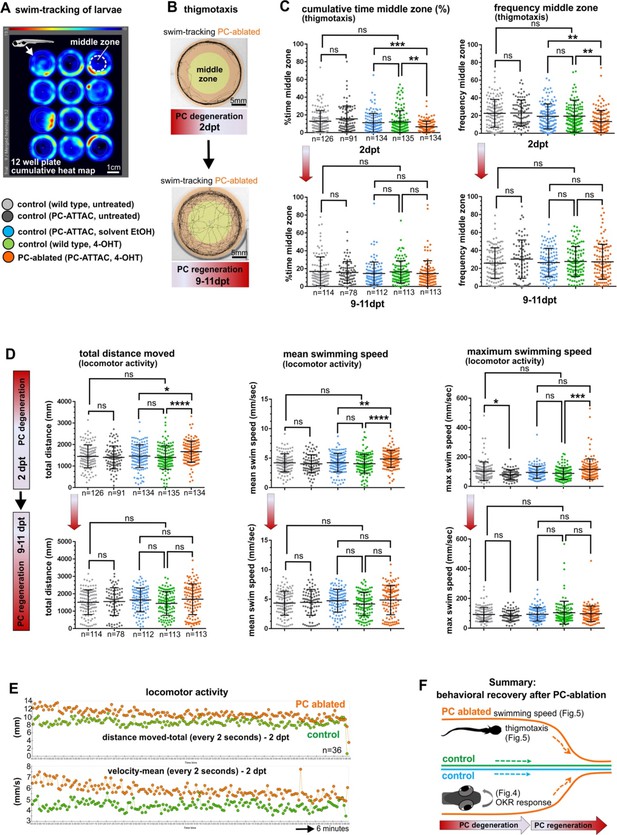
Swimming behavior analysis after induced Purkinje cell (PC) ablation in larvae.
(A) Heat map representing the location of zebrafish larvae in a 12-well plate during 6 min of swimming. (B) Example of swim track after PC ablation (2 dpt) and during regeneration (10 dpt) phases. (C) Quantitative analysis of swim preferences along the edge vs center zone of the arena (frequency of visits and percentage of time spent in the center zone) in PC-ablated larvae vs control groups. (D) Quantitative analysis of locomotor activity (total distance traveled, mean and maximum swimming speed) in PC-ablated larvae vs controls. (E) Graphs showing distance traveled and swim speed every 2 s during the tracking period, comparing PC-ablated vs control larvae. (F) Illustration summarizing impairment and recovery of optokinetic response (OKR) (Figure 4) and locomotor behavior during PC degeneration and early regeneration phases, respectively.The data from free swim tests correspond to the results from four independent ablations that were pooled. Statistical information: size sample n=78-135, statistical method=ANOVA Kruskal-Wallis, Dunn´s multiple comparisons test, levels of significance=P<0.05 (*), P<0.01 (**), P<0.001 (***), P<0.0001 (****). Additional information in Supplementary file 1.
-
Figure 5—source data 1
Datasets of thigmotaxis and swim speed analysis for Figure 5C, D.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig5-data1-v2.xlsx
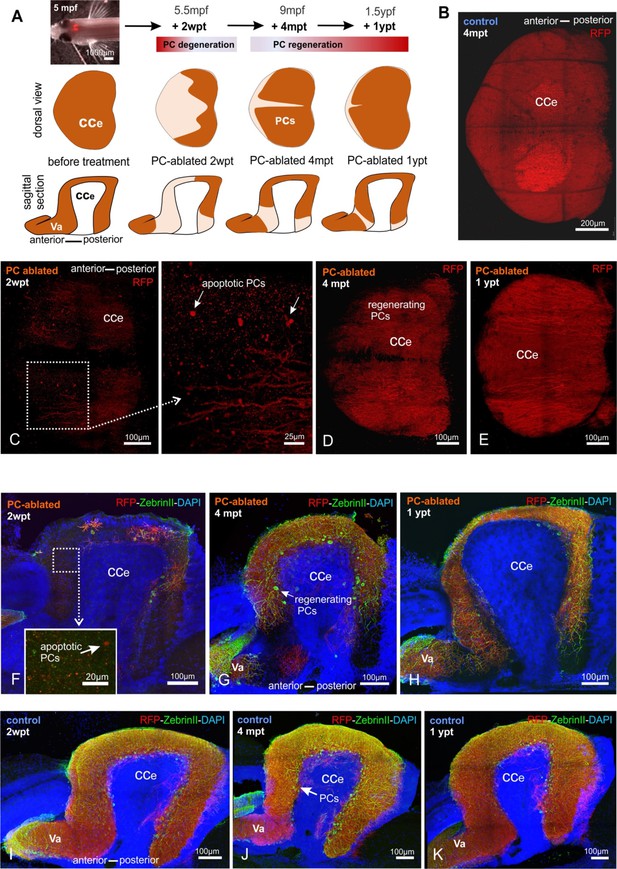
Monitoring of the Purkinje cell (PC) layer after induced PC ablation in adults.
(A) In vivo stereomicroscopy showing tagRFP-T fluorescence in the PC layer, and illustration of the time course of fluorescence recovery after PC ablation in adults (at 5 mpf) monitored until 1 ypt. Representative confocal images of whole mount cerebelli from dorsal view (B–E) and sagittal vibratome sections after immunostaining with the antibodies anti-tagRFP and anti-ZebrinII (F–K), comparing the cerebellum in ablated fish (endoxifen treated; C–E, F–H) vs control group (dimethyl sulfoxide [DMSO] treated; B, I–K). Arrows in C, F point out apoptotic bodies. Equivalent results were observed in two additional independent ablations in adult cerebelli (data not shown).
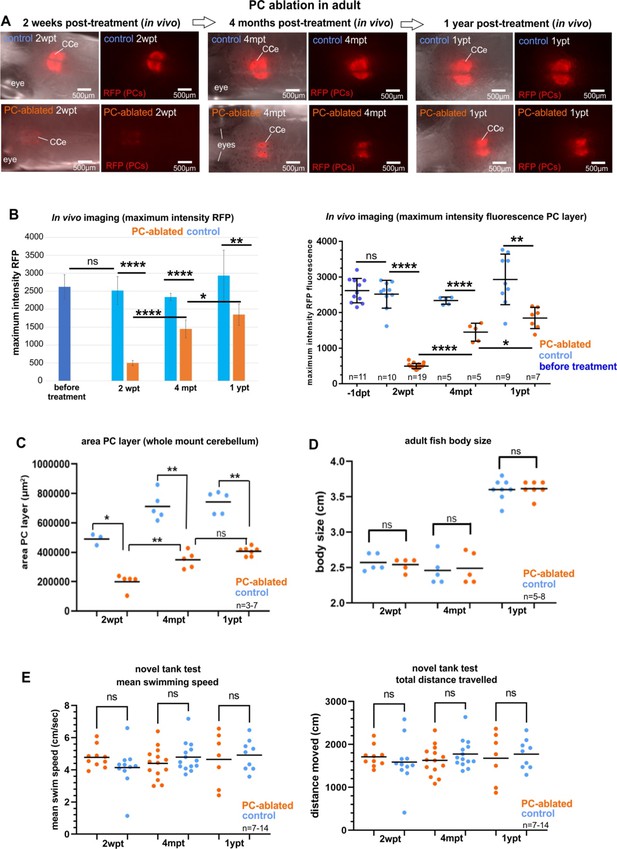
Imaging of the cerebellum in vivo, and quantification of the Purkinje cell (PC) area, body length, and swimming after PC ablation in adult zebrafish; related to Figures 6 and 7.
(A, B) RFP intensity analysis of adult regenerating PC layer. (C) Quantification of size of PC layer based on PC fluorescence. (D) Quantification of body size of adult PC-ablated and control fish. (E) Quantification of swim speed and traveled distance of adult PC-ablated zebrafish compared to controls during novel tank test. (A, B) In vivo imaging of ablated PC layer in adults. Images recorded in vivo by stereomicroscopy revealing PC-derived RFP fluorescence in the PC layer, and measurement of fluorescence intensity. All fishes treated with Endoxifen showed very faint fluorescence in the cerebellum 2 weeks post-ablation, verifying actual PC ablation in the entire Endoxifen-treated group. The recovery of the PC layer displayed a significant increase of fluorescence at 4 months and 1 year post-ablation. (C) Area of the PC layer from whole mount confocal imaging during PC degeneration and recovery. The significant difference of the area compared to control fish displays the efficiency of PC induced apoptosis, and the regrowth of the area within the ablated group 4 months after the ablation shows the progressive regeneration of the PC population. (D) Body length of adult fish after induced PC ablation. No differences between fish from PC-ablated and control groups were observed. (E) Novel tank test. Total distance traveled and mean swim speed after induced PC ablation in adults. No differences were observed between ablated and control fish during the PC degeneration nor during the regeneration periods were observed. Statistical information: statistical method=Student t-test was applied for two groups comparison (unpaired t-test two tailed for parametric, or Mann–Whitney two tailed for no parametric data), and ordinary one way, ANOVA Šídák's multiple comparisons test for multiple groups comparison, levels of significance=P<0.05 (*), P<0.01 (**), P<0.0001 (****). Additional information in Supplementary file 1.
-
Figure 6—figure supplement 1—source data 1
Measurements of maximum intensity of RFP fluorescence for Figure 6—figure supplement 1B, area size determinations of Purkinje cell (PC) layer for Figure 6—figure supplement 1C, adult zebrafish body size measurements for Figure 6—figure supplement 1D, mean swim speed and total distance values for behavioral analysis for Figure 6—figure supplement 1E.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig6-figsupp1-data1-v2.xlsx
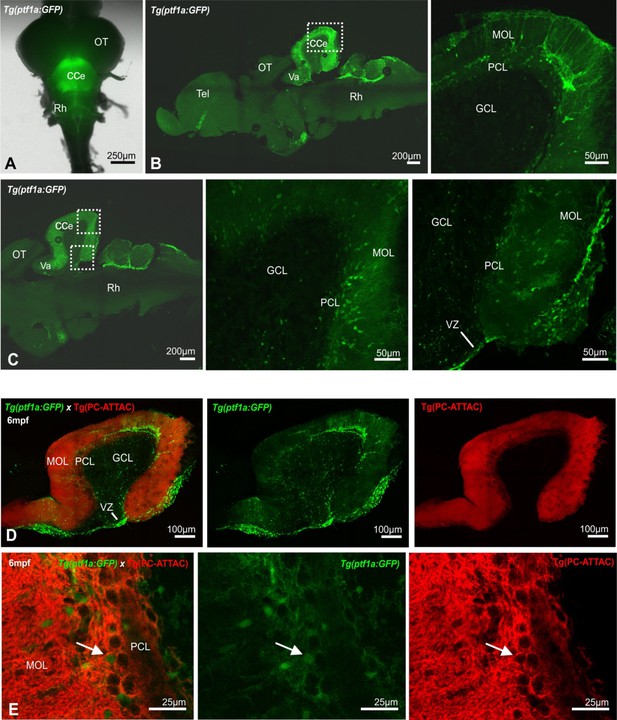
Distribution of ptf1a-expressing progenitors in the adult cerebellum (A–C).
Representative images of whole mount brain (A) and sagittal sections (B, C) from the transgenic Tg(ptf1a:EGFP) reporter line of a 3-year adult still revealing the presence of ptf1a-expressing progenitor cells in the cerebellar Purkinje cell (PC) and molecular layer in aged fish.
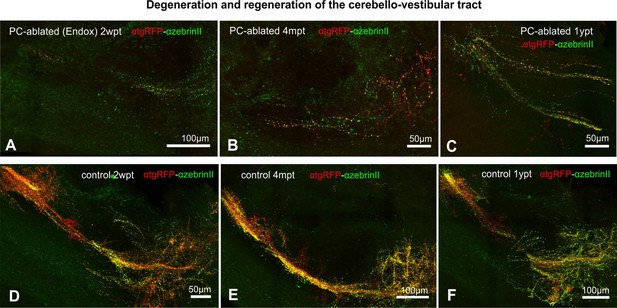
Degeneration and recovery of cerebello-vestibular tract after adult Purkinje cell (PC) ablation (A-F), related to Figures 6 and 7.
De- and regeneration of cerebello-vestibular tract in adult PC-ablated zebrafish compared to controls. (A–F) Images of the cerebello-vestibular tract after immunostaining with the antibodies anti-tagRFP and anti-ZebrinII on vibratome sections in PC-ablated fish (A-C) compared to control fish (D-F), show a vast tract degeneration at 2 weeks (A), and a partial recovery of this PC-derived axonal tract 4 months and 1 year after ablation (B, C). Rostral is to the left. Abbreviations: Cb cerebellum, CCe corpus cerebelli, GCL granular cell layer, MOL molecular cell layer, OT optic tectum, PCL Purkinje cell layer, Rh rhombencephalon, Tel telencephalon, VZ ventricular zone.
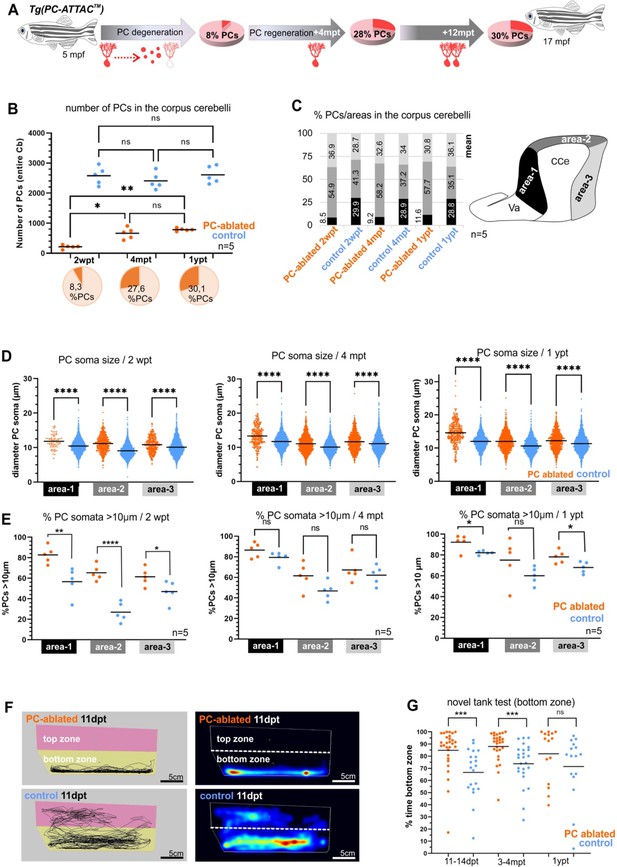
Quantitative analysis of Purkinje cells (PCs) and behavior test during degeneration and recovery after induced ablation in adults.
(A) Illustration of the percentage of PC layer replenishment during degeneration and regeneration phases. (B) Number and percentage of PCs after induced PC ablation monitored until 1 ypt. (C) Subdivision of the corpus cerebelli (CCe) into rostral, dorsal and caudal areas (areas 1–3), and the respective proportion of PCs in each area. (D) Quantification of the diameter of PC somata in the different areas of the CCe. (E) Percentage of PCs with somata larger than 10 µm from the total amount of PCs per area per brain. (F) Representative images of swim tracks and heat map of location of adult zebrafish in the novel tank test during 6 min. (G) Percentage of time spent in the bottom zone. The data correspond to the results from three independent PC ablations that were pooled. The cellular quantification in all graphs represent PCs from the entire CCe of 5 fishes per group per time point, as average from the whole PC population of each brain (B, C, E), or at single-cell level that were pooled (D). Statistical information: statistical method=analysis of variance (ANOVA) test for multiple groups comparison (ordinary one-way ANOVA- followed by Sidàks multiple comparison test), or Student’s t-test for two groups comparison (unpaired t-test two tailed for parametric, or Mann-–Whitney two tailed for no parametric data), levels of significance=P<0.05 (*), P<0.01 (**), P<0.001 (***), P<0.0001 (****). Additional information in Supplementary file 1.
-
Figure 7—source data 1
Purkinje cell (PC) quantification for Figure 7A-C, PC somata measurements for Figure 7D, quantification of PC subtypes für Figure 7E, and exploration behavior in the novel tank test assays for Figure 7G.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig7-data1-v2.xlsx
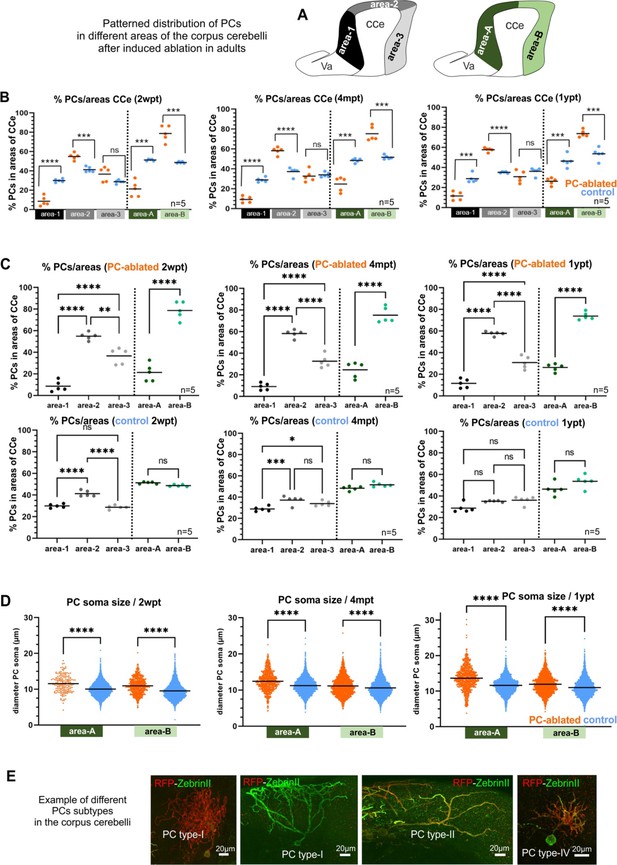
Quantification of patterned Purkinje cell (PC) death and recovery throughout the antero-posterior axis of the corpus cerebelli (CCe), related to Figure 7.
(A–C) Quantification of regional distribution of regenerating PCs after adult PC ablation. (D) Quantification of PC soma size of regenerating PCs at different time points after PC ablation in adult zebrafish compared to controls. (E) Examples of different PC subtype morphologies in the adult cerebellum. (A) Proposal of possible subdivisions of the CCe, into three areas (rostral = area 1, dorsal = area 2, and caudal = area 3) or into two halves (rostral = area A and caudal = area B). (B, C) Percentage of PCs present in the different subdivisions of the CCe comparing ablated and control groups (B), and among fishes of the same group (C). Both groups, ablated and controls, present a tendency of lower amounts of PCs in the most rostral area than caudal parts of the CCe (C). However, when comparing between groups, the percentage of PCs in those rostral areas is significantly smaller in PC-ablated fish than in control specimens (B). (D) Differences in the size of PC somata in the anterior and posterior halves of the CCe (areas A and B, respectively). (E) Example images of different PC subtypes, visualized individually after induced PC apoptosis, which show different soma sizes and morphologies of their dendritic tree as described by Chang et al., 2020. The quantification in all graphs represent PCs from the entire CCe of 5 fish per group per time point. Average values from the entire PC population of each brain (B, C), or from individual PCs were displayed together (D). Statistical information: statistical method=analysis of variance (ANOVA) test for multiple groups comparison (ordinary one-way ANOVA- followed by Sidàks multiple comparison test), or Student’s t-test for two groups comparison (unpaired t-test two tailed for parametric, or Mann-–Whitney two tailed for no parametric data), levels of significance=P<0.05 (*), P<0.01 (**), P<0.001 (***), P<0.0001 (****). Additional information in Supplementary file 1.
-
Figure 7—figure supplement 1—source data 1
Distribution values of Purkinje cells (PCs) for Figure 7—figure supplement 1B, C. Measurements of PC diameters for Figure 7—figure supplement 1D.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig7-figsupp1-data1-v2.xlsx
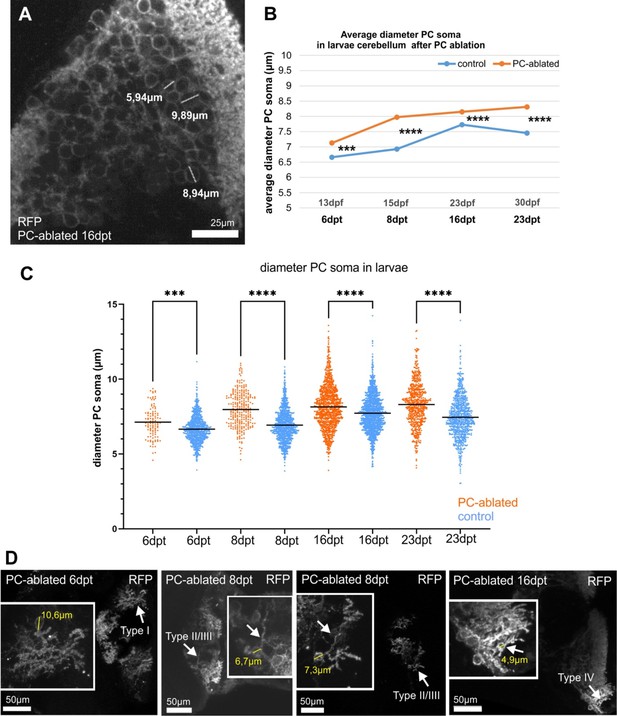
Diameter of Purkinje cell (PC) somata in larvae after PC ablation.
(A–D) Quantification of diameter of PC somata in larvae after PC ablation. (A) Detail showing PCs with different soma size, related to Figure 7. Quantitative analysis of the size of PC somata, average (B) and single PCs (C) after PC ablation in larvae, showing significantly larger diameter in PC-ablated larvae comparing to controls from beginning of regeneration phase. (D) Details of single PCs that correspond to different PC subtypes (according to descriptions by Chang et al., 2020). Statistical information: statistical method=ANOVA Kruskal-Wallis, Dunn´s multiple comparisons test, levels of significance=P<0.001 (***), P<0.0001 (****). Additional information in Supplementary file 1.
-
Figure 7—figure supplement 2—source data 1
Quantification of average Purkinje cell (PC) somata diameter for Figure 7—figure supplement 2B, and quantification of PC somata of individual PCs in larvae for Figure 7—figure supplement 2C.
- https://cdn.elifesciences.org/articles/79672/elife-79672-fig7-figsupp2-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-tagRFP (rabbit polyclonal) | Evrogen, Moscow, Russia | Catalog number: AB233 | IF (1:1000) |
| Antibody | anti-zebrinII (mouse monoclonal) | Donated by R. Hawkes, Univ. of Calgary, Canada | N/A | IF (1:500) |
| Antibody | anti-GFP (chick polyclonal) | Aves Labs, Davis, CA, USA | GFP-1010 | IF (1:1000) |
| Antibody | anti-BrdU (rat monoclonal) | Biozol GmbH, Eching, Germany | Abcam 6326 | IF (1:200) |
| Antibody | Alexa goat anti-rabbit IgG (H+L) 568 (goat polyclonal) | Invitrogen Inc, Carlsbad, CA, USA | A11011 | IF (1:500) |
| Antibody | Alexa goat anti-mouse IgG (H+L) 488 (goat polyclonal) | Invitrogen Inc, Carlsbad, CA, USA | A11001 | IF (1:1000) |
| Antibody | Donkey anti-chicken IgY (igG) (H+L) FITC (donkey polyclonal) | Jackson ImmunoResearch, West Grove, PA, USA | 703-545-155 | IF (1:1000) |
| Antibody | Alexa goat anti-rat IgG (H+L) 488 (goat polyclonal) | Invitrogen Inc, Carlsbad, CA, USA | A11006 | IF (1:1000) |
| Chemical compound/drug | cis/trans-4-Hydroxy-tamoxifen (4-OHT) | Sigma-Aldrich, St. Louis, MO, USA | H6278 | |
| Chemical compound/drug | Endoxifen | Sigma-Aldrich, St. Louis, MO, USA | E8284 | |
| Chemical compound/drug | BrdU | Sigma-Aldrich, St. Louis, MO, USA | B50021-G | |
| Commercial assay, kit | EdU (Click-EdU Alexa Fluor 647 Imaging kit) | Thermo Fisher Scientific, Waltham, MA, USA | C10340 | |
| Chemical compound/drug | d-Tubocurare hydrochloride-pentahydrate | Sigma-Aldrich, St. Louis, MO, USA | T2379 | |
| Chemical compound/drug | Tricaine, MS-222 | Sigma-Aldrich, St. Louis, MO, USA | E10521 | |
| Chemical compound/drug | DAPI | Thermo Fisher Scientific, Waltham, MA, USA | 62247 | |
| Chemical compound/drug | Normal Goat Serum, NGS | Vector Laboratories, Burlingame, CA, USA | S-1000 | |
| Chemical compound/drug | Albumin Fraction V | Carl Roth GmbH, Karlsruhe, Germany | 80076.2 | |
| Chemical compound/drug | DMSO | AppliChem GmbH, Darmstadt, Germany | 67-68-5 | |
| Strain, strain background (Danio rerio) | AB wild-type zebrafish | N/A | Fish facility TU-Braunschweig | |
| Strain, strain background (Danio rerio) | Brass pigmentation mutant zebrafish | N/A | Fish facility TU-Braunschweig | |
| Genetic reagent (Danio rerio) | Tg[ca8:-FynTagRFP-T2A-Casp8-ERT2 (PC-ATTAC)]bz11 | (Weber et al., 2016) 10.1242/dev.122721 | Fish facility TU-Braunschweig | |
| Genetic reagent (Danio rerio) | Tg(–7.5ca8:EGFP)bz12 | (Namikawa et al., 2019b) 10.1177/1179069519880515 | Fish facility TU-Braunschweig | |
| Genetic reagent (Danio rerio) | Tg(olig2:EGFP)vu12 Tg(olig2:dsRed)vu19 | (Shin et al., 2003) doi: 10.1023/B:MICS.0000006847.09037.3a. (Almeida et al., 2011) doi:10.1242/dev.071001 | Fish facility TU-Braunschweig | |
| Genetic reagent (Danio rerio) | Tg(ptf1a:EGFP) | (Godinho et al., 2005) doi:10.1242/dev.02075 | Fish facility TU Braunschweig | |
| Genetic reagent (Danio rerio) | Tg(gfap:EGFP)mi2001 | (Bernardos and Raymond, 2006) doi:https://doi.org/10.1016/j.modgep.2006.04.006 | Fish facility TU Braunschweig | |
| Software, algorithm | Patchmaster Software | HEKA Elektronik GmbH, Reutlingen, Germany | ||
| Software, algorithm | IGOR Pro 6.37 | Wavemetrics Inc, Portland, OR, USA | ||
| Software, algorithm | Leica LAS X | Leica Microsystems GmbH, Wetzlar, Germany | ||
| Software, algorithm | Prism9 Graph Pad | GraphPad Software, San Diego, CA, USA | ||
| Software, algorithm | Excel Microsoft | Microsoft Inc, Redmond, WA, USA | ||
| Software, algorithm | Free Imaging software FIJI | https://imagej.net | ||
| Software, algorithm | Ethovision Noldus software | Noldus Inc, Wageningen, The Netherlands | ||
| Software, algorithm | Corel Draw software | Corel Corporation, Ottawa, Ontario, Canada |
Additional files
-
Supplementary file 1
Statistical data related to Figures 1—7 and respective figre supplements.
Mean, standard deviation, statistic test applied, ‘n’ number, p value, and level of significance are indicated. The reference to the different figures and groups of the comparisons performed in each figure are highlighted in bold. Abbreviations: 4-OHT 4-hydroxy-tamoxifen, BrdU Bromodeoxyuridine, DMSO dimethyl sulfoxide, dpf days post-fertilization, dpt days post-treatment, EdU Ethynyl-2′-deoxyuridine, Endox Endoxifen, EtOH ethanol, gfap glial fibrillary acidic protein, mpf months post-fertilization, mpt months post-treatment, NTT novel tank test, OKR optokinetic response, PC Purkinje cells, PCL Purkinje cell layer, ptf1a pancreas-associated transcription factor 1a, Tg transgenic, wpt weeks post-treatment, WT wild-type, ypf years post-fertilization, ypt years post-treatment.
- https://cdn.elifesciences.org/articles/79672/elife-79672-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/79672/elife-79672-mdarchecklist1-v2.pdf





