Promoter sequence and architecture determine expression variability and confer robustness to genetic variants
Figures
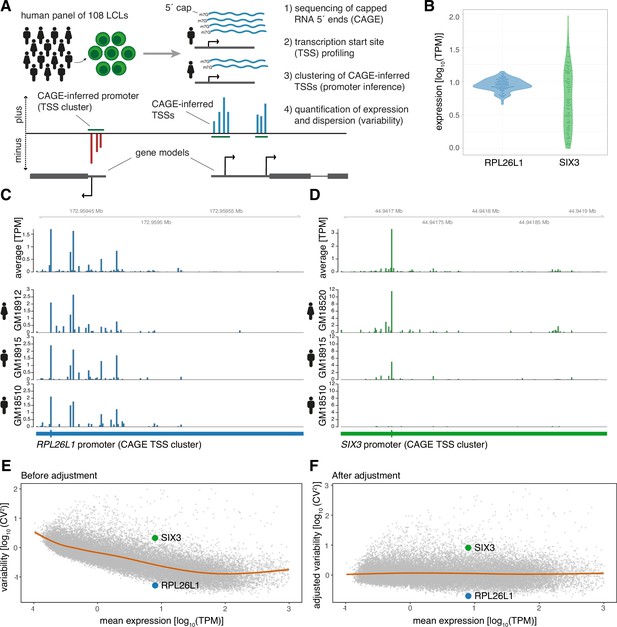
CAGE profiling of TSSs reveals diverse promoter variability across individuals.
(A) Illustration of the experimental design and approach for measuring promoter activity and variability. Capped 5’ ends of RNAs from LCLs derived from 108 individuals were sequenced with CAGE, followed by individual-agnostic positional clustering of proximal CAGE-inferred TSSs (first 5’ end bp of CAGE reads). The expression level of the resulting CAGE-inferred promoters proximal to annotated gene TSSs were quantified in each individual and used to measure promoter variability. (B) Example of promoter activity (TPM normalized count of CAGE reads) across individuals for a low variable promoter (gene RPL26L1) and a highly variable promoter (gene SIX3) with similar average expression across the panel. (C–D) Genome tracks for two promoters showing average TPM-normalized CAGE data (expression of CAGE-inferred TSSs) across individuals (top track) and TPM-normalized CAGE data for three individuals (bottom tracks) for a low variable promoter (panel C, gene RPL26L1) and a highly variable promoter (panel D, gene SIX3). (E–F) The CV2 (squared coefficient of variation) and mean expression relationship of 29,001 CAGE-inferred promoters across 108 individuals before (E) and after (F) adjustment of the mean expression-dispersion relationship. The CV2 and mean expression are log10 transformed, orange lines show loess regression lines fitting the dispersion to the mean expression level, and example gene promoters from B-D are highlighted in colors.
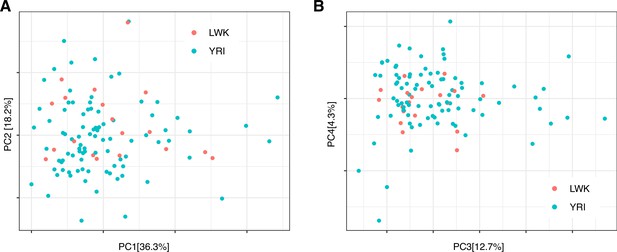
PCA plot of promoter expression (CAGE) across the LCL panel.
(A-B) 1st and 2nd (A), and 3rd and 4th (B) principal components (PCs), colored according to population (YRI and LWK). PCA was performed using TPM-normalized expression for all 29,001 considered promoters. Percentage of variation accounted for by each principal component is shown in brackets with the axis label.
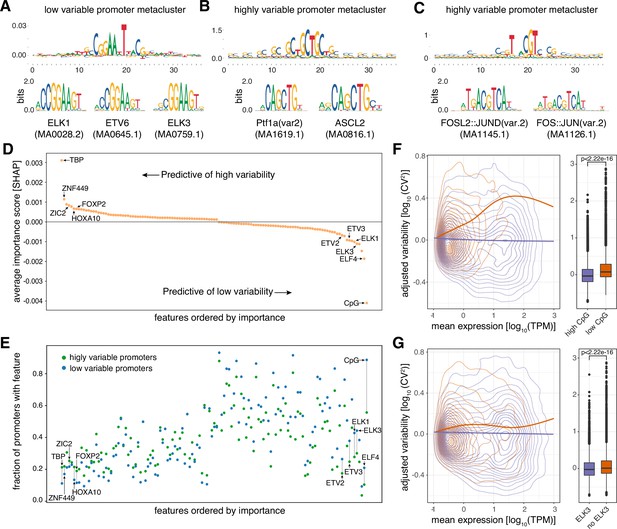
Promoter sequence features are highly predictive of promoter variability.
(A) Sequence logo of a metacluster (top) identified for low variable promoter sequences that matches known TF motifs (bottom) for ETS factors ELK1, ETV6, and ELK3. (B–C) Sequence logos of two metaclusters (top) identified for highly variable promoter sequences that match known TF motifs (bottom) for PTF1A and ASCL2 (B) and FOSL2-JUND and FOS-JUN heterodimers (C). (D) Average contribution (SHAP values) of CpG content and each of the 124 TFs identified as important for predicting promoter variability. Features are ordered by their average contribution to the prediction of highly variable promoters and selected TFs are highlighted. For a full version of the plot see Figure 2—figure supplement 3A. (E) The frequency of predicted TF binding sites (presence/absence) in highly variable (green) and low variable (blue) promoters. TFs are ordered as in D. For a full version of the plot see Figure 2—figure supplement 3B and C. (F–G) Promoters split into groups based on the presence/absence of high CpG content (F), and predicted binding sites of ELK3 (G). For both features displayed in panels F and G, the left subpanels display the relationship between log10-transformed mean expression levels and adjusted log10-transformed CV2 with loess regression lines shown separately for each promoter group. The right subpanels display box-and-whisker plots of the differences in adjusted log10-transformed CV2 between the two promoter groups (central band: median; boundaries: first and third quartiles; whiskers:+/-1.5 IQR). p-values were determined using the Wilcoxon rank-sum test.
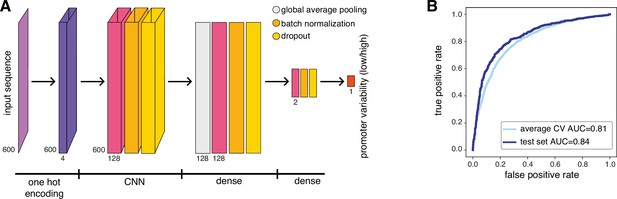
Neural network model and performance.
(A) Neural network architecture used for learning promoter variability from promoter sequence. The architecture is composed of one convolutional layer with 128 hidden units, followed by global average pooling and two dense layers with 128 and 2 nodes, respectively. (B) Receiver-operating curves (ROC) for average cross validation (light blue, AUC = 0.81) and the test set (dark blue, AUC = 0.84).
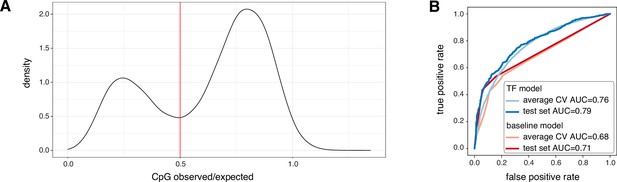
Random forest features and performance.
(A) Observed / expected CpG ratio calculated in windows covering +/-500 bp around the CAGE summit position of considered promoters. Red vertical line marks the threshold (0.5) between low and high CpG content. (B) Random forest model (TF model) receiver-operating curves (ROC) for average cross validation (light blue, AUC = 0.76) and the test set (dark blue, AUC = 0.79). Shown are also the ROC for a decision tree (baseline) model based on CpG content and TBP binding sites alone (average cross validation AUC = 0.68, orange; test set AUC = 0.71, red).
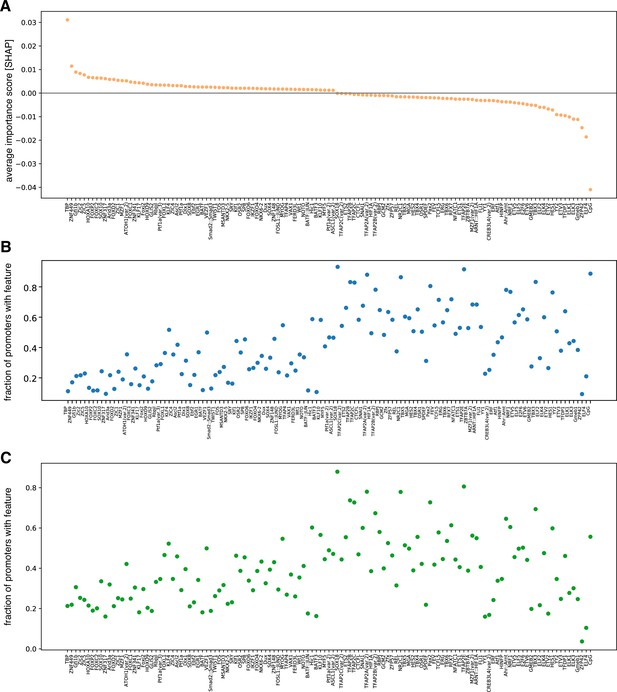
Full panel of features found to be predictive of promoter variability.
(A) Average contribution (SHAP values) of CpG content and each of the 124 TFs identified as important for predicting promoter variability. TFs are ordered by their average contribution to the prediction of highly variable promoters. (B–C) The frequency of predicted TF binding sites (presence/absence) in low variable (B) and highly variable (C) promoters. TFs follow the same order as in panel A.
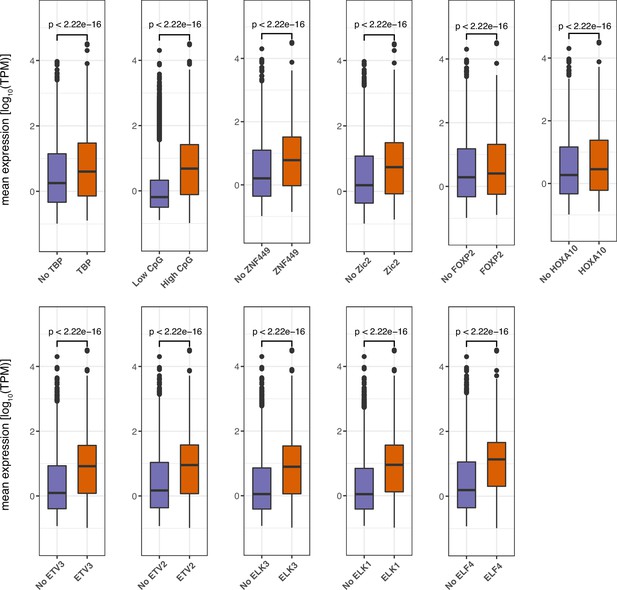
Association between TF binding sites and promoter expression level.
Box-and-whisker plots displaying the difference in TPM normalized promoter expression between in the absence (blue) or presence (orange) of TF binding sites. For all box-and-whisker plots, central band: median; boundaries: first and third quartiles; whiskers:+/-1.5 IQR. p-values were determined using the Wilcoxon rank-sum test.
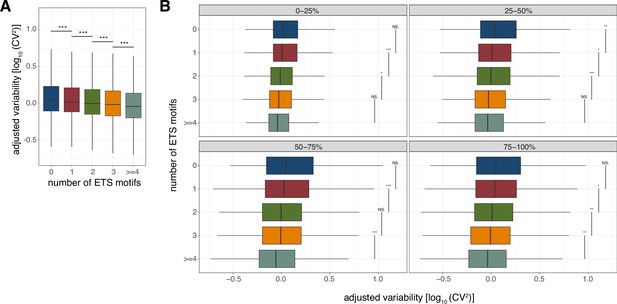
Association between the number of ETS binding sites and promoter variability.
(A) Variability of promoters grouped by their number of predicted non-overlapping ETS binding sites. (B) Variability of promoters grouped by their number of predicted non-overlapping ETS binding sites split based on promoter expression level quartiles. For all box-and-whisker plots, central band: median; boundaries: first and third quartiles; whiskers:+/-1.5 IQR. In both panels, outliers were not plotted. p-values were determined using the Wilcoxon rank-sum test (*: p<0.05; **: p<0.01; ***: p<0.001; NS.: non-significant).
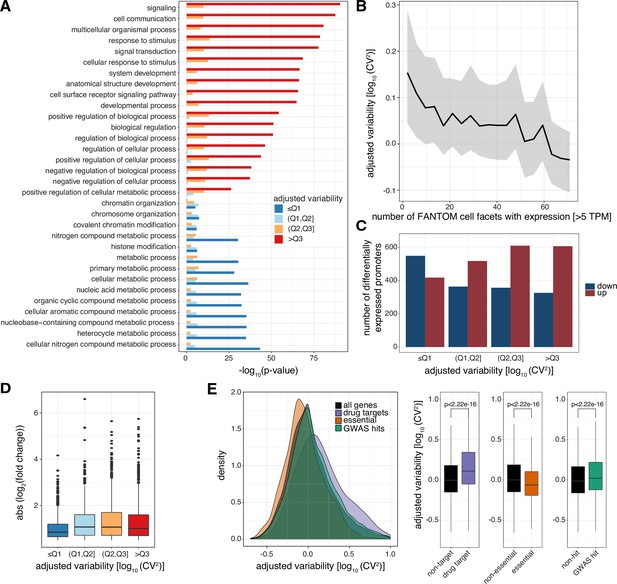
Levels of promoter variability are reflective of distinct biological processes and a selective trade-off between robustness and plasticity.
(A) GO term enrichment, for biological processes, of genes split by associated promoter variability quartiles (Q1, Q2, Q3). Top 10 GO terms of all groups are displayed and ranked based on p-values of the >Q3 variability group. (B) Median promoter variability (line) and interquartile range (shading), as a function of the number of FANTOM cell facets (grouping of FANTOM CAGE libraries associated with the same Cell Ontology term) that the associated gene is expressed in (mean facet expression >5 TPM). (C) The number of differentially expressed promoters, split by variability quartiles, after 6 h TNFα treatment. Promoters are separated into down-regulated (blue) and up-regulated (red). p-values were calculated using Fisher’s exact test. (D) Absolute log2 fold change of differentially expressed promoters, split by variability quartiles, after 6 h of TNFα treatment. (E) Distribution of promoter variability associated with drug-targets (purple), essential (orange), or GWAS hits (green) genes, compared to all promoters (black). Left: density plot of promoter variability per gene category. Right: Box-and-whisker plots of promoter variability split by each category of genes. p-values were determined using the Wilcoxon rank-sum test. For all box-and-whisker plots, central band: median; boundaries: first and third quartiles; whiskers:+/-1.5 IQR.
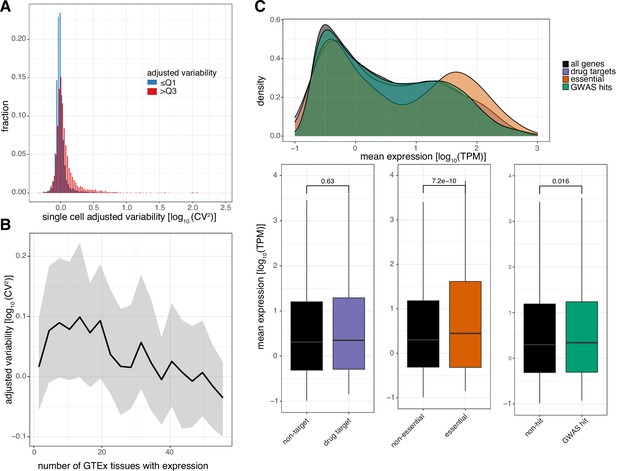
Levels of promoter variability are reflective of distinct biological processes.
(A) Distribution of single cell adjusted variability [log10(CV2)] of genes for low variable promoters (blue) and highly variable promoters (red). (B) Median promoter variability (line) and interquartile range (shading), as a function of the number of GTEx tissues the associated gene is expressed in (median tissue expression >5 RPKM). (C) Distribution of promoter expression associated with drug-targets (purple), essential (orange), or GWAS hits (green) genes, compared to all promoters (black). Top: density plot of promoter expression per gene category. Bottom: Box-and-whisker plots of promoter expression split by each category of genes. p-values were determined using the Wilcoxon rank-sum test. For all box-and-whisker plots, central band: median; boundaries: first and third quartiles; whiskers:+/-1.5 IQR.
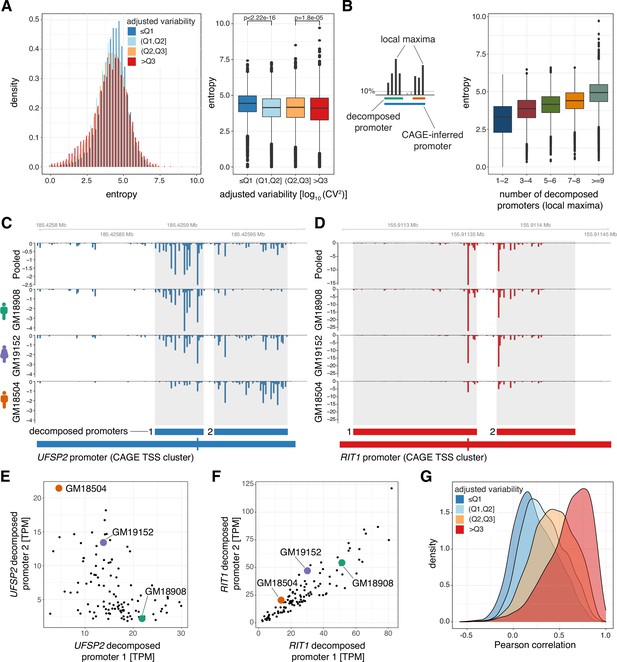
Low variable promoters exhibit flexibility in transcription initiation architecture.
(A) Promoter shape entropy for promoters split by variability quartiles, displayed as densities (left) and in a box-and-whisker plot (right). (B) Illustration of the local maxima decomposition approach (left; see Materials and methods) and box-and-whisker plot displaying the relationship between the Shannon entropy and the number of local maxima-inferred decomposed promoters. (C–D) Examples of two promoters each containing two decomposed promoters exhibiting low correlation across individuals (panel C, gene UFSP2) and high correlation across individuals (panel D, gene RIT1). Both panels display genome tracks of average, TPM-normalized CAGE-inferred TSS expression levels across the panel (Pooled, top track) and for three individuals (GM18908, GM19152, GM18504, lower tracks). Below the genome tracks, the original promoter and resulting decomposed promoters (shaded in genome tracks) are shown. (E–F) Relationship between TPM-normalized CAGE expression of decomposed promoter 1 (x-axis) and 2 (y-axis) across all 108 LCLs for example genes UFSP2 (E) and RIT1 (F). The expression values for individuals included in panels B and C are highlighted. (G) Densities of the lowest Pearson correlation between all pairs of decomposed promoters originating from the same promoter across all CAGE-inferred promoters with at least two decomposed promoters. For all box-and-whisker plots, central band: median; boundaries: first and third quartiles; whiskers:+/-1.5 IQR.
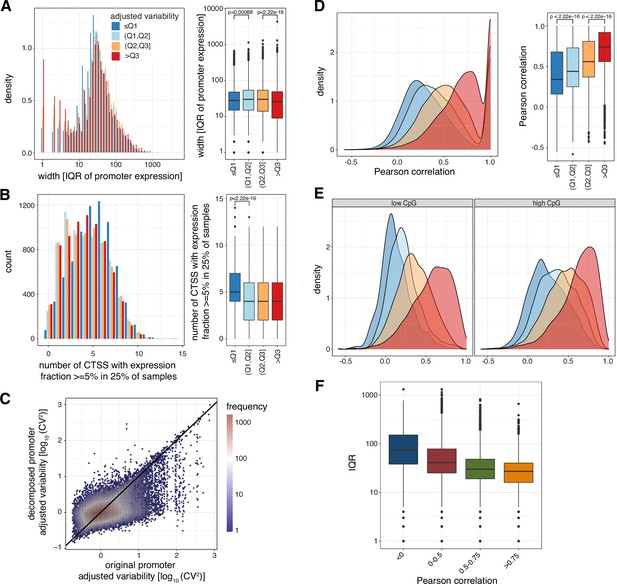
Low variable promoters tend to be composed of multiple clusters of TSSs rather than broader TSS signatures.
(A-B) Promoter shape metrics for promoters split by variability quartiles. The left subpanels display the distribution of IQRs (widths containing the 25th to 75th percentiles of contained CAGE signal) (A) and the number of TSSs with expression fraction ≥ 5% in 25% of samples (B). The right subpanels display box-and-whisker plots of the differences in these metrics across promoters split by variability quartiles (central band: median; boundaries: first and third quartiles; whiskers:+/-1.5 IQR.). (C) The relationship between adjusted log10-transformed CV2 of the original promoter and adjusted log10-transformed CV2 of local-maxima decomposed promoters as a 2D density chart. (D) Densities (left) and box-and-whisker plots (right) of the Pearson correlation between all possible pairs of decomposed promoters originating from the same promoter across all CAGE-inferred promoters with at least two decomposed promoters (central band: median; boundaries: first and third quartiles; whiskers:+/-1.5 IQR.). (E) Densities of the lowest Pearson correlation between any pair of decomposed promoters originating from the same promoter across all CAGE-inferred promoters with at least two decomposed promoters, split based on CpG content levels. (F) Box-and-whisker plots of the IQRs (widths containing the 25th to 75th percentiles of contained CAGE signal) split based on levels of lowest Pearson correlation between any pair of decomposed promoters originating from the same promoter (central band: median; boundaries: first and third quartiles; whiskers:+/-1.5 IQR).
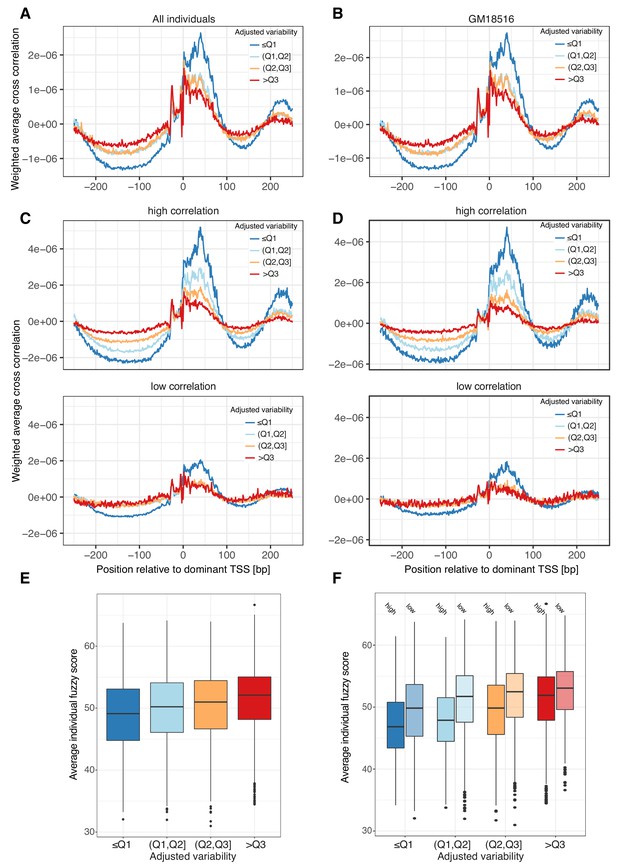
Low variable promoters with highly correlated decomposed promoters tend to have a fixed +1 nucleosome position across individuals.
(A-D) Weighted average cross correlation of MNase-seq read 5’ ends versus CAGE-inferred TSS expression profiles around promoter CAGE summit positions, split by variability quartiles, for pooled CAGE data (across all 108 LCLs) versus pooled MNase-seq data (across 7 LCLs, panels: A, C) or using only CAGE and MNase-seq data from one LCL (GM18516, panels: B, D). Weighted average cross correlations were calculated either across all promoters (A-B) or separately for promoter groups split based on their minimum correlation between contained decomposed promoter pairs (high Pearson’s correlation >0.39: top panel, low Pearson’s correlation <= 0.39: bottom panel) (C-D). (E-F) Average nucleosome fuzziness score across all 7 LCLs, split by variability quartiles (E) and minimum correlation between contained decomposed promoter pairs (high Pearson’s correlation >0.39: solid colors, low Pearson’s correlation <= 0.39: faded colors) (F).
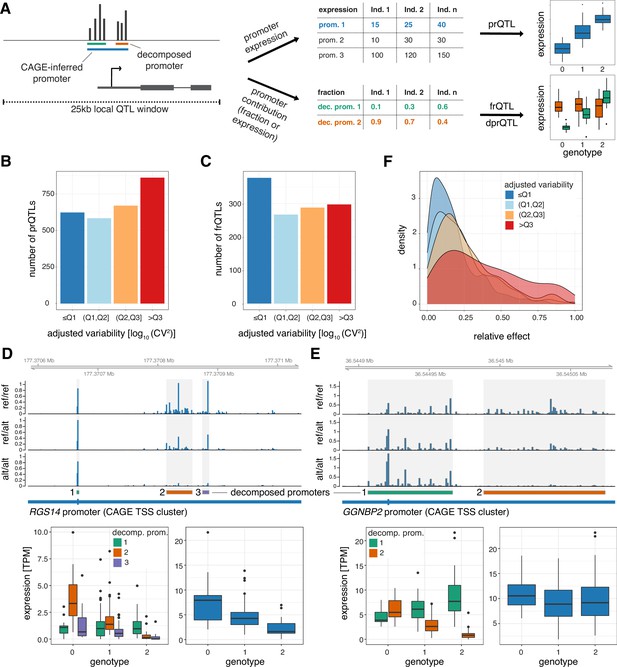
Plasticity in TSS usage is linked with increased mutational robustness.
(A) Illustration of the strategy for testing the effects of genetic variants on promoter expression (prQTLs, TPM-normalized CAGE counts), decomposed promoter expression (dprQTLs, TPM-normalized CAGE counts), and decomposed promoter contribution to the encompassing promoter expression (frQTLs, ratios of TPM-normalized CAGE counts between decomposed and encompassing promoters). For both approaches only SNPs within 25 kb of the promoter CAGE signal summit were tested. (B) Number of prQTLs detected (FDR < 0.05), split by promoter variability quartiles. (C) Number of encompassing promoters with at least one frQTL detected for a contained decomposed promoter (FDR < 0.05), split by encompassing promoter variability quartiles. (D–E) Examples of two promoters associated with frQTLs for a highly variable promoter with limited buffering of promoter expression (panel D, gene RGS14) and for a low variable promoter with strong buffering of promoter expression (panel E, gene GGNBP2). Upper panels display genome tracks showing average TPM-normalized CAGE data across homozygous individuals for the reference allele (top track), heterozygous individuals (middle track), and homozygous individuals for the variant (alternative) allele (bottom track). The bottom left subpanels display box-and-whisker plots of the differences in TPM-normalized CAGE data between genotypes for each decomposed promoter. The bottom right subpanels display box-and-whisker plots of the differences in TPM-normalized CAGE data between the three genotypes for the original encompassing promoter. For all box-and-whisker plots, central band: median; boundaries: first and third quartiles; whiskers:+/-1.5 IQR. (F) Density plot of the maximal relative change in expression between reference and variant alleles (relative effect size) for the most significant frQTL of each broad promoter with FDR ≥ 5%, split by variability quartiles.
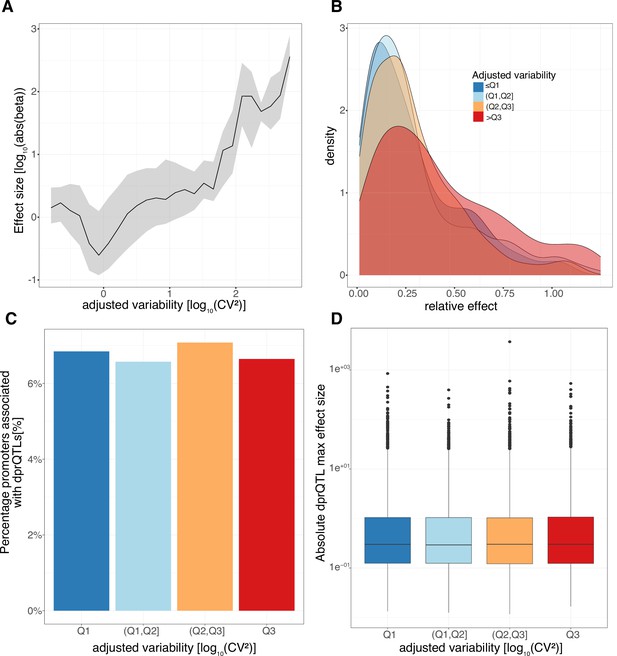
Low variable promoters with highly correlated decomposed promoters tend to have a fixed +1 nucleosome position across individuals.
(A-D) Weighted average cross correlation of MNase-seq nucleosome occupancy profiles relative to promoter CAGE summit positions, split by variability quartiles, for pooled CAGE data (across all 108 LCLs) versus pooled MNase-seq data across 7 LCLs, panels: (A), (C) or using only CAGE and MNase data from one LCL (GM18516), panels: (B), (D). Weighted average cross correlations were calculated either across all promoters (A,B) or separately for promoter groups split based on their minimum correlation between contained decomposed promoter pairs (high Pearson’s correlation >0.39: top panel, low Pearson’s correlation ≤ 0.39: bottom panel) (C,D). (E–F) Average nucleosome fuzzy score across all 7 LCLs, split by variability quartiles (E) and minimum correlation between contained decomposed promoter pairs (high Pearson’s correlation >0.39: solid colors, low Pearson’s correlation ≤ 0.39: faded colors) (F).
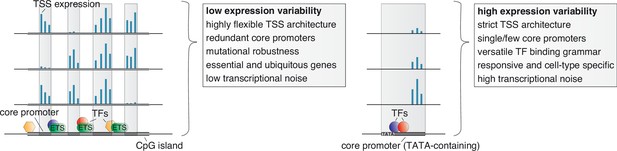
Unifying mechanisms influencing the variability in expression across individuals, the specificity in expression across cell types, and the stochasticity in expression across individual cells.
Low variable promoters (left) are frequently associated with high CpG content (CpG islands), multiple binding sites of ETS factors, and a highly flexible transcription initiation architecture arising from multiple redundant core promoters (decomposed promoters) in a permissive nucleosome positioning environment. These stabilizing features along with a less complex TF binding grammar likely also act to buffer transcriptional noise across single cells and cause ubiquitous expression across cell types. The flexibility in redundant core promoter activities confers a novel layer of mutational robustness to genes. Highly variable promoters (right), on the other hand, are associated with a highly versatile TF regulatory grammar, TATA boxes, and low flexibility in TSS usage. These features likely cause, in addition to high expression variability between individuals, a responsiveness to external stimuli, cell-type restricted activity, high transcriptional noise across single cells, and less tolerance for genetic variants.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18505 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18507 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19238 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19239 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18879 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18501 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18876 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18877 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18878 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19206 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19043 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18487 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18486 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19209 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19153 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18881 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18517 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19144 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19210 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18508 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19099 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18489 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19223 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18853 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18916 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19147 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19257 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19131 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19119 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19201 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19204 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19092 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19130 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19137 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19102 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19159 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18871 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19200 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19171 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19207 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18516 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18499 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19143 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19093 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19172 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19098 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18520 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19152 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19116 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19138 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18504 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19036 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18870 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19310 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18511 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19222 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19038 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19046 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19314 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19313 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19044 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19020 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18873 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18907 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18909 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18868 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18910 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18908 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19095 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19107 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18867 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19108 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19121 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19117 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19175 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19184 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19213 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18519 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18502 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19113 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19028 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19041 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19307 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19031 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18874 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19118 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19190 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19149 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19248 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18934 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19114 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19146 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18923 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18924 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18933 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18917 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19214 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19185 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19027 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19225 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19198 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19035 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19197 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19235 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18858 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19026 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18865 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19025 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18915 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19030 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19037 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19024 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19019 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18864 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18523 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19017 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18522 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18488 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19247 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18510 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18856 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18912 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM18861 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19141 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM19160 | |
| cell line (Homo-sapiens) | Lymphoblastoid cell line | Coriell | GM12878 | |
| sequence-based reagent | MPG beads, 10 ml | Pure biotech | MSTR0510 | |
| sequence-based reagent | AMPure, 60 ml | Ramcon | A63881 | |
| sequence-based reagent | RNAClean, 40 ml | Ramcon | A63987 | |
| sequence-based reagent | Phusion | Th.Geyer, Finnzymes | M0530L | |
| sequence-based reagent | PrimeScript | TaKaRa | 2680 A | |
| sequence-based reagent | RNAseONE | Th.Geyer | M4265 | |
| sequence-based reagent | ddH2O | VWR | ||
| commercial assay or kit | MinElute PCR purification kit (250 columns) | Qiagen | #28006 | |
| sequence-based reagent | LA Taq | TaKaRa | RR002A | |
| sequence-based reagent | DNA1000 kit | Agilent | 5067–1504 | |
| sequence-based reagent | RNA pico kit | Agilent | 5067–1513 | |
| sequence-based reagent | Biotin (long arm) hydrazide, 50 mg | VWR | Vectsp-1100 | |
| sequence-based reagent | E-gel sizeselect | Lifetech | G6610-02 | |
| commercial assay or kit | PureLink Dnase set | Lifetech | #12185010 | |
| sequence-based reagent | EcoP15I, 2500 U | Th.Geyer, NEB | N/R0646L | |
| sequence-based reagent | Sinefungin, 2 mg | Merck, Calbiochem-Novabiochem International | #567051 | |
| sequence-based reagent | Antarctic phosphatase, 5000 U | Bionordika, NEB | M0289L | |
| sequence-based reagent | Trehalose dihydrat | Sigma | Y0001172-1EA | |
| sequence-based reagent | d-Sorbitol | VWR | 85529–250 G | |
| sequence-based reagent | NaIO4, 5 g | Sigma | 311448–5 G | |
| sequence-based reagent | E. coli tRNA, 500 U | Sigma | R1753-500UN | ribonucleic acid, transfer from Escherichia coli Type XX, Strain W, lyophilized powder |
| sequence-based reagent | RQ1 RNase-free DNase | Promega | M6101 | |
| sequence-based reagent | Proteinase K | Lifetech | 25530–049 | |
| sequence-based reagent | ATP, 10 mM | Bionordika, NEB | P0756S | |
| sequence-based reagent | Trizol LS, 100 ml | Lifetech, Invitrogen | 10296–010 | |
| sequence-based reagent | DNA ligation kit, Mighty Mix | TaKaRa | 00006023 TAKARA | |
| sequence-based reagent | T4 DNA ligase, 20000 U | Th.Geyer, NEB | N/M0202L | |
| sequence-based reagent | Exonuclease I, 3.000 U | Th.Geyer | N/M0293S | |
| sequence-based reagent | dNTPs, 10 mM, 1 ml | Kælder | ||
| sequence-based reagent | Sodium acetate | Sigma | S2889-250G | |
| sequence-based reagent | Sodium citrate, 500 g | Sigma, MP Biomedicals | W302600-1KG-K | |
| sequence-based reagent | EDTA (4x100 ml, 0.5 M pH = 8.0) | Lifetech | 15575–020 | |
| sequence-based reagent | PCR SYBR mix 2*5 mL | Lifetech | 4364344 | |
| other | Penicillin-Streptomycin | ThermoFisher Scientific | 15140122 | Cell culture supplement |
| other | L-Glutamine | ThermoFisher Scientific | A2916801 | Cell culture supplement |
| other | RPMI 1640 | ThermoFisher Scientific | 11875083 | Cell culture media |
| other | Fetal bovine serum | ThermoFisher Scientific | A3840302 | Cell culture supplement |
| peptide, recombinant protein | TNFa | R&DSystems | P01375 | 25 ng/ml |
Additional files
-
Supplementary file 1
LCL sample and CAGE library information.
[tab-delimited]
Row names: Cell line IDs
Columns:
- https://cdn.elifesciences.org/articles/80943/elife-80943-supp1-v2.zip
-
Supplementary file 2
CAGE-inferred promoters associated with GENCODE-annotated TSSs.
[tab-delimited]
Row names: genomic coordinates of promoters provided as chromosome:start-end;strand
Columns:
- https://cdn.elifesciences.org/articles/80943/elife-80943-supp2-v2.zip
-
Supplementary file 3
Gene level expression characterization across GM12878 single cells.
[tab-delimited]
Row names: Ensembl IDs
Columns:
- https://cdn.elifesciences.org/articles/80943/elife-80943-supp3-v2.zip
-
Supplementary file 4
Promoter differential expression results using DESeq2 in GM12878 after TNFα treatment.
[tab-delimited]
Row names: genomic coordinates of promoters provided as chromosome:start-end;strand
Columns:
- https://cdn.elifesciences.org/articles/80943/elife-80943-supp4-v2.zip
-
Supplementary file 5
Decomposed promoter expression characterization and its association with original promoter expression variability.
[tab-delimited]
Row names: genomic coordinates of decomposed tag clusters provided as chromosome:start-end;strand
Columns:
- https://cdn.elifesciences.org/articles/80943/elife-80943-supp5-v2.zip
-
Supplementary file 6
Lead prQTL hit for each promoter.
[tab-delimited]
Row names: row number
Columns:
- https://cdn.elifesciences.org/articles/80943/elife-80943-supp6-v2.zip
-
Supplementary file 7
Lead frQTL hit for each promoter.
[tab-delimited]
Row names: row number
Columns:
- https://cdn.elifesciences.org/articles/80943/elife-80943-supp7-v2.zip
-
Supplementary file 8
Gene level expression characterization across LCLs using RNA-seq (Geuvadis).
[tab-delimited]
Row names: Ensembl ID of the associated gene
Columns:
- https://cdn.elifesciences.org/articles/80943/elife-80943-supp8-v2.zip
-
Supplementary file 9
Promoters associated with stabilizing frQTLs.
[tab-delimited]
Row names: row number
Columns:
- https://cdn.elifesciences.org/articles/80943/elife-80943-supp9-v2.zip
-
MDAR checklist
- https://cdn.elifesciences.org/articles/80943/elife-80943-mdarchecklist1-v2.docx





