Recombinant origin and interspecies transmission of a HERV-K(HML-2)-related primate retrovirus with a novel RNA transport element
Figures
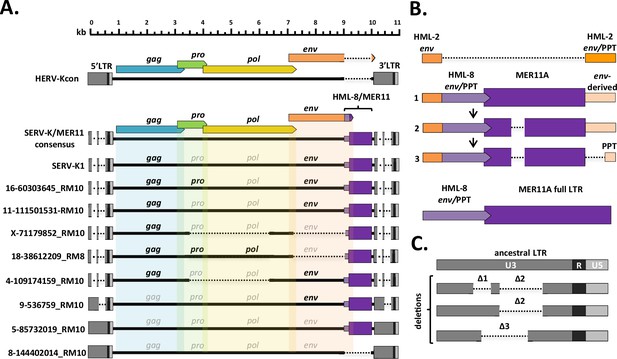
Structural features of rhesus macaque HML-2 proviruses.
(A) Overall structure of selected rhesus proviruses compared to consensus human-specific HML-2. HERVK11 HML-8-derived region shown in purple, with light purple indicating the HERVK11-derived region of env and dark purple indicating the long terminal repeat (LTR)-derived MER11 element. Intact open reading frames (ORFs) shown in bold, defective or missing ORFs indicated by grayed out font. Proviruses were selected to show notable structural features such as intact ORFs, shared internal deletions, and distinct LTR types. Deletions or gaps are indicated by dotted lines. (B) Structural evolution of recombinant region, including parental HML-2 and HML-8 regions for comparison. 1: recombinant region in oldest proviruses, with no MER11 deletion and intact env-derived non-coding sequence; 2: younger recombinants with MER11 deletion; 3: youngest recombinants with MER11 deletion and deletion of env-derived non-coding sequence outside the polypurine tract (PPT). Bright orange = HML-2-derived env coding sequence; pale orange = HML-2 env-derived non-coding sequence (including PPT); pale purple = HML-8-derived env coding sequence; dark purple = HML-8 LTR-derived non-coding sequence (MER11 element). (C). SERV-K/MER11 LTR types, showing the accumulation of deletions in U3 in younger LTRs compared to the ancestral SERV-K/MER11 LTR. U3, R, and U5 regions of LTRs indicated by dark gray, black, and light gray coloration, respectively.
-
Figure 1—source data 1
SERV-K/MER11 proviruses in rhesus macaques.
Table with genomic coordinates, assembly each provirus was identified in, orientation, intact open reading frames (ORFs), oldest common ancestor insertion was identified in, and presence of shared deletions for each locus analyzed in this paper. Incomplete ORFs longer than 90% of full-length ORF marked with an asterisk.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Non-recombinant HML-2-like proviruses in rhesus macaques.
Table with genomic coordinates for the non-recombinant HML-2-like proviruses (i.e. non-SERV-K/MER11) present in the rhesus macaque reference genome.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig1-data2-v2.xlsx
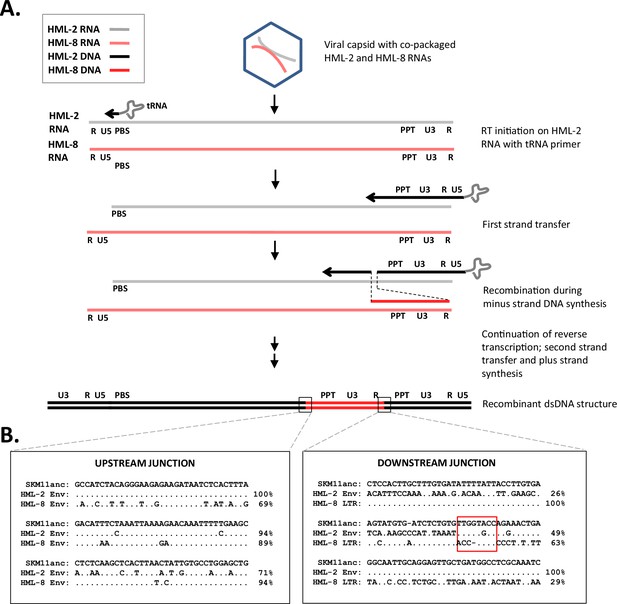
Origin of SERV-K/MER11 via RT-mediated recombination between co-packaged HERV-K (HML-2) and HERVK11 (HML-8) genomic RNAs.
(A) Recombination via two RT jumps during minus strand synthesis. (B) Sequence alignment of recombination breakpoints, comparing a reconstructed ancestral recombinant SERV-K/MER11 (SKM11anc) with the consensus human HML-2 HERV-Kcon and a rhesus macaque HML-8 sequence (rheMac10 chr2:185106142–185107371). Percent nucleotide sequence identity of HML-2 and HML-8 sequences to SKM11anc is noted to the right of each line. Red box in downstream junction outlines possible microhomology between HML-2 and HML-8 sequences.
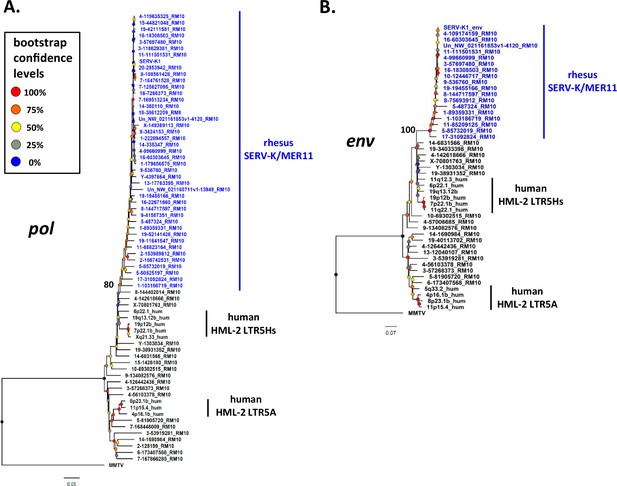
Phylogeny of SERV-K/MER11 pol and env.
Maximum likelihood phylogenies for SERV-K/MER11 pol (A) and env (B) with 1000 bootstrap replicates. SERV-K/MER11 sequences are in blue. In black, sequences from non-recombinant rhesus HML-2, human HML-2 LTR5Hs, human HML-2 LTR5A included for comparison, along with MMTV pol or env as outgroups. Nodes colored according to bootstrap replicate percentages; basal node of SERV-K/MER11 clade marked with exact bootstrap value. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site.
-
Figure 2—source data 1
Pol alignment.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig2-data1-v2.zip
-
Figure 2—source data 2
Env alignment.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig2-data2-v2.zip
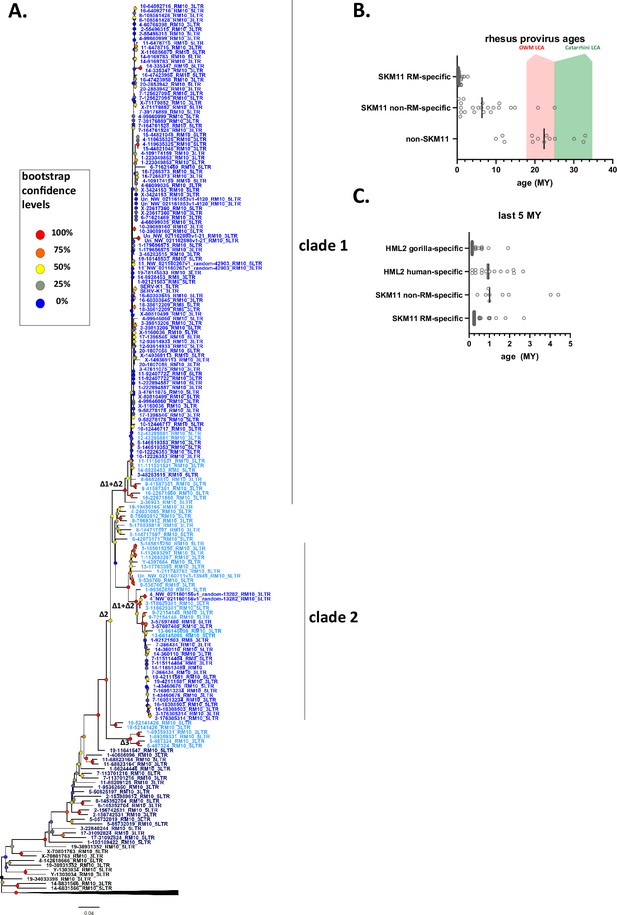
Phylogeny of SERV-K/MER11 long terminal repeats (LTRs) and LTR-based age estimates.
(A) Maximum likelihood phylogeny of rhesus macaque HML-2 LTRs with 100 bootstrap replicates. SERV-K/MER11 clade colored blue, and shaded according to species specificity; dark blue proviruses are found in all Old World monkeys (OWM), light blue are cercopithecine specific, and royal blue are rhesus macaque specific. One clade of non-recombinant rhesus proviruses has been collapsed for clarity. Nodes colored according to bootstrap replicate values. Δ1, Δ2, Δ1 + Δ2, and Δ3 mark estimated last common ancestor for each LTR deletion or combination of LTR deletions (see Figure 1C). (B) Proviral integration times estimated using the sequence divergence between cognate 5′ and 3′ LTRs. Proviruses with no differences between their LTRs were plotted at 250,000 years. Red and green shaded areas mark range of estimated ages for the last common ancestors of the OWM and Catarrhine primates, respectively. SKM11 = SERV K/MER11. (C). Proviral integration times in the past 5 million years. Estimated ages of human and gorilla-specific HML-2s included for reference.
-
Figure 3—source data 1
Long terminal repeat (LTR) alignment.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig3-data1-v2.zip
-
Figure 3—source data 2
Table of estimated provirus ages.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig3-data2-v2.xlsx
-
Figure 3—source data 3
Rhesus macaque and crab-eating macaque genomic DNA samples used for PCR screening.
Table with Coriell ID numbers, sex and species of origin for each genomic DNA sample used for PCR screening.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig3-data3-v2.xlsx
-
Figure 3—source data 4
Primers used for PCR screening.
Table with sequences of primers to the flanking regions of each rhesus macaque proviral insertion tested for insertional polymorphism, and the sequence of the primer specific to the SERV-K/MER11 5′ UTR used to detect full-length proviruses.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig3-data4-v2.xlsx
-
Figure 3—source data 5
Genotypes and allele frequencies of screened proviruses.
Table with genotype results in the screened samples for each screened provirus, with calculated allele frequencies in the 14 rhesus macaques screened.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig3-data5-v2.xlsx
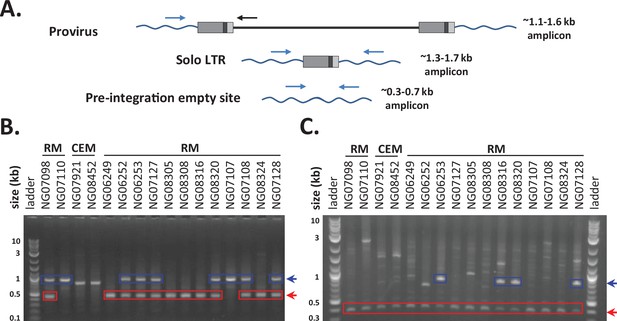
Allele-specific PCR screening.
(A) 3 primer allele-specific PCR for amplifying both full-length provirus, solo long terminal repeat (LTR), and pre-integration empty site alleles for specific proviral insertions. Primers specific to the 5′ and 3′ flanking sequences for 23 proviruses were designed, and used along with a primer specific to the 5′ UTR of SERV-K/MER11 proviruses to screen 14 rhesus macaque (RM) and 2 crab-eating macaque (CEM) genomic DNA samples for the presence or absence of proviral insertions. Examples of PCR screening results for 19-18145532_RM10 (B) and SERV-K1 (C). Gel electrophoresis of PCR products for each sample, with proviral amplicons outlined in blue and empty site amplicons outlined in red. No solo LTR alleles were identified for these proviruses.
-
Figure 3—figure supplement 1—source data 1
Original file for the PCR gel electrophoresis analysis in Figure 3—figure supplement 1B.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig3-figsupp1-data1-v2.zip
-
Figure 3—figure supplement 1—source data 2
Original file for the PCR gel electrophoresis analysis in Figure 3—figure supplement 1C.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig3-figsupp1-data2-v2.zip
-
Figure 3—figure supplement 1—source data 3
PDF containing Figure 3—figure supplement 1B and uncropped image of the corresponding PCR gel with relevant bands labeled.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig3-figsupp1-data3-v2.zip
-
Figure 3—figure supplement 1—source data 4
PDF containing Figure 3—figure supplement 1C and uncropped image of the corresponding PCR gel with relevant bands labeled.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig3-figsupp1-data4-v2.zip
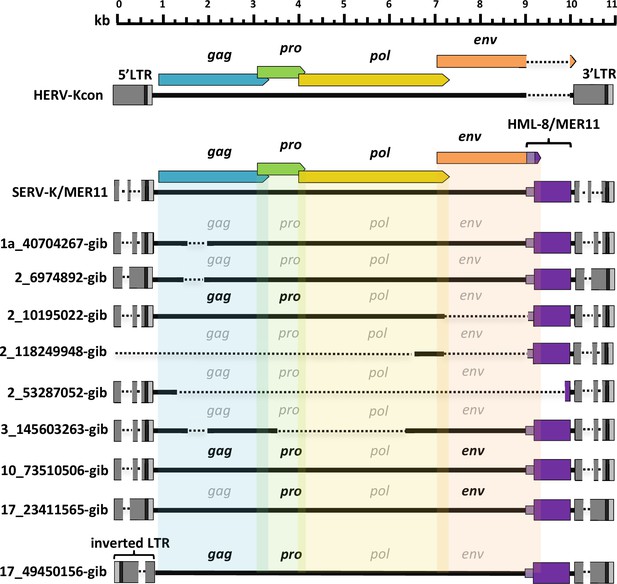
Structures of gibbon proviruses.
Structure of gibbon SERV-K/MER11 provirus compared to rhesus SERV-K/MER11 and human HML-2. HERVK11 HML-8-derived region shown in purple, with light purple indicating the HERVK11-derived region of env and dark purple indicating the long terminal repeat (LTR)-derived MER11 element. Intact open reading frames (ORFs) shown in bold, defective or missing ORFs indicated by grayed out font. Deletions or gaps in alignment shown by dotted lines.
-
Figure 4—source data 1
SERV-K/MER11 proviruses in northern white-cheeked gibbon assembly.
Table with genomic coordinates, orientation and intact open reading frames (ORFs) for SERV-K/MER11 proviruses identified in the northern white-cheeked gibbon genome assembly.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig4-data1-v2.xlsx
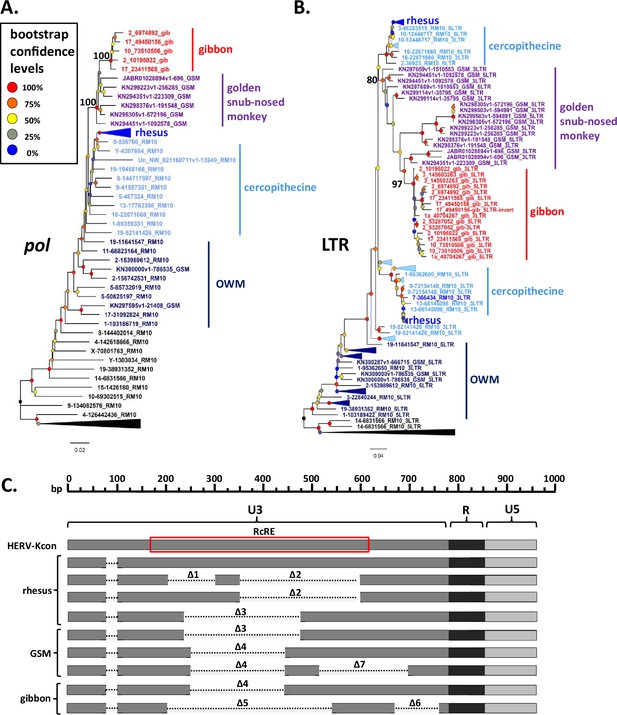
Phylogeny and long terminal repeat (LTR) deletions of gibbon and golden snub-nosed monkey (GSM) SERV-K/MER11 proviruses.
(A, B) Pol and LTR maximum likelihood phylogenies of gibbon and GSM SERV-K/MER11 proviruses with 100 bootstrap replicates. Rhesus SERV-K/MER11 and non-recombinant proviruses were included for comparison; large clades of rhesus proviruses collapsed for clarity. Nodes colored by bootstrap confidence level, branches colored by provirus type and species specificity. Black = non-recombinant rhesus, dark blue = OWM SERV-K/MER11, light blue = cercopithecine specific, royal blue = rhesus specific, purple = GSM, red = gibbon. (C) Schematic of deletions in U3 of rhesus, GSM and gibbon SERV-K/MER11 LTRs, compared with a human HML-2 full-length 968 bp LTR (HERV-Kcon). Dotted lines mark deletions relative to the HERV-Kcon LTR. Approximate location of 433 bp HML-2 Rec Response Element (RcRE) noted by red box.
-
Figure 5—source data 1
SERV-K/MER11 proviruses in golden snub-nosed monkey assembly.
Table with genomic coordinates and orientation of SERV-K/MER11 proviruses identified in the golden snub-nosed monkey genome assembly.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Pol alignment.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig5-data2-v2.zip
-
Figure 5—source data 3
Long terminal repeat (LTR) alignment.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig5-data3-v2.zip
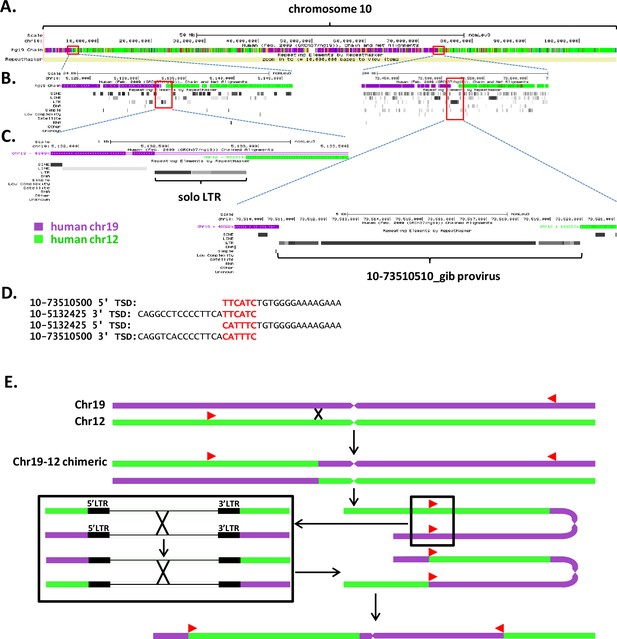
10-73510510 provirus is the result of intra- or interchromosomal recombination.
(A) Screenshot of chromosome 10 of the northern white-cheeked gibbon genome in the UCSC Genome Browser, demonstrating synteny between chr10 and human chr12 and chr19. This is a visualization of the alignment of human chr12 and chr19 to gibbon chr10 in the hg19 Chain/Net track. Purple regions align with chr19, and green regions align with chr12. Red boxes outline regions containing two SERV-K/MER11 insertions. (B) Regions containing proviral insertions expanded to show local genomic context, with flanking regions aligning to different human chromosomes. (C) SERV-K/MER11 solo long terminal repeat (LTR) 10-5132425_gib and provirus 10-73510510_gib, with flanking sequence. (D) Mismatched target site duplications of 10-5132425_gib and 10-73510510_gib. (E) Potential recombination scenario: chr10 of the white-cheeked gibbon initially composed of two regions, one syntenic to human chr19, and one to human chr12. Homologous recombination between two proviruses in the opposite orientation to each other, one in the chr19 region and one in the chr12 region, leads to a reciprocal translocation on chr10, resulting in the current structure, with two regions syntenic to chr19 and two to chr12.
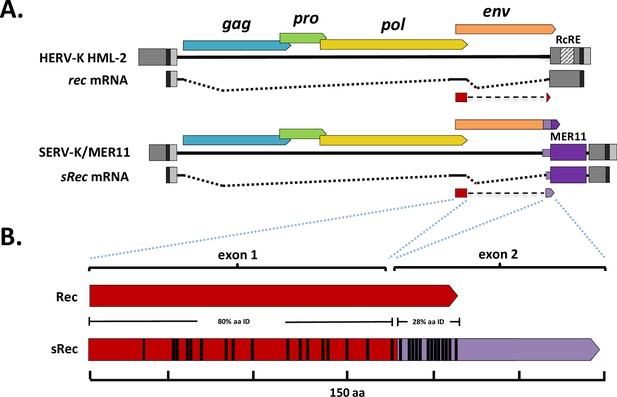
SERV-K/MER11 chimeric rec-like transcript and Rec response element deletions.
(A) Comparison of human HML-2 rec transcript to SERV-K/MER11 sRec transcript. Both are doubly spliced sub-genomic transcripts. The second coding exon of sRec is encoded by the HML-8-derived region, in purple, with light purple denoting the HML-8 env-derived region, and dark purple the HML-8 long terminal repeat (LTR)-derived region. The HML-2 Rec Response Element (RcRE) in the U3 region of the LTR is also marked. (B) HML-2 Rec protein compared to chimeric SERV-K/MER11 sRec. Red region of sRec is homologous to Rec, with black bars denoting amino acid differences. Purple region is the 62 aa HML-8-derived exon, with black bars again showing amino acid differences.
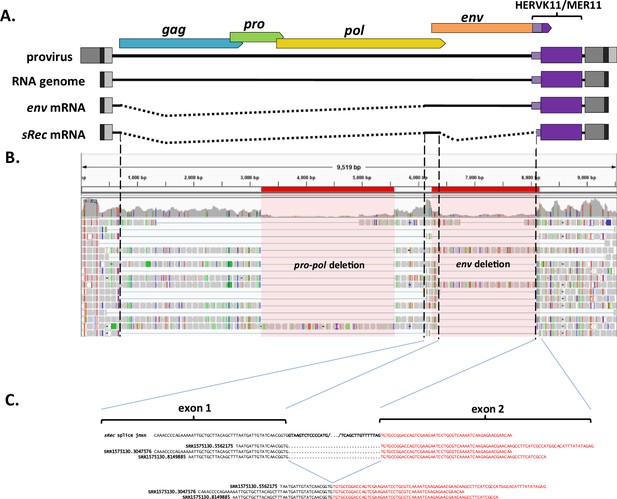
Identification of rec-like transcript from rhesus macaque RNAseq data.
(A) Diagram of SERV-K/MER11 provirus, full-length genomic RNA and env and sRec sub-genomic mRNAs. (B) Screen shot of the Integrative Genomics Viewer showing rhesus macaque induced pluripotent stem cell (iPSC) RNAseq reads aligned to the SERV-K1 genome. Red highlighted regions mark the locations of two large deletions shared by multiple proviruses. Dotted lines mark the splice acceptors and donors for the two introns that are spliced out to make sRec. Multiple reads cross both splice junctions. (C) Examples of reads crossing the second splice junction between the two coding exons of sRec. Exon 1 in black, exon 2 in red, with intronic portions of splice site bolded in black.
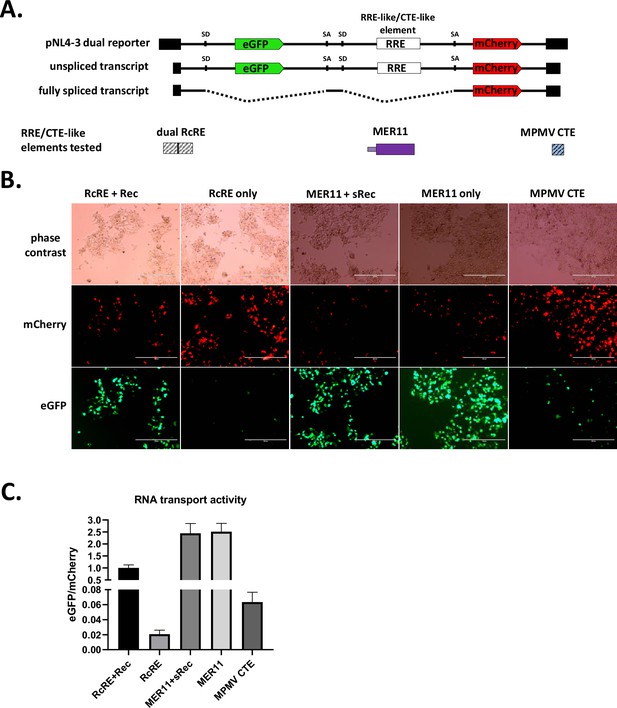
Constitutive transport element (CTE) activity of HERVK11 HML-8-derived region in SERV-K/MER11.
(A) Schematics of dual color lentiviral reporter and transport elements tested. Base construct is an NL4-3 HIV provirus modified to express eGFP from an unspliced transcript and mCherry from a fully spliced transcript. Transport element of interest replaces RRE. SD = splice donor, SA = splice acceptor. HML-8-derived region was tested for unspliced RNA transport function by transfection with and without sRec; dual HERV-Kcon Rec Response Element (RcRE) with and without Rec was used as a control for RcRE-like activity, and MPMV CTE as a control for CTE-like activity. (B) Fluorescent imaging of transfected cells. mCherry = transport element-independent signal, eGFP = transport element-dependent signal. Scale bar = 400 μm. (C) Quantification of RNA transport activity using flow cytometry. eGFP and mCherry mean fluorescent intensity of transfected cell populations were measured, and the ratio of eGFP/mCherry plotted for each construct as a measure of RNA transport activity. The mean and standard deviation of three replicates are plotted for each condition. See Figure 7—figure supplement 1 for gating strategy and flow dot plots for each condition.
-
Figure 7—source data 1
DNA sequences of the SERV-K/MER11 consensus putative transport elements tested.
Sequence of the 1545 bp consensus MER11U3R insert initially tested for RNA transport activity, comprising the last 283 nucleotides of the SERV-K/MER11 env open reading frame (ORF), followed by 726 nucleotides of sequence homologous to HML-8 long terminal repeats (LTRs), aka MER11 elements, followed by 42 nucleotides of HML-2 env-derived sequence, including the polypurine tract (PPT), followed by the U3 and R regions of the SERV-K/MER11 LTR, 494 nucleotides in length combined. This sequence was cloned into the pNL4-3 dual reporter vector described in the methods section. MER11 and MER11-Δenv were derived by truncating MER11U3R; MER11 by deleting the last 513 bp of MER11U3R, and MER11-Δenv by deleting the last 513 bp and first 258 bp of MER11U3R. The remaining 25 bp of env-derived sequence were retained in MER11-Δenv as they overlap with the HML-8 LTR.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig7-data1-v2.zip
-
Figure 7—source data 2
DNA sequence of consensus sRec open reading frame (ORF) tested for RNA transport activity.
Sequence of the consensus sRec ORF tested for RNA transport activity, with the first 261 nucleotides comprising the first, HML-2-derived coding exon, with the remaining 189 nucleotides comprising the HML-8-derived exon. This sequence was cloned into a pCMV expression vector.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig7-data2-v2.zip
-
Figure 7—source data 3
Flow cytometry data.
Table with flow cytometry data including eGFP and mCherry mean fluorescence intensities used to calculate RNA transport activity.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig7-data3-v2.xlsx
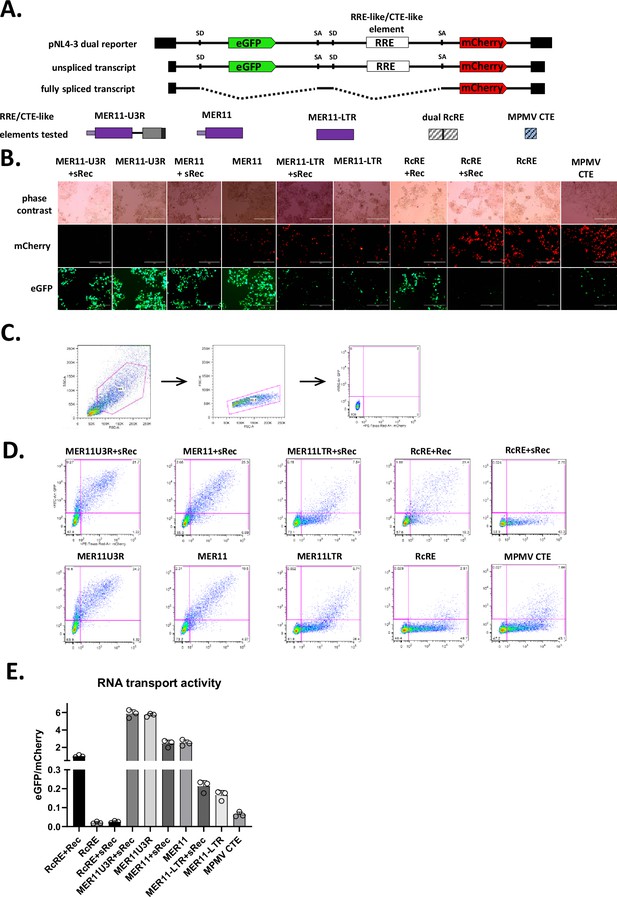
Constitutive transport element (CTE) activity of MER11 element with SERV-K/MER11 U3R and without HML-8 env region.
(A) Schematics of dual color lentiviral reporter and transport elements tested as in Figure 7. Three SERV-K/MER11 constructs were tested for unspliced RNA transport function by transfection with and without Rec or sRec. Dual HML-2 Rec Response Element (RcRE) and MPMV CTE as controls. (B) Fluorescent imaging of transfected cells. mCherry = transport element-independent signal, eGFP = transport element-dependent signal. Scale bar = 400 μm. (C) Gating scheme for quantification of RNA transport activity using flow cytometry. Cells were first gated on a forward scatter vs side scatter plot, followed by gating for single cells using forward scatter height vs forward scatter area. Lastly, untransfected cells were used to gate out the eGFP mCherry double negative population; the eGFP+, mCherry+, and eGFP+mCherry+ populations were combined for further analysis. (D) eGFP vs mCherry plots for each experimental condition, showing the gate used to define the combined single and double positive populations. (E) RNA transport activity of each element. eGFP and mCherry mean fluorescent intensity of transfected cell populations were measured, and the ratio of eGFP/mCherry plotted for each construct as a measure of RNA transport activity. The mean and standard deviation of three replicates are plotted for each condition.
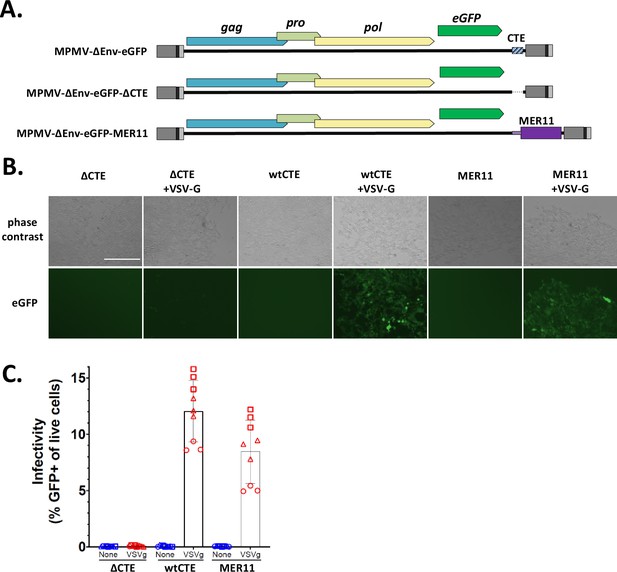
MER11 constitutive transport element (CTE) can functionally replace the MPMV CTE in the context of a single round vesicular stomatitis virus glycoprotein G (VSV-G) pseudotyped viral infection.
(A) MPMV proviral constructs expressing eGFP in place of Env, with either the wild-type MPMV CTE (wtCTE), no CTE (ΔCTE), or the MER11 CTE replacing wtCTE (MER11), were transfected into HEK293T cells with and without VSV-G. Viral supernatants were harvested 72 hr after transfection and used to infect HEK293T target cells. Three infections were performed, each in triplicate. Infectivity was assayed after 72 hr via fluorescent imaging (B) with one representative image shown per condition, and flow cytometry (C) using % GFP expressing cells as a measure of infectivity. For images, the scale bar = 400 μm. For flow cytometry, each data point represents one experimental replicate, with different shapes corresponding to different infection rounds.
-
Figure 8—source data 1
Flow cytometry data.
Table with flow cytometry data including percent eGFP expressing cells for each experimental replicate.
- https://cdn.elifesciences.org/articles/80216/elife-80216-fig8-data1-v2.xlsx
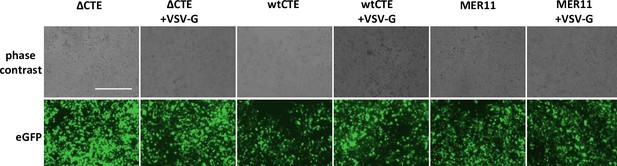
Fluorescent imaging of producer cells for infection assay.
MPMV proviral constructs expressing eGFP in place of Env, with either the wild-type MPMV constitutive transport element (wtCTE), no CTE (ΔCTE), or the MER11 CTE replacing wtCTE (MER11), were transfected into HEK293T cells with and without vesicular stomatitis virus glycoprotein G (VSV-G). Cells were imaged 72 hr after infection and viral supernatant was harvested for infections. Scale bar = 400 μm.





