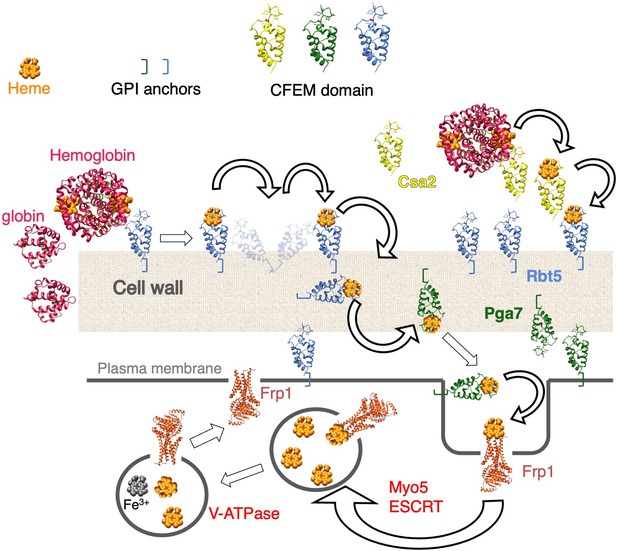
Ferric reductase-related proteins mediate fungal heme acquisition
Figures
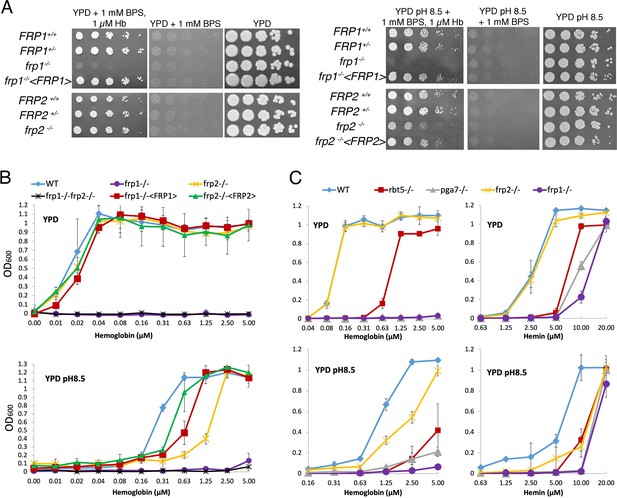
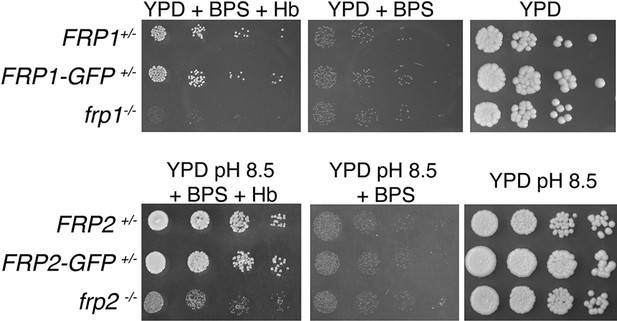
FRP1 is essential for hemoglobin-iron acquisition, whereas FRP2 contributes to growth on hemoglobin at alkaline pH.
(A) Fivefold dilutions of cultures of the indicated strains were spotted on YPD or YPD pH 8.5, with the indicated supplements, and incubated for 3 days (Hb and BPS plates) or 2 days (YPD plates) at 30°C. WT = KC2, FRP1+/-=KC859, frp1-/-=KC870, frp1-/-<FRP1 >= KC1024, FRP2+/-=KC901, frp2-/-=KC912, frp2-/-<FRP2 >= KC1379. (B) The strains with indicated genotypes in the ccc2-/- background were grown in triplicates in YPD or YPD pH 8.5 media supplemented with 1 mM ferrozine and the indicated amounts of hemoglobin, and incubated at 30°C for 3 days. Each result is the average of three cultures. Standard deviations are indicated by vertical bars. WT = KC811, frp1-/-=KC1146, frp2-/-=KC1414, frp1-/- frp2-/-=KC1412, frp1-/-<FRP1 >= KC1146, frp2-/-<FRP2>=KC1411. (C) The frp1-/- and frp2-/- heme-iron utilization phenotype was compared to that of the CFEM protein mutants rbt5-/- and pga7-/-. The strains were grown in YPD or YPD pH 8.5, with 1 mM ferrozine and the indicated concentrations of hemoglobin or hemin, and grown and measured as in B. Wild type = KC68, rbt5-/-=KC139, pga7-/-=KC485, frp1-/-=KC923, frp2-/-=KC913. All strains in B and C carry a deletion of the CCC2 gene, which causes a defect in high-affinity iron import and prevents growth in the presence of ferrozine.
-
Figure 1—source data 1
Excel file with data used to make Figure 1B.
- https://cdn.elifesciences.org/articles/80604/elife-80604-fig1-data1-v2.zip
-
Figure 1—source data 2
Excel file with data used to make Figure 1C.
- https://cdn.elifesciences.org/articles/80604/elife-80604-fig1-data2-v2.zip
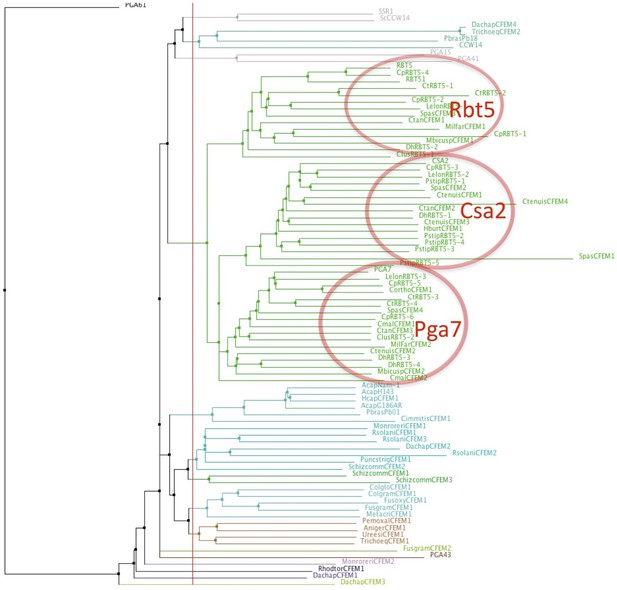
Proximity tree of Ascomycete CFEM protein sequences.
The genomes of ascomycete species were screened for sequences similar to Candida albicans Rbt5 by BLAST. Eighty-six sequences were aligned using the MAFFT G-INS-i algorithm (Katoh et al., 2005) and a tree was built on this alignment using the NJ method. The 46 Candiaceae CFEM proteins from 14 species (green lines) cluster in three groups, with homology to Rbt5, Csa2, and Pga7, respectively. Species prefixes of proteins in the three clusters most similar to C. albicans Rbt5, Csa2, and Pga7: no prefix – C. albicans; Cp – Candida parapsilosis; Ct – Candida tropicalis; Dh – Debaryomyces hansenii; Lelon – Lodderomyces elongisporus; Pstip – Pichia stipitis; Ctan – Candida tanzawaensis; Spas – Spathaspora passalidarum; Ctenuis – Candida tenuis; Clus – Candida lusitaniae; Mbicus – Metschnikowia bicuspidate; Milfar – Millerozyma farinosa; Cmal – Candida maltosa; Hburt – Hypopichia burtonii. Eight species show one apparent ortholog each of Rbt5, Csa2, and Pga7, 4 species show orthologs of two of the three at least, and two species show orthologs of only one. Additional CFEM proteins (bottom part of the figure) are from more distantly related species and may not participate in heme acquisition.
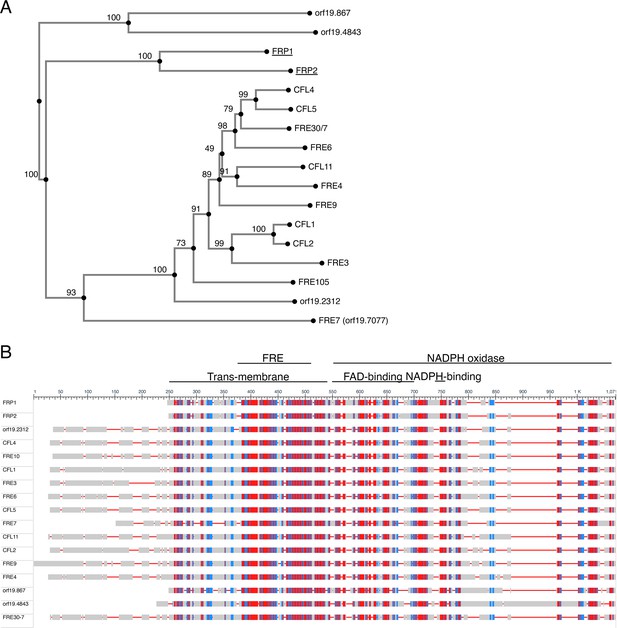
Similarity between the Frp1 and Frp2 sequences and the predicted C. albicans ferric reductases sequences.
(A) Proximity tree of Candida albicans ferric reductase-related protein sequences. Seventeen protein sequences most related to Fre10 were aligned using the MAFFT G-INS-i algorithm (Katoh et al., 2005) and a tree was built on this alignment using the NJ method with bootstrap resampling to estimate the confidence of each branching (indicated as percentages near each node). (B) Schematic alignment of the 17 C. albicans ferric reductase-like protein sequences. The level of sequence conservation is color-coded red-blue-gray from highest to lowest. The alignment was obtained using NCBI’s ‘Cobalt’ multiple alignment tool. The conserved domains are indicated on top. They were assigned using NCBI’s conserved domains database. The standard orf19 names of the named proteins are: FRP1 – orf19.5634; FRP2 – orf19.7112; CFL4 – orf19.1932; FRE10 – orf19.1415; CFL1 – orf19.1263; FRE3 – orf19.1270; FRE6 – orf19.6138; CFL5 – orf19.1930; FRE7 – orf19.7077; CFL11 – orf19.701; CFL2 – orf19.1264; FRE9 – orf19.3538; FRE4 – orf19.1844; FRE30-7 is the combined reading frames of orf19.6139 and orf19.6140, which were alternatively assigned a single or two distinct ORFs in different versions of the C. albicans genome database. Our re-sequencing indicates that they in fact form a single ORF.
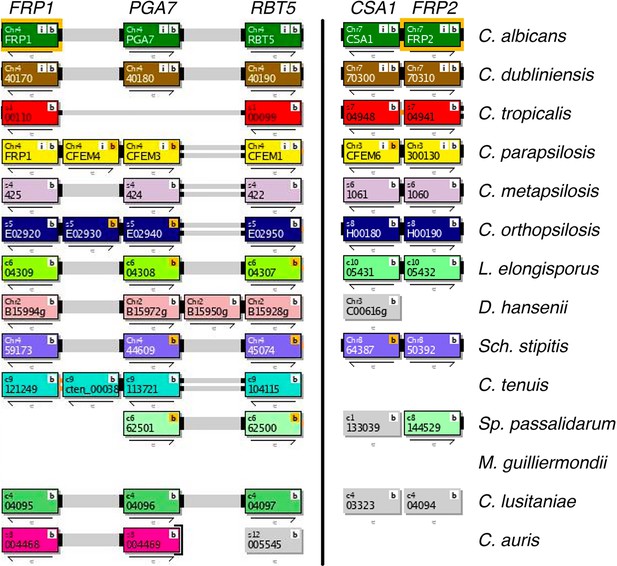
Synteny of the FRP1-PGA7 (left) and CSA1-FRP2 (right) genomic regions across Saccharomycetales species listed in the Candida Gene Order Browser database (cgob3.ucd.ie; Fitzpatrick et al., 2010; Maguire et al., 2013).
Both regions are syntenic in Candida albicans, Candida dubliniensis, Candida parapsilosis, Candida orthopsilosis, Lodderomyces elongisporus and Scheffersomyces stipitis, and only FRP1-PGA7 in Candida lusitaniae. FRP1 is absent in Spathaspora passalidarum, FRP2 is absent in Debaryomyces hansenii, Candida tenuis, and Candida auris. Both are absent in Meyerozyma guilliermondii. Overall, the FRP1-PGA7 synteny is conserved in 75% and the CSA1-FRP2 synteny in 70% of genomes where they are present. Note that RBT5, encoding another CFEM protein that participates in heme acquisition, is also part of the FRP1-PGA7 region.
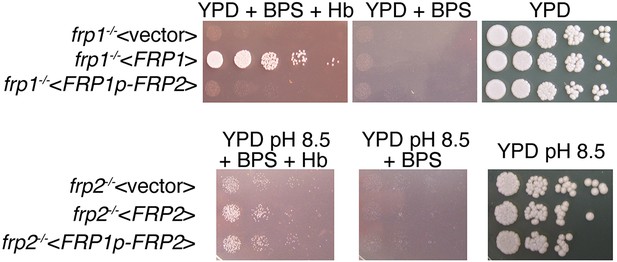
FRP2 under the control of the FRP1 promoter is unable to complement the frp1-/- mutant but does complement the frp2-/- mutant.
(Top) The frp1-/- mutant strain KC870 was transformed either with the vector plasmid BES116, with the FRP1-containing plasmid KB2546 or with an FRP2 open reading frame fused to the FRP1 promoter, KB2575. The cells were spotted on YPD, on YPD with 1 mM BPS, with or without 1 μM hemoglobin, as indicated. (Bottom) The frp2-/- mutant strain KC912 was transformed either with the vector plasmid BES116, with the FRP2-containing plasmid KB2576, or with an FRP2 open reading frame fused to the FRP1 promoter, KB2575. The cells were spotted on YPD, on YPD with 1 mM BPS, with or without 1 μM hemoglobin, all at pH 8.5, as indicated.
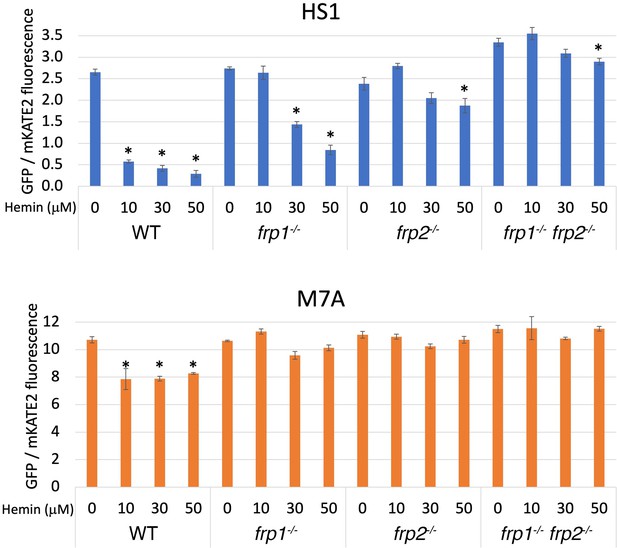
The frp1-/- and frp2-/- mutants are defective in heme uptake from the cytoplasm.
The high-affinity HS1 and low-affinity M7A ratiometric heme sensors were used to monitor heme influx into the cytoplasm in the wild-type (KC2), frp1-/-(KC870), frp2-/-(KC912), and frp1-/- frp2-/- strains (KC1410) grown for 4 hr in YPD medium at pH 8.5 with 1 mM ferrozine, and with the indicated concentrations of hemin chloride. Each data point is the average of three different cultures, each measured twice. Vertical bars indicate standard deviations, and the asterisks indicate measurements that are significantly different from the 0 μM hemin reading with p≤0.0001.
-
Figure 2—source data 1
Excel file with data used to make Figure 2C.
- https://cdn.elifesciences.org/articles/80604/elife-80604-fig2-data1-v2.zip
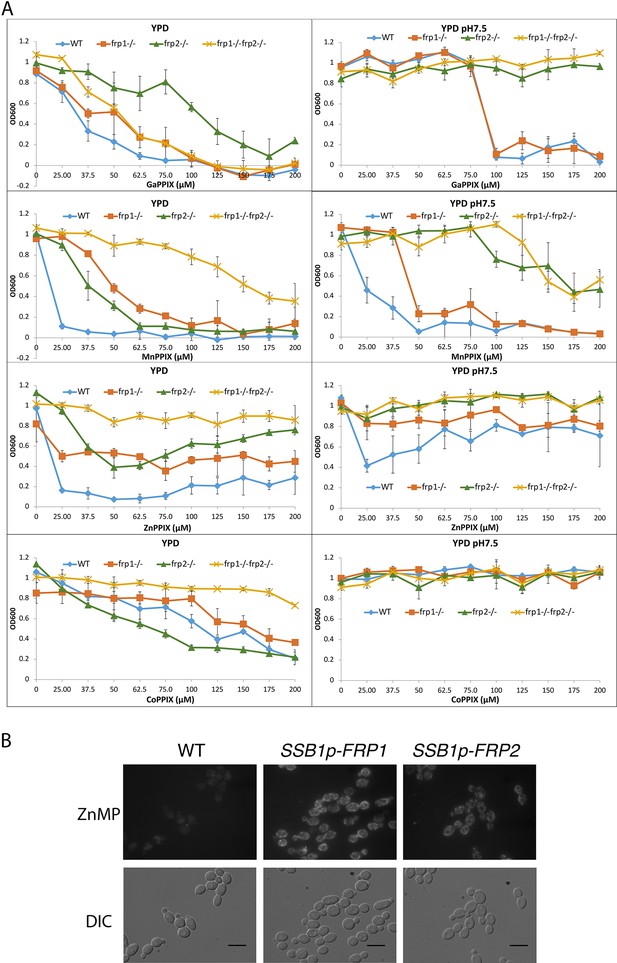
Frp1 and Frp2 participate in the uptake of heme homologs.
(A) FRP1 and FRP2 are differentially required for sensitivity to toxic heme homologs. The indicated strains were diluted in YPD medium with different concentrations of metal-protoporphyrin IX compounds, as indicated, and grown in 96-well plates at 30°C for 2 days. The graph points indicate the averages of triplicate cultures, and the standard deviations are indicated by vertical bars. The strains used are KC590 (WT), KC966 (frp1-/-), KC1053 (frp2-/-), KC1061 (frp1-/- frp2-/-). (B) Expression of either FRP1 or FRP2 is sufficient to enable ZnMP uptake. Wild-type strain KC2 (WT), strain KC1080 that has a single FRP1 gene under the SSB1 promoter (SSB1p-FRP1) and strain KC1244 that has a single FRP2 gene under the SSB1 promoter (SSB1p-FRP2) were grown in YPD to log phase, then exposed to 1 mM ZnMP for 10 min, washed and visualized by epifluorescence microscopy. Scale bar = 5 μm.
-
Figure 3—source data 1
Excel file with data used to make Figure 3A.
- https://cdn.elifesciences.org/articles/80604/elife-80604-fig3-data1-v2.zip
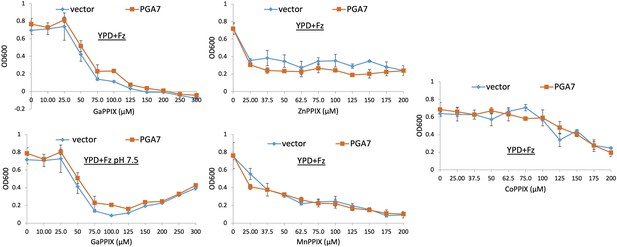
Pga7 is not required for sensitivity to non-iron metalloprotoporphyrins (MPPs).
A pga7-/- strain was transformed with either a vector plasmid (KC646) or with a PGA7-containing plasmid (KC647). The strains were diluted in YPD medium supplemented with 1 mM ferrozine, and with the MPP at the indicated concentrations, and grown in triplicate cultures at 30°C. Each datapoint indicates the average of the triplicate cultures, and the vertical bars indicate the standard deviations.
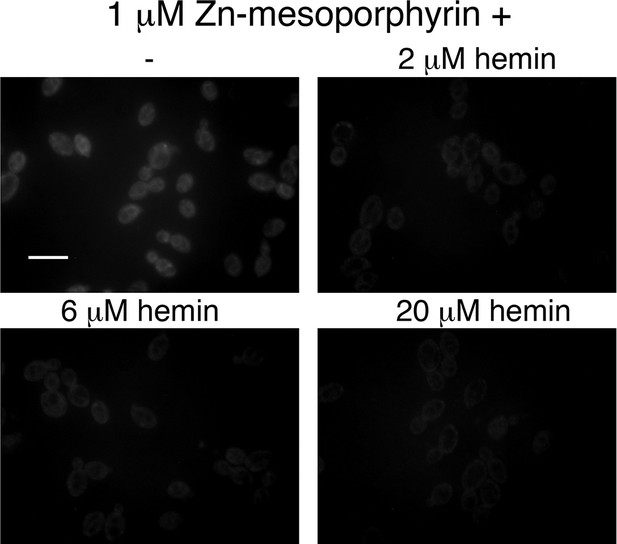
Hemin competes with zinc-mesoporphyrin for uptake by Candida albicans cells.
A wild-type strain was grown for 4 hr in YPD + 1 mM ferrozine to induce expression of the heme uptake system, then the cells were exposed for 10 min to 1 μM zinc-mesoporphyrin, without or with the indicated amounts of hemin. The cells were then washed twice in phosphate-buffered saline and visualized by epifluorescence microscopy with a rhodamine filter set. Scale bar = 5 μm.
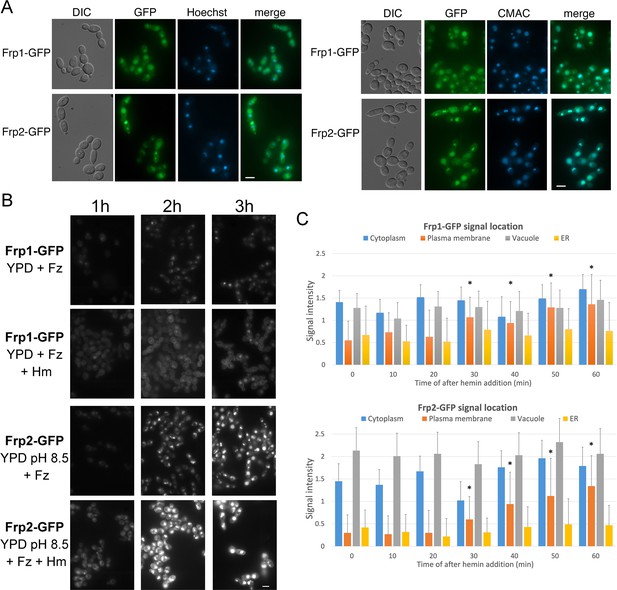
Subcellular localization of Frp1-GFP and Frp2-GFP fusion proteins.
(A) The cells (Frp1-GFP=KC914, Frp2-GFP=KC1405) were grown in iron-limited medium for 3 hr. Left panels: Localization of the Frp-GFP proteins vs. the nuclear stain Hoechst 33324. Right panels: Localization of the Frp-GFP proteins vs. the vacuole stain CMAC. Scale bars = 5 μm. (B) Location of Frp1-GFP and Frp2-GFP after induction by iron starvation, without and with added 50 μM hemin. The cells were grown to late-log phase in YPD, then shifted to the indicated media, and visualized at the indicated times by epifluorescence microscopy. Scale bar = 5 μm. (C) Kinetics of Frp1/2-GFP relocation after exposure to hemin. The cells were grown in iron-limited medium for 3 hr and then 50 µM hemin was added. The graphs describe quantitation of subcellular localization of the Frp1-GFP and Frp2-GFP signals after exposure to hemin. At least 100 cells were observed for each timepoint, and the signal intensity at each subcellular location was assigned a value from 0 to 3. The graph indicates the average intensities at each of four cellular locations. Note that ‘ER’ denotes location on the perinuclear membrane and its projections, whereas ‘plasma membrane’ could possibly also include cortical ER, which cannot be differentiated at this level of resolution. The asterisks indicate the plasma membrane values that differ statistically from t=0’ with p<0.00001 by Mann-Whitney’s U test.
-
Figure 4—source data 1
Excel file with data used to make Figure 4C.
- https://cdn.elifesciences.org/articles/80604/elife-80604-fig4-data1-v2.zip
FRP1-GFP and FRP2-GFP fusion proteins retain heme uptake activity and support growth on hemoglobin.
The strains (FRP1+/-=KC859, FRP1+/--GFP=KC916, frp1-/-=KC870, FRP2+/-=KC901, FRP2+/--GFP=KC1245, frp2-/-=KC912) were spotted on YPD, on YPD with 1 mM BPS, with or without 1 μM hemoglobin, for FRP1. For FRP2 the cells were spotted on YPD pH 8.5, on YPD pH 8.5 with 1 mM BPS, with or without 1 μM hemoglobin as indicated, and incubated for 3 days (Hb and BPS plates) or 2 days (YPD plates) at 30°C.
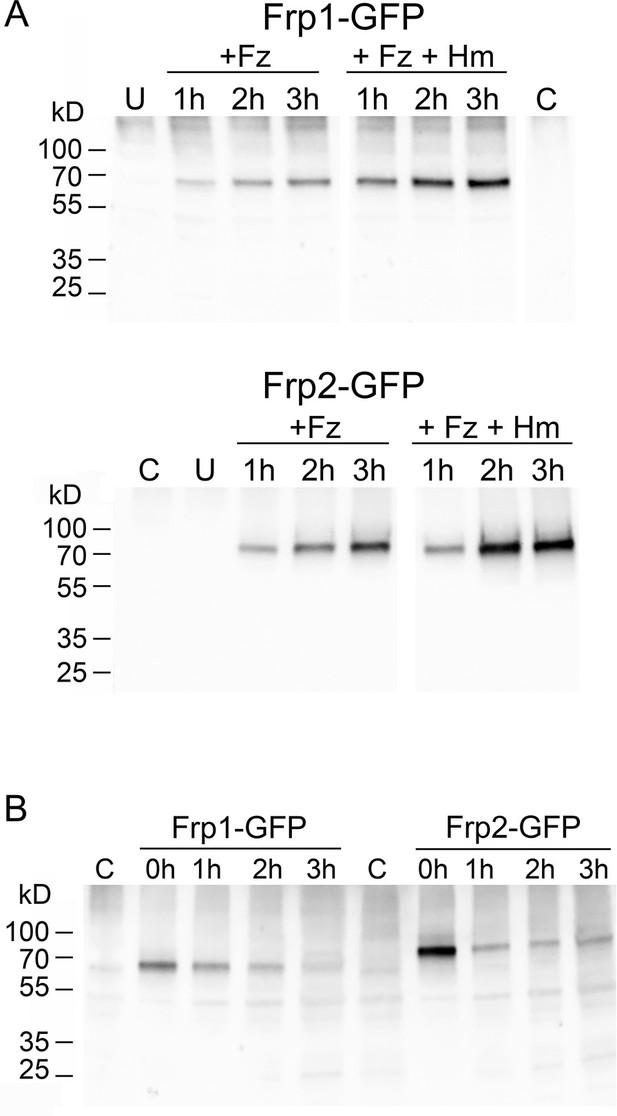
Only full-length Frp1-GFP and Frp2-GFP proteins are detectable under all conditions.
(A) The cultures depicted in Figure 4B were sampled at the same timepoints for protein extraction. Equal protein amounts were loaded on gels, and the GFP fusion proteins were detected after Western blotting with a rabbit anti-GFP antibody. The time after shift to YPD+1 mM ferrozine±50 μM hemin (Frp1-GFP) or to YPD pH 8.5+1 mM ferrozine ±50 μM hemin, as indicated, is indicated above the lanes. U=uninduced starting culture, C=control strain without GFP. (B) The same strains were grown overnight in iron-limited medium (Frp1: YPD+1 mM ferrozine, Frp2: YPD pH 8.5+1 mM ferrozine), diluted and grown another 2 hr in the same media, then washed and resuspended in iron satiation medium (YPD). The indicated timepoints are after shift to YPD. To detect decay rather than dilution of the protein pool present at the shift, each lane contains the same volume of cell culture.
-
Figure 4—figure supplement 2—source data 1
Original Western blots used to make Figure 4—figure supplement 2 A-B.
- https://cdn.elifesciences.org/articles/80604/elife-80604-fig4-figsupp2-data1-v2.zip
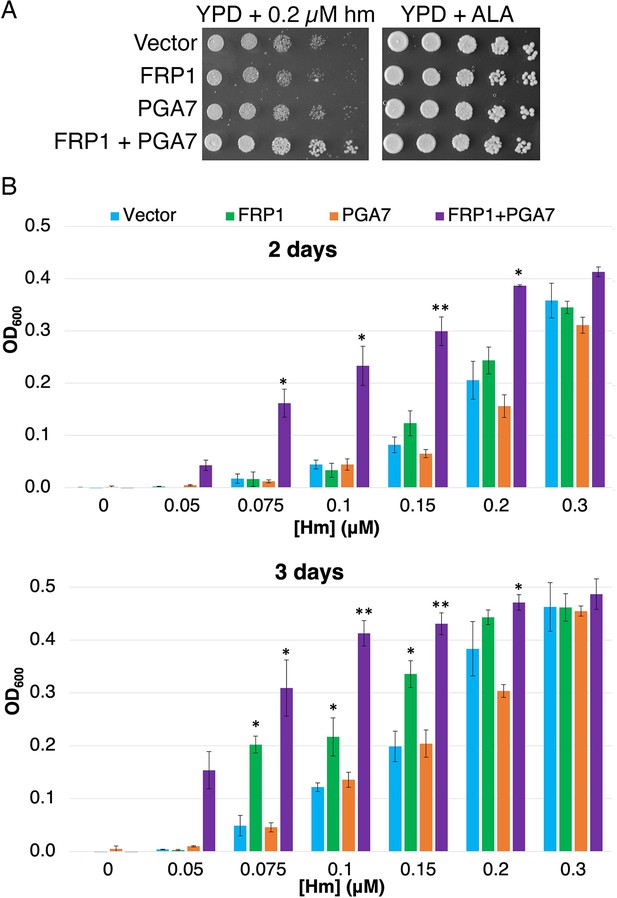
Frp1 and Pga7 collaborate in heme uptake.
(A) Saccharomyces cerevisiae hem1Δ cells (KY1498) were transformed with a vector plasmid, or with plasmids HTB2p-FRP1 (KB2569), HTA2p-PGA7(KB2789) or HTB2p-FRP1 HTA2p-PGA7 (KB2566) and drop-diluted on SC-HIS plates supplemented with either 0.2 μM hemin or 50 μg/ml δ-aminolevulinic acid (ALA), as indicated. The plates were incubated for 2 days at 30°C. (B) The same strains were diluted in SC-HIS medium supplemented with the indicated amounts of hemin, and incubated at 30°C for 2 or 3 days, as indicated. For each plasmid, three independent transformant colonies were grown. The data indicate the average of the three cultures, and the error bars indicate the standard deviations. Statistically significant differences compared to vector control are indicated with one asterisk (p<0.05) or two asterisks (p<0.001) (Student’s t-test).
-
Figure 5—source data 1
Excel file with data used to make Figure 5B.
- https://cdn.elifesciences.org/articles/80604/elife-80604-fig5-data1-v2.zip
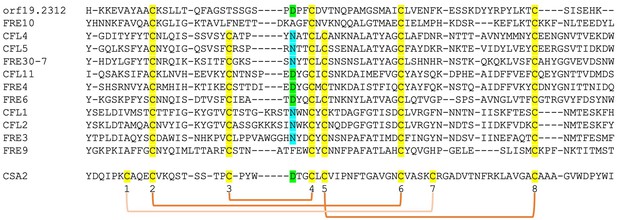
Alignment by MAFFT (Katoh et al., 2005) of the N-termini of 12 FRE-like proteins.
The alignment with Csa2 was superimposed based on the structural alignment shown in Figure 6—figure supplement 1. The eight CFEM cysteines are highlighted in yellow and numbered below the sequence. The orange linkers connect between the cysteines that form disulfide bonds in the Csa2 structure. The Csa2 heme iron-coordinating Asp residue and its homologs in ferric reductases (FREs) are highlighted in green, and the corresponding Asn residues in blue.
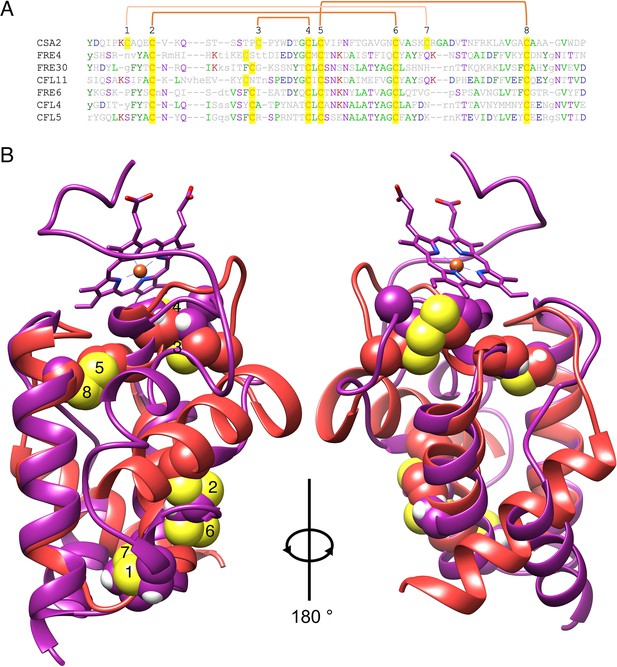
Alignment between the Csa2 CFEM structure and predicted FRE N-termini.
(A) Alignment of the Csa2 sequence with the six FREs that appear in Supplementary file 1. The eight CFEM cysteines are highlighted and numbered above the sequence. The orange linkers connect between the cysteines that form disulfide bonds in the Csa2 structure and the FRE N-terminus predicted structures. (B) Alignment of a ribbon representation of the Csa2 structure (PDB 4y7s; purple) with that of the Cfl11 N-terminus predicted structure (Alphafold PDB Q59PZ9; red) (Varadi et al., 2022). The cysteines are rendered as spheres, and the sulfur atoms are yellow. The numbering is the same as in A. The two views of the structure are rotated 180°. The structures were aligned and rendered with Chimera (Pettersen et al., 2004).
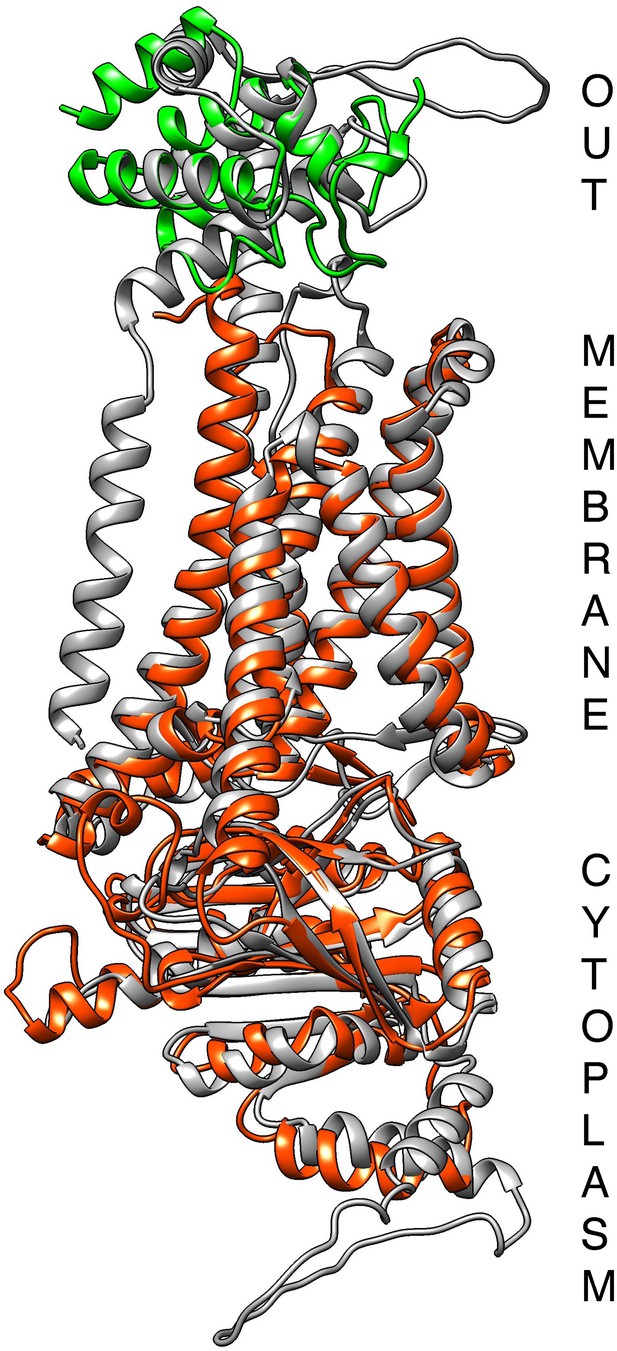
Alignment of the Alphafold-predicted structures of the ferric reductase Cfl11 (grey), of Frp1 (orange-red), and of the Pga7 CFEM domain (green).
The structures were aligned and rendered with Chimera (Pettersen et al., 2004).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Candida albicans) | WT | Fonzi and Irwin, 1993 | KC2=CAF3-1 | ura3Δ::imm434/ura3Δ::imm434 |
| Strain, strain background (Candida albicans) | ccc2-/- | Weissman et al., 2002 | KC68 | KC2 ccc2Δ::hisG/ccc2Δ::hisG |
| Strain, strain background (Candida. albicans) | rbt5-/- | Weissman and Kornitzer, 2004 | KC139 | KC68 rbt5Δ/rbt5Δ |
| Strain, strain background (Candida. albicans) | pga7-/- | Kuznets et al., 2014 | KC485 | KC68 pga7Δ/pga7Δ |
| Strain, strain background (Candida albicans) | WT | Fonzi and Irwin, 1993 | KC590=CAI4 | ura3Δ::imm434/ura3Δ::imm434 |
| Strain, strain background (Candida albicans) | pga7-/- | Kuznets et al., 2014 | KC646 | KC590 pga7Δ/pga7Δ ADE2/ade2::URA3 |
| Strain, strain background (Candida. albicans) | pga7-/- PGA7 URA3 | Kuznets et al., 2014 | KC647 | KC590 pga7Δ/pga7Δ ADE2/ade2::PGA7 URA3 |
| Strain, strain background (Candida albicans) | pga7-/- URA3 | This work | KC811 | KC68 ADE2/ade2::URA3 |
| Strain, strain background (Candida albicans) | FRP1+/- | This work | KC859 | KC2 FRP1/frp1Δ |
| Strain, strain background (Candida albicans) | frp1-/- | This work | KC870 | KC2 frp1Δ/frp1Δ |
| Strain, strain background (Candida albicans) | FRP1+/- | This work | KC901 | KC2 FRP2/frp2Δ |
| Strain, strain background (Candida albicans) | frp2-/- | This work | KC912 | KC2 frp2Δ/frp2Δ |
| Strain, strain background (Candida albicans) | frp2-/- | This work | KC913 | KC68 frp2Δ/frp2Δ |
| Strain, strain background (Candida albicans) | FRP1-GFP | This work | KC914 | KC2 FRP1/FRP1-GFP URA3 |
| Strain, strain background (Candida albicans) | FRP1-GFP | This work | KC916 | KC2 frp1Δ/FRP1-GFP URA3 |
| Strain, strain background (Candida albicans) | frp1-/- | This work | KC923 | KC68 frp1Δ/frp1Δ |
| Strain, strain background (Candida albicans) | frp1-/- | This work | KC966 | KC590 frp1Δ/frp1Δ |
| Strain, strain background (Candida albicans) | frp1-/- URA3 | This work | KC1023 | KC870 ADE2/ade2::URA3 |
| Strain, strain background (Candida albicans) | frp1-/- FRP1 URA3 | This work | KC1024 | KC870 ADE2/ade2::FRP1 URA3 |
| Strain, strain background (Candida albicans) | frp2-/- | This work | KC1053 | KC590 frp2Δ/frp2Δ |
| Strain, strain background (Candida albicans) | frp1-/- frp2-/- | This work | KC1061 | KC590 frp1Δ/frp1D frp2Δ/frp2Δ |
| Strain, strain background (Candida albicans) | frp1-/- FRP1p-FRP2 | This work | KC1064 | KC870 ADE2/ade2::FRP1p-FRP2 URA3 |
| Strain, strain background (Candida albicans) | SSB1p-FRP1 | This work | KC1080 | KC870 ADE2/ade2::SSB1p-FRP1 URA3 |
| Strain, strain background (Candida albicans) | frp1-/- URA3 | This work | KC1146 | KC923 ADE2/ade2::URA3 |
| Strain, strain background (Candida albicans) | SSB1p-FRP2 | This work | KC1244 | KC912 ADE2/ade2::SSB1p-FRP2 URA3 |
| Strain, strain background (Candida albicans) | FRP2-GFP | This work | KC1245 | KC912 frp2Δ/FRP2-GFP URA3 |
| Strain, strain background (Candida albicans) | frp2-/- URA3 | This work | KC1246 | KC912 ADE2/ade2::URA3 |
| Strain, strain background (Candida albicans) | frp2-/- FRP2 URA3 | This work | KC1379 | KC912 ADE2/ade2::FRP2 URA3 |
| Strain, strain background (Candida albicans) | FRP2-GFP | This work | KC1405 | KC2 FRP2/FRP2-GFP URA3 |
| Strain, strain background (Candida albicans) | frp1-/- frp2-/- | This work | KC1410 | KC2 frp1Δ/frp1D frp2Δ/frp2Δ |
| Strain, strain background (Candida albicans) | frp2-/- FRP2 URA3 | This work | KC1411 | KC913 ADE2/ade2::FRP2 URA3 |
| Strain, strain background (Candida albicans) | frp2-/- URA3 | This work | KC1412 | KC923 frp2Δ/frp2Δ::hisG-URA3-hisG |
| Strain, strain background (Candida albicans) | frp2-/- URA3 | This work | KC1414 | KC913 ADE2/ade2::URA3 |
| Strain, strain background (Candida albicans) | frp2-/- FRP1p-FRP2 | This work | KC1447 | KC912 ADE2/ade2:: FRP1p-FRP2 URA3 |
| Strain, strain background (Sandida cerevisiae) | hem1 | This work | KY1498 | ura3-1 can1-100 GAL+leu2-3,112 trp1-1 ade2-1 his3-11,15 hem1Δ::KanMX |
| Antibody | Anti-GFP (Rabbit polyclonal) | Abcam | Cat # ab290 | Use at 1:5000 |
| Antibody | Anti-rabbit IgG, HRP-conjugated (Goat polyclonal) | Sigma | Cat # A9169 | Use at 1:5000 |
| Recombinant DNA reagent | FRP1 blaster plasmids | This work | KB2392 KC2393 | Digest SacI-KpnI for transformation |
| Recombinant DNA reagent | FRP2 blaster plasmids | This work | KB2395 KB2396 | Digest SacI-KpnI for transformation |
| Recombinant DNA reagent | FRP1 reintegrant | This work | KB2546 | Contains the FRP1 region from –395 to +1877 |
| Recombinant DNA reagent | FRP2 reintegrant | This work | KB2576 | Contains the FRP2 region from –895 to +1982 |
| Recombinant DNA reagent | FRP1-GFP | This work | KB2431 | Contains FRP1 (+3 to+1662) fused to eGFP |
| Recombinant DNA reagent | FRP2-GFP | This work | KB2695 | Contains FRP2 (–969 to +1776) fused to eGFP |
| Recombinant DNA reagent | FRP1p-FRP2 | This work | KB2575 | Contains the FRP1 promoter (−395 to –1) fused to the FRP2 (+1 to +1878) |
| Recombinant DNA reagent | SSB1p-FRP1 | This work | KB2450 | Contains FRP1 (–2 to +1826) under SSB1 promoter (−400 to –1) |
| Recombinant DNA reagent | SSB1p-FRP2 | This work | KB2696 | Contains FRP2 (–3 to +1933) under SSB1 promoter (−400 to –1) |
| Recombinant DNA reagent | PGA7 FRP1 | This work | KB2566 | H2Ap-PGA7, H2Bp-FRP1 codon corrected |
| Recombinant DNA reagent | FRP1 | This work | KB2569 | H2Bp-FRP1 codon corrected |
| Recombinant DNA reagent | PGA7 | This work | KB2789 | H2Ap-PGA7 |
| Sequence-based reagent | FRP1 codon-corrected | This work | B35984-1/M131416 | FRP1 codon-corrected in pUC57 |
| Sequence-based reagent | SacI-FRP1 5’ (–795) | This work | PCR primer 1503 | GCGAGCTCCCAGCAGCACTTCCTG |
| Sequence-based reagent | FRP1 5’ (–1) – SpeI | This work | PCR primer 1504 | ggactaGTTGAAAGTTAAACTTGGTTA |
| Sequence-based reagent | HindIII-FRP1 3’ (+1662) | This work | PCR primer 1505 | GGGAAGCTTAGGGTATATAGGATAAAT |
| Sequence-based reagent | FRP1 3’ (+2258) - KpnI | This work | PCR primer 1506 | gcggtACCCAAATGCATGGGTAAAC |
| Sequence-based reagent | FRP1 test (–810) | This work | PCR primer 1507 | CACTTGCACTACCAGTTTCG |
| Sequence-based reagent | SacI-FRP2 5’ (–730) | This work | PCR primer 1508 | GGGAGCTCGGAAAATAAGTTGTTCTTTG |
| Sequence-based reagent | FRP2 5’ (+3) – SpeI | This work | PCR primer 1509 | CGACTAGTCCATGGCTGATAAGTTG |
| Sequence-based reagent | HindIII-FRP2 3’ (+1753) | This work | PCR primer 1510 | GGGAAGCTTCTATAACGAGTCGTACGA |
| Sequence-based reagent | FRP2 3’ (+2394) – KpnI | This work | PCR primer 1511 | CCGGTACCTGATCCTTGGATGCCA |
| Sequence-based reagent | FRP2 test (–750) | This work | PCR primer 1512 | GTAACAAACCCGAGAACACC |
| Sequence-based reagent | RI-FRP1 | This work | PCR primer 1513 | CCGAATTCAACCATGGCTATTCCAT |
| Sequence-based reagent | FRP1(+1740)-XhoI | This work | PCR primer 1514 | cgctcgaGGTGTGTCCTTACGTACAG |
| Sequence-based reagent | RI-FRP2 | This work | PCR primer 1515 | gcGAATTCCATGGACGAAGAACTTCAG |
| Sequence-based reagent | FRP2 (+1872)-XhoI | This work | PCR primer 1516 | ggctcGAGAGTGCTGTGAGGTTATG |
| Sequence-based reagent | BamHI-FRP1(+3) | This work | PCR primer 1522 | gcggatccgctattccatttgatcaacag |
| Sequence-based reagent | FRP1(+1662)-XhoI | This work | PCR primer 1523 | cgctcgagaaacgactctgtataacaatac |
| Sequence-based reagent | SpeI-FRP1 5' (–395) | This work | PCR primer 1541 | ccACTAGTCGTAATCAGCAGCAGATAC |
| Sequence-based reagent | FRP1 3' (+1877) - KpnI | This work | PCR primer 1542 | ccGGTACCGCACAAGCGGGTACT |
| Sequence-based reagent | FRP1 (+1550)-XhoI | This work | PCR primer 1543 | GGCTCGAGTTTGTGAATGATGGCGA |
| Sequence-based reagent | PGA7 (+1087)-HindIII | This work | PCR primer 1544 | GCAAGCTTGGCATACTCAATTTGATG |
| Sequence-based reagent | 5’-F1HEM1 | This work | PCR primer | CCCTCAATAATCATAACAGTACTTAGGTTTTTTTTTCAGTCGGATCCCCGGGTTAATTAA |
| Sequence-based reagent | 3’-R1HEM1 | This work | PCR primer | CCTTGTACCTCTATCTCAGCCCATGCATATATTGGTTGTTGAATTCGAGCTCGTTTAAAC |
| Sequence-based reagent | Promoter 5’ (HTA2) | This work | PCR primer | TATATATTAAATTTGCTCTTGTTC |
| Sequence-based reagent | Promoter 3’ (HTB2) | This work | PCR primer | TAGTTGTAGAGTAAGTTGTTG |
| Sequence-based reagent | SacI-HTB2p –684 | This work | PCR primer | gcgaGCTCTTGTTCTGTACTTTCC |
| Sequence-based reagent | PGA7 5’ | This work | PCR primer | GAACAAGAGCAAATTTAATATATAATGCATTTCATATTCTACTTGA |
| Sequence-based reagent | Pga7-SacI (+697) | This work | PCR primer | Pga7-SacI (+697) |
| Sequence-based reagent | FRP1 5’ | This work | PCR primer | CAACAACTTACTCTACAACTAATGGCTATTCCATTTGATCAA |
| Sequence-based reagent | FRP1-SalI | This work | PCR primer | Ccccgtcgacggtatcga |
| Chemical compound, drug | Hemin | Frontier Scientific | H651-9 | Hemin chloride |
| Chemical compound, drug | Bovine hemoglobin | Sigma-Aldrich | H2500 | |
| Chemical compound, drug | Ferrozine | Sigma-Aldrich | P9762 | 3-(2-Pyridyl)–5,6-diphenyl-1,2,4-triazine-4’,4”-disulfonic acid sodium salt |
| Chemical compound, drug | BPS | Sigma-Aldrich | B1375 | Bathophenanthroline sulfonate |
| Chemical compound, drug | ALA | Merck | 08339 | δ-Aminolevulinic acid |
| Chemical compound, drug | GaPPIX | Frontier Scientific | P40167 | Ga3+-protoporphyrin IX chloride |
| Chemical compound, drug | CoPPIX | Frontier Scientific | Co654-9 | Co3+-protoporphyrin IX chloride |
| Chemical compound, drug | MnPPIX | Frontier Scientific | MnP562-9 | Mn3+-protoporphyrin IX chloride |
| Chemical compound, drug | ZnPPIX | Frontier Scientific | Zn625-9 | Zn2+-protoporphyrin IX |
| Chemical compound, drug | ZnMP | Chem-Cruz | Sc-396862 | Zn2+-mesoporphyrin |
| Other | Alignment summary of 40 saccharomycetales species for phylogenetic profiling | This work | github.com/BKU-Technion/FRP |
List of Candida albicans strains.
| Name | Genotype | Origin |
|---|---|---|
| KC2=CAF3-1 | ura3Δ::imm434/ura3Δ::imm434 | Fonzi and Irwin, 1993 |
| KC68 | KC2 ccc2Δ::hisG/ccc2Δ::hisG | Weissman et al., 2002 |
| KC139 | KC68 rbt5Δ/rbt5Δ | Weissman and Kornitzer, 2004 |
| KC485 | KC68 pga7Δ/pga7Δ | Kuznets et al., 2014 |
| KC590=CAI4 | ura3Δ::imm434/ura3Δ::imm434 | Fonzi and Irwin, 1993 |
| KC646 | KC590 pga7Δ/pga7Δ ADE2/ade2::URA3 | Kuznets et al., 2014 |
| KC647 | KC590 pga7Δ/pga7Δ ADE2/ade2::PGA7 URA3 | Kuznets et al., 2014 |
| KC811 | KC68 ADE2/ade2::URA3 | This work |
| KC859 | KC2 FRP1/frp1Δ | This work |
| KC870 | KC2 frp1Δ/frp1Δ | This work |
| KC901 | KC2 FRP2/frp2Δ | This work |
| KC912 | KC2 frp2Δ/frp2Δ | This work |
| KC913 | KC68 frp2Δ/frp2Δ | This work |
| KC914 | KC2 FRP1/FRP1-GFP URA3 | This work |
| KC916 | KC2 frp1Δ/FRP1-GFP URA3 | This work |
| KC923 | KC68 frp1Δ/frp1Δ | This work |
| KC966 | KC590 frp1Δ/frp1Δ | This work |
| KC1023 | KC870 ADE2/ade2::URA3 | This work |
| KC1024 | KC870 ADE2/ade2::FRP1 URA3 | This work |
| KC1053 | KC590 frp2Δ/frp2Δ | This work |
| KC1061 | KC590 frp1Δ/frp1Δ frp2Δ/frp2Δ | This work |
| KC1064 | KC870 ADE2/ade2::FRP1p-FRP2 URA3 | This work |
| KC1080 | KC870 ADE2/ade2::SSB1p-FRP1 URA3 | This work |
| KC1146 | KC923 ADE2/ade2::URA3 | This work |
| KC1244 | KC912 ADE2/ade2::SSB1p-FRP2 URA3 | This work |
| KC1245 | KC912 frp2Δ/FRP2-GFP URA3 | This work |
| KC1246 | KC912 ADE2/ade2::URA3 | This work |
| KC1379 | KC912 ADE2/ade2::FRP2 URA3 | This work |
| KC1405 | KC2 FRP2/FRP2-GFP URA3 | This work |
| KC1410 | KC2 frp1Δ/frp1Δ frp2Δ/frp2Δ | This work |
| KC1411 | KC913 ADE2/ade2::FRP2 URA3 | This work |
| KC1412 | KC923 frp2Δ/frp2Δ::hisG-URA3-hisG | This work |
| KC1414 | KC913 ADE2/ade2::URA3 | This work |
| KC1447 | KC912 ADE2/ade2:: FRP1p-FRP2 URA3 | This work |
Additional files
-
Supplementary file 1
List of Alphafold 2-predicted Candida albicans protein structures most similar to the Csa2 CFEM hemophore domain structure.
- https://cdn.elifesciences.org/articles/80604/elife-80604-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/80604/elife-80604-mdarchecklist1-v2.docx