Structural features stabilized by divalent cation coordination within hepatitis E virus ORF1 are critical for viral replication
Figures

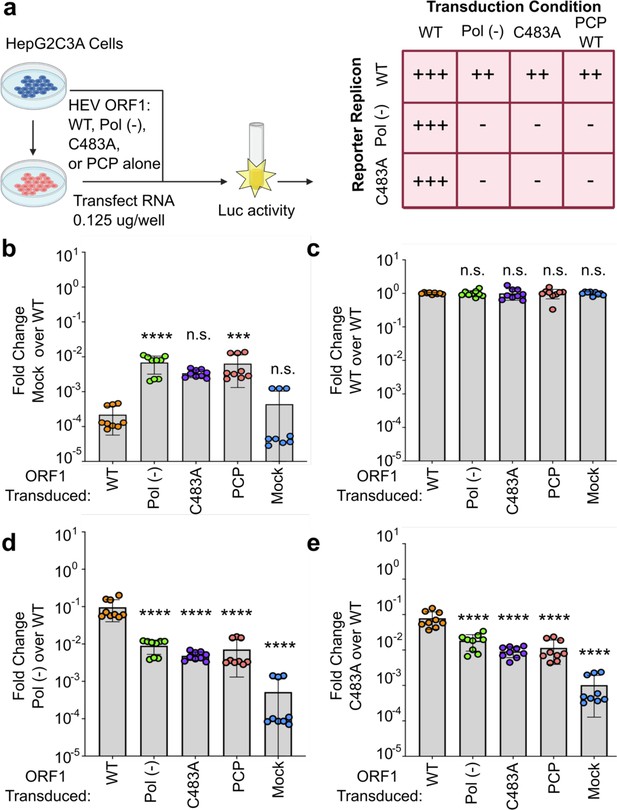
Mutations within the hepatitis E virus (HEV) putative protease domain render the virus replication incompetent.
(a) Genome organization of HEV and Kernow strain genotype 3 reporter replicon. ORF2 and ORF3 were replaced by a Gaussia Luciferase reporter in frame with the subgenomic promoter and translation start site. MeT – methyltransferase; Y – Y-domain; PCP – putative papain-like cysteine protease; HVR – hypervariable region; X – macro-domain; Hel – helicase; RdRp – RNA-dependent RNA polymerase. The putative catalytic dyad of HEV within the reporter replicon pPCP is denoted (residues C483 and Y590). GDD – catalytic triad of RdRp. Gluc – Gaussia Luciferase. (b), Replication kinetics of Kc1/p6 WT HEV Gluc RNA or HEV pol (-) Gluc, or HEV RNA bearing mutations in the putative PCP transfected into HUH7.5 cells. Cell culture supernatants were collected from transfected cells at time points indicated (drop in signal at D2 indicates wash step to eliminate signal from input RNA). (c) End point analysis (day 4) of data from panel b. (d–g) End point analysis (4 days post transfection) comparison of C483A mutant replication kinetics to WT and pol (-) in (d), Kernow strain (genotype 3) (e), SHEV (genotype 3) strain (f), SAR55 (genotype 1) and (g), TW6196E (genotype 4) of HEV when transfected into HepG2C3A human hepatoma cells. (h) Alternate cysteine and alanine codons at C483 show viral replication deficiency caused by amino acid substitution and not RNA base substitution. These data represent an end point analysis (4 days post transfection) when HEV RNA replicons are transfected into HUH7 human hepatoma cells. One-way ANOVA with Dunnett’s multiple comparison analysis were conducted to determine significance. * - p<0.05, ** - p<0.01, *** - p<0.001, and **** - p<0.0001. Data shown in d–g are from experiments done in technical and biological triplicate. Data shown in b, c, and h are from experiments done in technical triplicate and biological duplicate. Raw Gluc data provided in file Figure 1—source data 1.
-
Figure 1—source data 1
Mutations within the hepatitis E virus (HEV) putative protease domain render the virus replication incompetent.
Spreadsheet of raw Gaussia luciferase (GLuc) data kinetics for (b–c) point and double mutant bearing replicons, (d) Kc1/p6 genotype 3 HEV replicons, (e) SHEV3 genotype 3 replicons, (f) SAR55 genotype 1 HEV replicons, (g) Tw6196E genotype 4 replicons, and (h) alternate codons of Kc1/p6 genotype 3 HEV replicons.
- https://cdn.elifesciences.org/articles/80529/elife-80529-fig1-data1-v1.xlsx

Alignment of one or more representative strain(s) from each known hepatitis E virus (HEV) genotype putative papain-like cysteine protease (PCP) domain with rubella virus (RUBV) protease reveals highly conserved octa-cysteine motif in HEV.
Black – HEV putative PCP. Brown – upstream flanking 32 AAs – downstream flanking 8 AAs. (*) Catalytic dyad of RUBV protease at putative catalytic dyad positions within HEV putative PCP domain. Bold – eight highly conserved cysteines across all known HEV genotypes. TW4 - HEV strain TW6196E. Alignments generated using ClustalW algorithm within the genome sequence viewing program SnapGene.
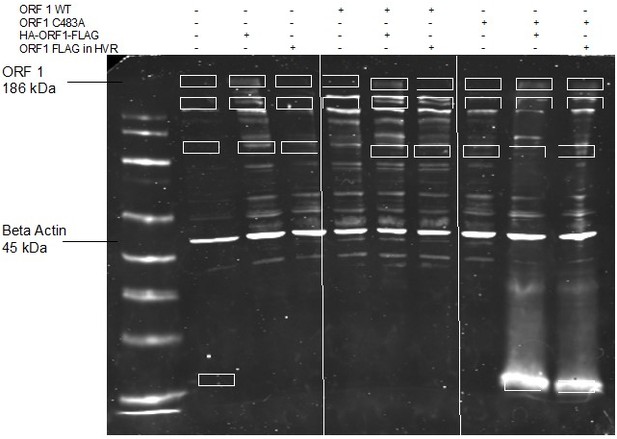
Hepatitis E virus (HEV) ORF1 putative papain-like cysteine protease (pPCP) cannot function outside of the context of the full-length protein, and C483A replication deficiency can be rescued in trans.
(a) Schematic of the transcomplementation assay. HepG2C3A human hepatoma cells were transduced with lentivirus expressing HEV ORF1 wt, Pol (-), C483A, or pPCP only (ORF1 AAs 433–592 in Kc1/p6) and subsequently transfected with in vitro transcribed RNA of wt, pol (-), or C483A replicons. End point analysis was conducted 4 days post transfection with Gaussia luciferase quantification. Cells transduced with WT ORF1 can rescue luciferase expression in pol (-) and C483A mutants in trans, whereas all other conditions, including the pPCP out of context of ORF1, cannot. (b–e) End point analysis of fold change luciferase signal over WT replicon signal of (b), Mock (c), WT (d), Pol (-), or (e), C483A. One-way ANOVA with Dunnett’s multiple comparison analysis were conducted to determine significance. * - p<0.05, ** - p<0.01, *** - p<0.001, and **** - p<0.0001. Schematic in panel a generated with BioRender. Pol (-) – replication incompetent replicon due to mutation in RNA dependent RNA polymerase. Gluc – Gaussia luciferase. Data shown in b–e are from experiments done in technical and biological triplicate. Raw Gluc data provided in file Figure 2—source data 1.
-
Figure 2—source data 1
Hepatitis E virus (HEV) ORF1 putative papain-like cysteine protease (pPCP) cannot function outside of the context of the full-length protein, and C483A replication deficiency can be rescued in trans.
Spreadsheet of raw Gaussia luciferase (GLuc) data kinetics for transduced HepG2C3A cells (mock, ORF1 WT, ORF1 Pol [-], ORF1 C483A, and HEV pPCP domain) transfected with HEV Kc1/p6 replicons (WT, Pol [-], and C483A). Each sheet corresponds to different transduction condition.
- https://cdn.elifesciences.org/articles/80529/elife-80529-fig2-data1-v1.xlsx
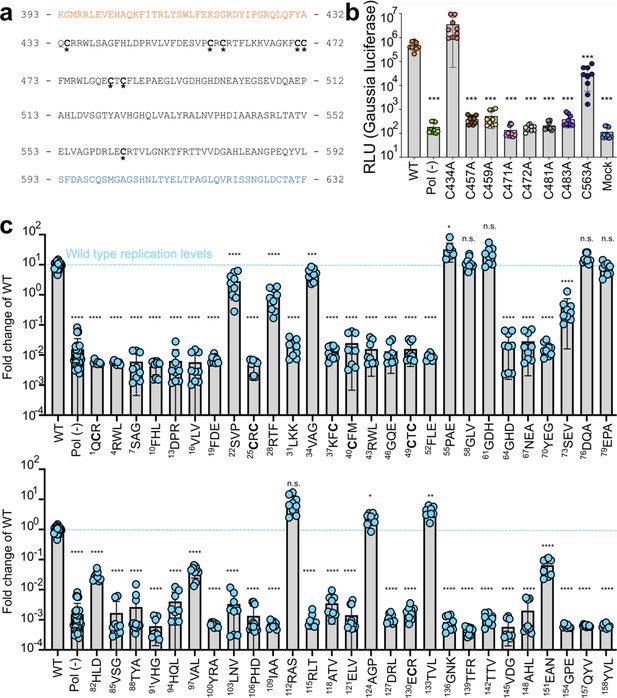
Point mutations of highly conserved cysteines and alanine scanning mutagenesis within putative papain-like cysteine protease (pPCP) identifies residues and regions indispensable for viral replication.
(a) Partial sequence of Kc1/p6 hepatitis E virus (HEV) ORF1 including pPCP that shows eight cysteines that are highly conserved across all eight known HEV genotypes. Orange – upstream 40 amino acids within Y domain prior to pPCP. Blue – downstream 40 amino acids within HVR after pPCP. (b), HepG2C3A cells were transfected with in vitro transcribed RNA of WT, polymerase deficient, or point mutants in one of the conserved cysteines within the pPCP. Cell culture supernatants were collected for 4 days post transfection prior to Gaussia luciferase (Gluc) quantification. (c) Unbiased triplet alanine scanning mutagenesis of entire pPCP region. HepG2C3A cells were transfected with in vitro transcribed RNA of triplet alanine scanning mutant replicons to assess viral replication capacity. Data shown are fold change of wild type replicon. Brown-Forsythe one-way ANOVA with Dunnett’s T3 multiple comparison analysis were conducted to determine significance. * - p<0.05, ** - p<0.01, *** - p<0.001, and **** - p<0.0001. HVR – hypervariable region. Raw Gluc data provided in file Figure 3—source data 1.
-
Figure 3—source data 1
Point mutations of highly conserved cysteines and alanine scanning mutagenesis within putative papain-like cysteine protease (pPCP) identifies residues and regions indispensable for viral replication.
Spreadsheet of raw Gaussia luciferase (GLuc) data kinetics for Kc1/p6 conserved cysteine point mutant replicons and alanine scanning mutagenized triplets.
- https://cdn.elifesciences.org/articles/80529/elife-80529-fig3-data1-v1.xlsx
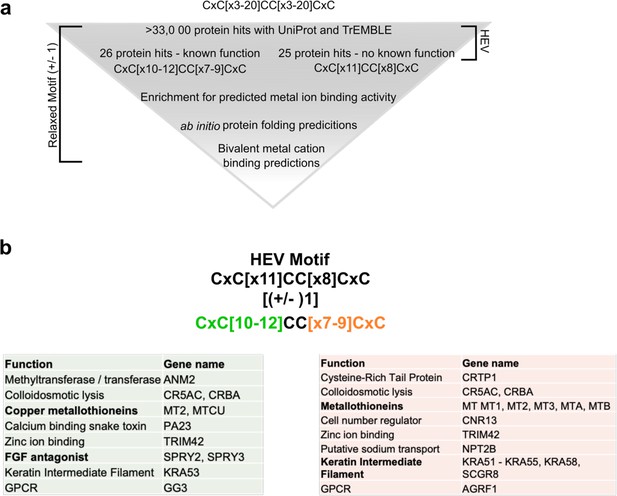
Bioinformatic analysis (PROSITE) predicts CxC[X11]CC[X8]CxC motif (CxC motif) to be necessary for divalent metal ion coordination.
(a) Bioinformatic pipeline depicting hexa-cysteine motif search across all deposited proteins in UniProt and TrEMBLE databases. Exact hepatitis E virus (HEV) motif search yielded 25 protein hits, all with unknown functions (right side). 11 amino acids upstream of core CC motif and eight amino acid motif downstream of core CC were relaxed by (±) 1 to yield 26 protein hits, all with known functions and an enrichment for divalent ion binding activity. (b) Utilizing motif searches with ScanProSite (de Castro et al., 2006) and TrEMBLE (Bairoch and Apweiler, 2000; Boeckmann et al., 2003; O’Donovan et al., 2002), we used a relaxed expression of the CxC motif where each stretch of [x] could be (±) 1: (CxC[x](10-12)CC[x](7-9)CxC). This analysis brought forth 26 proteins with known functions, enriched for proteins with divalent metal ion binding activity. Green: proteins that matched the first stretch of [x 10–12]. Orange: proteins that matched the second stretch of [x 7–9].
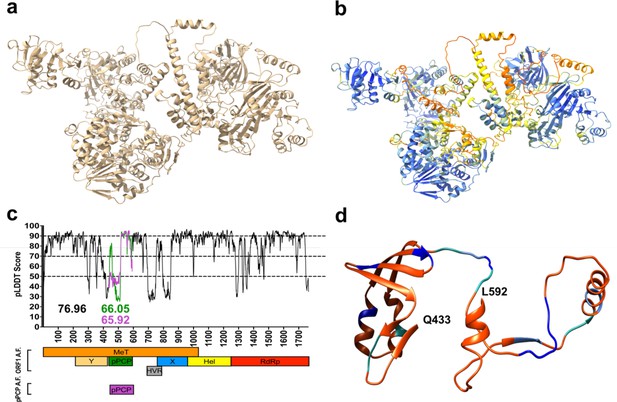
Structural prediction models of hepatitis E virus (HEV) putative papain-like cysteine protease (pPCP) demonstrates low-confidence scores, suggesting lack of highly ordered secondary structure.
(a) HEV ORF1-WT AlphaFold structure prediction. (b) HEV ORF1-WT AlphaFold structure prediction pseudo-colored by pLDDT score gradation (darker blue – higher pLDDT Score, darker red – lower pLDDT score). (c) pLDDT score of AlphaFold prediction of HEV ORF1 across HEV genome organization for all of ORF1 (black, average 76.96), pPCP when measured with entirety of ORF1 (green, average 66.05), and pPCP when predicted by AlphaFold alone (purple, average 65.92). (d) HEV ORF1 pPCP AlphaFold prediction pseudo-colored by alanine scanning mutagenesis ORF1 replication tolerance. Color based on fold change of WT in (Figure 3C). Orange – below (–2) Sea green – between (–2) and (–1). Cornflower blue – between (–1) and 0. Dark blue – replicated above WT levels. Beginning and end residues of pPCP noted in bold.
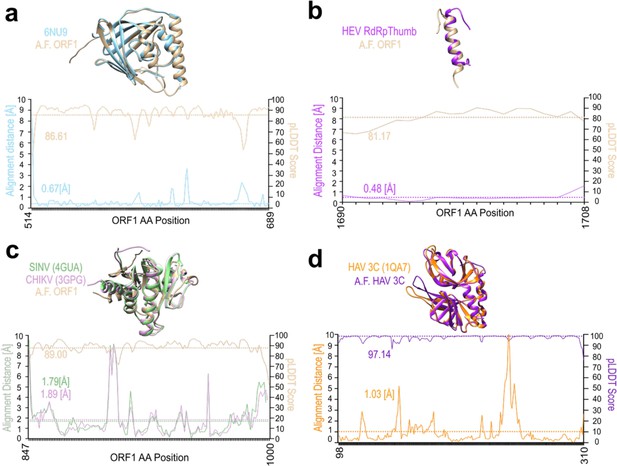
AlphaFold predicts structured domains of hepatitis E virus (HEV) ORF1 and viral protease of hepatitis A virus (HAV) with high confidence.
(a–c) Superimposition of AlphaFold (A.F.) prediction of corresponding Kernow C1/p6 ORF1 regions (Bronze) plus distance plot in (Å) and pLDDT score of A.F. prediction with: (a) the crystal structure of HEV SAR-55 ORF1 amino acids 510–691 (Blue; PDB: 6NU9; Proudfoot et al., 2019). (b) Amphipathic ‘thumb’ of HEV gt3 strain 83-2-27 RNA dependent RNA Polymerase (amino acids 1628–1647; ORF1 kc1/p6 residues 1684–1709; Purple; Oechslin et al., 2022). (c) Macro domains of Sindbis virus (Green; SINV amino acids 1342–1,509 of PDB: 4GUA; Shin et al., 2012) and Chikungunya virus (Pink; CHIKV; PDB: 3GPG; Malet et al., 2009). (d) Superimposition of HAV 3 C protease with corresponding A.F. prediction plus distance plot in (Å) and pLDDT score of A.F. prediction. Orange – HAV 3 C protease (PDB: 1QA7; Bergmann et al., 1999) and Purple – HAV 3 C protease A.F. prediction. Hatched lines show averages of corresponding measurement.

Sequence alignment of Kc1/p6 macrodomain with macrodomains of Sindbis virus (SINV) and Chikungunya virus (CHIKV).
Alignment of ORF1 macrodomain (AAs 857–1000) with SINV macrodomain (AAs 1342–1,509 of PDB: 4GUA; Shin et al., 2012) and CHIKV macrodomain (PDB: 3DPG; Malet et al., 2009). Dash – gap in alignment. Colon - similar amino acid. Period – dissimilar amino acid. Line – identical amino acid.
-
Figure 4—figure supplement 2—source data 1
Similarity and identity percentages of Kc1/p6 macrodomain with macrodomains of Sindbis virus (SINV) and Chikungunya virus (CHIKV).
Left: percent similarity and identity with SINV macrodomain (AAs 1342–1,509 of PDB: 4GUA; Shin et al., 2012). Right: percent similarity and identity with CHIKV macrodomain (PDB: 3DPG; Malet et al., 2009).
- https://cdn.elifesciences.org/articles/80529/elife-80529-fig4-figsupp2-data1-v1.xlsx
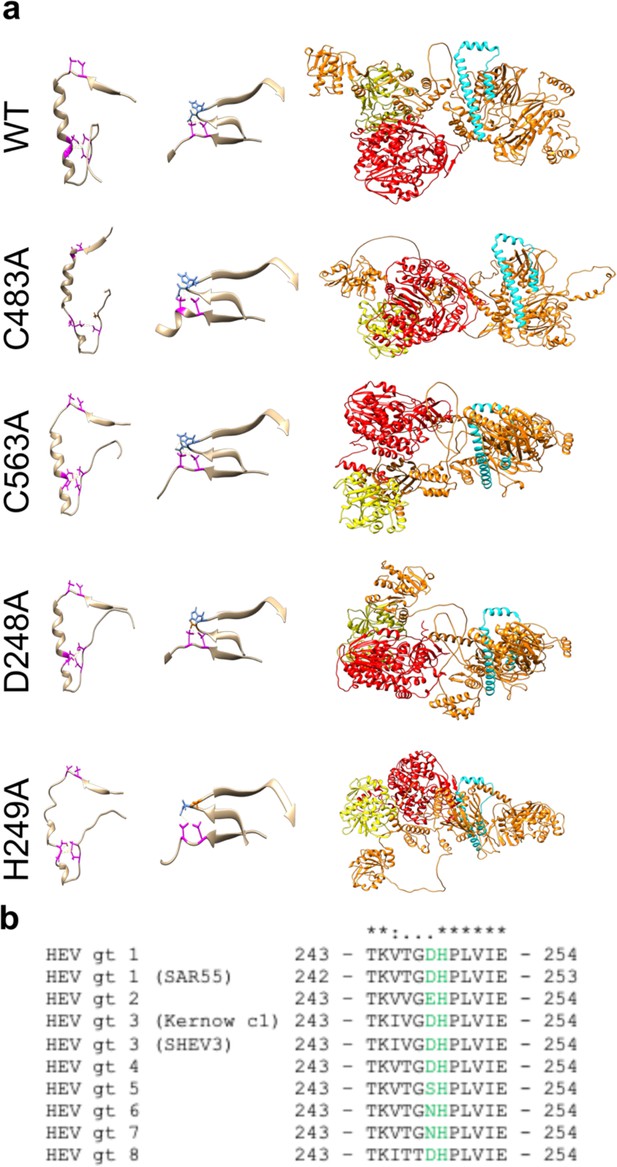
Structural prediction models suggest conserved cysteines within CxC[x11]CC[x8]CxC motif form divalent ion coordination pockets and novel domain-domain interaction with upstream Y-domain.
(a) Alphafold structural predictions of domains within hepatitis E virus (HEV) ORF1. Left: pseudo zinc-finger (amino acids 451–493 within putative papain-like cysteine protease [pPCP]). Magenta: conserved cysteines C457, C459, C471, C472, C481, and C483A. Potential divalent ion coordination tetrahedron outlined in yellow hatched line. Middle: amino acids 242–259 of HEV ORF1 Y-domain, and amino acids 451–462 of HEV pPCP. Conserved cysteines C457 and C459 are outlined in magenta. D248 and H249 of upstream Y-domain highlighted in blue. Novel interdomain divalent ion coordination domain outlined in yellow hatched line. Right: HEV ORF1 protein demonstrating folding of WT and point mutant proteins. Orange (AAs 1–1036): methyltransferase (Magden et al., 2001). Yellow (AAs 1018–1262): Helicase (Devhare et al., 2014; Karpe and Lole, 2010). Red (AAs 1257–1709): RNA dependent RNA polymerase (Koonin et al., 1992; Oechslin et al., 2022). Cyan: Putative membrane association domain (Parvez, 2017). (b) Multiple sequence alignment of HEV genotypes 1–8 of partial Y-domain containing variable residue D248 and highly conserved residue H249. (*) identical residue. (:) similar residue. (.) dissimilar residue. Yellow hatched line – bonds between coordinating amino acids.
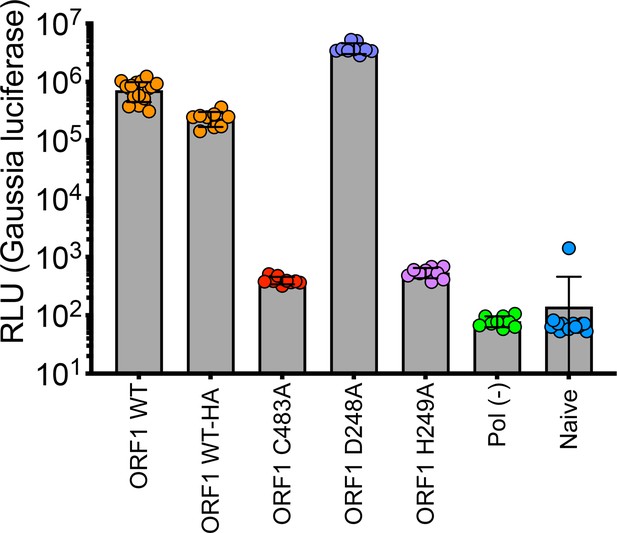
Replication end point analysis of WT-HA, D248A, and H249A Gaussia luciferase (Gluc) constructs.
HepG2C3A cells were transfected with in vitro transcribed RNA of WT, polymerase deficient, WT-HA-tagged, or point mutants in one of the upstream interacting Y-domain (D248A and H249A). Cell culture supernatants were collected for 4 days post transfection prior to Gluc quantification. Experiments were conducted in technical and biological triplicate.
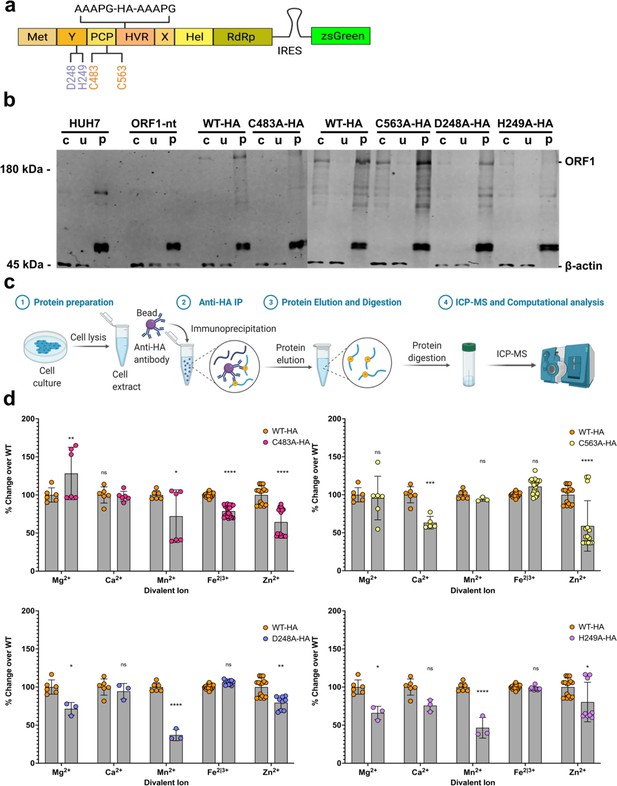
Inductively coupled plasma mass spectrometry (ICP-MS) shows divalent cation coordination by hepatitis E virus (HEV) ORF1.
(a) HA-epitope tag flanked by linker sequence inserted into hypervariable region (HVR) of HEV ORF1 lentiviral construct. (b) Western blot analysis of HUH7 human hepatoma cell produced epitope tagged ORF1 purification (crude cell lysate, unbound fraction, purified ORF1). (c) Workflow of protein sample preparation, purification, and HNO3/H2O2 digestion for ICP-MS. IP- Immuno-purification WT HA-tagged ORF1 (left), C483A HA-tagged ORF1 (middle), and C563A HA-tagged ORF1 (right). (d) Fold-change over WT of biologically relevant divalent metal ions (Zheng et al., 2008) bound by purified mutant epitope tagged ORF1. Any trace metal not measured as a value above a MilliQ water only purification control was excluded from this analysis on a run-by-run basis due to not being above IP buffer elution conditions. Two-way ANOVA with Dunnett’s multiple comparison analysis were conducted to determine significance. * - p<0.05, ** - p<0.01, *** - p<0.001, and **** - p<0.0001. Schematics in a and c were created with Biorender.com. Unedited western blots included in file Figure 6—source data 1.
-
Figure 6—source data 1
Inductively coupled plasma mass spectrometry (ICP-MS) shows bivalent cation coordination by hepatitis E virus (HEV) ORF1.
- https://cdn.elifesciences.org/articles/80529/elife-80529-fig6-data1-v1.xlsx
Inductively coupled plasma mass spectrometry (ICP-MS) shows divalent cation coordination by hepatitis E virus (HEV) ORF1 disrutped when mutated at residue C483.
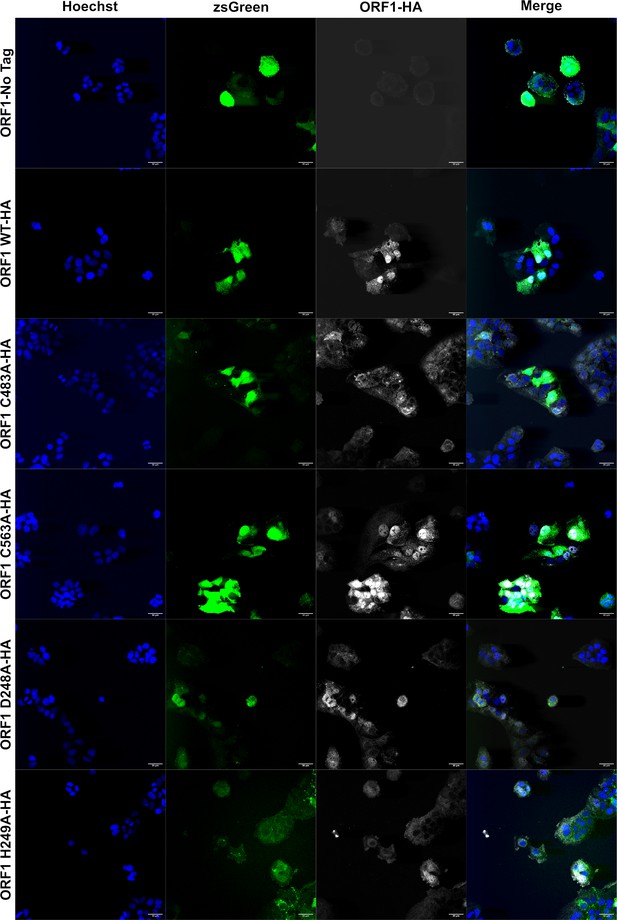
Immunofluorescence of epitope tagged ORF1 demonstrates loss of membrane association when divalent ion coordination residues are mutated.
Confocal microscopy images of HUH7 cells stably expressing ORF1 bicistronic for zsGreen. ORF1-HA tagged proteins were imaged using a rabbit-anti-HA antibody and AlexaFluor647 (goat anti-rabbit IgG [H+L]) secondary antibody. Nuclei were visualized with Hoechst 33342 stain. All images taken at 40× magnification and analyzed using Fiji (ImageJ2) image analysis software. Source files of all TIFF images included in Figure 7—source data 1.
-
Figure 7—source data 1
Immunofluorescence of epitope tagged ORF1 demonstrates loss of membrane association when divalent ion coordination residues are mutated.
TIFF files of confocal microscopy used in subcellular localization analysis.
- https://cdn.elifesciences.org/articles/80529/elife-80529-fig7-data1-v1.xlsx
Videos
AlphaFold Prediction of ORF1 WT - putative papain-like cysteine protease (pPCP) Alanine Scan Gradation.
Best ranked model of ORF1 WT AlphaFold prediction algorithm that transitions to a visual representation of the alanine scanning mutagenesis data across the pPCP. Color gradation corresponds fold change of WT of alanine triplet. Below (–2) fold change is orange. Between (–2) and (–1) fold change is sea green. Between (–1) and 0 fold change is cornflower blue, Above WT replication levels is dark blue.
AlphaFold Prediction of ORF1 WT.
Best ranked model of AlphaFold prediction algorithm of ORF1 WT, exhibiting the conserved hexa-cysteine motif (magenta), interacting Y-domain residues D248 and H249 (cornflower blue), methyltransferase domain (orange), putative papain-like cysteine protease (pPCP; green), helicase (yellow), RNA dependent RNA polymerase (red), and putative intracellular membrane association site (cyan). Hatched yellow lines – measured distances between sulfur atoms of cysteines, second oxygen of aspartic acid, or second nitrogen of histidine. Orientation of each video is with the putative membrane association site pointing up, with the pseudo zinc-finger composed of cysteines within the hexa-cysteine motif on the right.
AlphaFold Prediction of ORF1 C483A.
Best ranked model of AlphaFold prediction algorithm of ORF1 C483A, exhibiting the conserved hexa-cysteine motif (magenta), interacting Y-domain residues D248 and H249 (cornflower blue), methyltransferase domain (orange), putative papain-like cysteine protease (pPCP; green), helicase (yellow), RNA dependent RNA polymerase (red), and putative intracellular membrane association site (cyan). Hatched yellow lines – measured distances between sulfur atoms of cysteines, second oxygen of aspartic acid, or second nitrogen of histidine. Orientation of each video is with the putative membrane association site pointing up, with the pseudo zinc-finger composed of cysteines within the hexa-cysteine motif on the right. Mutation of C483A is predicted and shown here to disrupt the molecular architecture of a pseudo-zinc-finger, likely causing a relaxation of several predicted bond lengths to beyond biological relevance (Zheng et al., 2008).
AlphaFold Prediction of ORF1 C563A.
Best ranked model of AlphaFold prediction algorithm of ORF1 C563A, exhibiting the conserved hexa-cysteine motif (magenta), interacting Y-domain residues D248 and H249 (cornflower blue), methyltransferase domain (orange), putative papain-like cysteine protease (pPCP; green), helicase (yellow), RNA dependent RNA polymerase (red), and putative intracellular membrane association site (cyan). Hatched yellow lines – measured distances between sulfur atoms of cysteines, second oxygen of aspartic acid, or second nitrogen of histidine. Orientation of each video is with the putative membrane association site pointing up, with the pseudo zinc-finger composed of cysteines within the hexa-cysteine motif on the right. Mutation of C563A is predicted to cause a relaxation of several predicted bond lengths to beyond biological relevance (Zheng et al., 2008).
AlphaFold Prediction of ORF1 D248A.
Best ranked model of AlphaFold prediction algorithm of ORF1 D248A, exhibiting the conserved hexa-cysteine motif (magenta), interacting Y-domain residues D248 and H249 (cornflower blue), methyltransferase domain (orange), putative papain-like cysteine protease (pPCP; green), helicase (yellow), RNA dependent RNA polymerase (red), and putative intracellular membrane association site (cyan). Hatched yellow lines – measured distances between sulfur atoms of cysteines, second oxygen of aspartic acid, or second nitrogen of histidine. Orientation of each video is with the putative membrane association site pointing up, with the pseudo zinc-finger composed of cysteines within the hexa-cysteine motif on the right.
AlphaFold Prediction of ORF1 H249A.
Best ranked model of AlphaFold prediction algorithm of ORF1 H249A, exhibiting the conserved hexa-cysteine motif (magenta), interacting Y-domain residues D248 and H249 (cornflower blue), methyltransferase domain (orange), putative papain-like cysteine protease (pPCP; green), helicase (yellow), RNA dependent RNA polymerase (red), and putative intracellular membrane association site (cyan). Hatched yellow lines – measured distances between sulfur atoms of cysteines, second oxygen of aspartic acid, or second nitrogen of histidine. Orientation of each video is with the putative membrane association site pointing up, with the pseudo zinc-finger composed of cysteines within the hexa-cysteine motif on the right.
Additional files
-
Supplementary file 1
Source data for inductively coupled plasma mass spectrometry (ICP-MS) experimental runs and primer sequences used in plasmid generation.
Spreadsheet tabs 1–3: raw ion reads for inductively coupled plasma mass spectrometry data for each of the independent experimental runs, organized by date of run. Spreadsheet tab 4: primer sequences for each construct generated for use int his manuscript.
- https://cdn.elifesciences.org/articles/80529/elife-80529-supp1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/80529/elife-80529-mdarchecklist1-v1.docx
-
Source data 1
AlphaFold Predictions Figures 4 and 5.
These files are the best ranked (ranked 0) predictions generated by AlphaFold of HEV ORF1, its associated point mutants, and the hepatitis A virus (HAV) 3 C protease.
- https://cdn.elifesciences.org/articles/80529/elife-80529-data1-v1.zip