Single-cell transcriptomics identifies Keap1-Nrf2 regulated collective invasion in a Drosophila tumor model
Figures
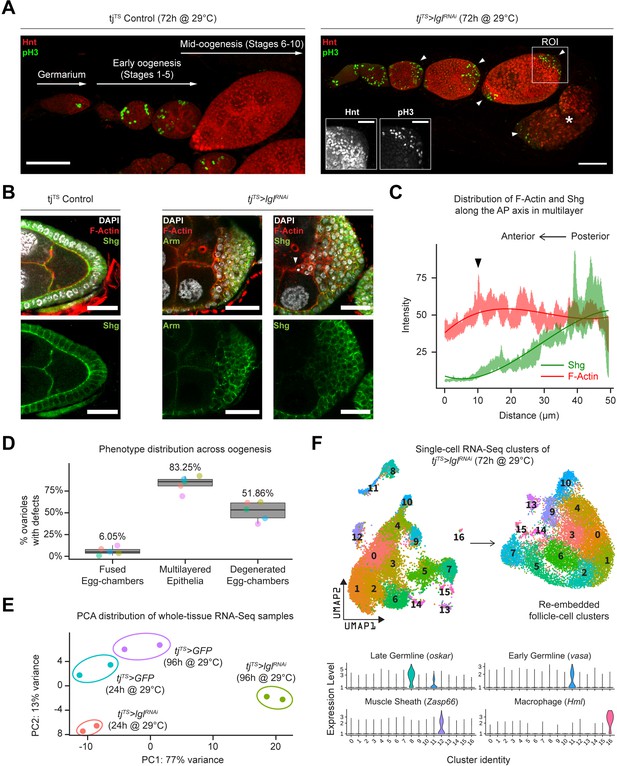
Inducing Lgl knockdown in follicle cells causes distinct phenotypic and transcriptomic changes.
(A) Left: orthogonal projection of a single ovariole displaying individual egg chambers containing experimental control follicle cells at early and midoogenesis. Follicle cells at mitotic stages are infrequently detected by pH3 staining (green), while endocycling follicle cells at midoogenesis are labeled by Hnt staining (red). Right: ovariole containing egg chambers with Lgl-KD in follicle cells exhibit continued cell division (marked by pH3 staining in green) in cells that accumulate at egg chamber termini (arrowheads) at early-to-midoogenesis developmental transition and midoogenesis. Degenerated egg chambers containing dying germline cells are marked by asterisks (*). Scale bars: 50 µm. Distinct Hnt and pH3 staining within the Lgl-KD multilayers are highlighted for the region of interest (ROI) within the image. ROI scale bar: 20 µm. (B) Left: cross-section of the posterior egg chamber epithelia containing experimental control follicle cells that exhibit intact Shg (DE-Cad) staining at cell junctions. Middle and right: posterior multilayers of egg chambers containing Lgl-KD in follicle cells show declining enrichment (green) of Armadillo (Arm; middle panel) and Shg (DE-Cad; right panel) along the anterior–posterior (AP) axis (right to left). F-actin (red) is found enriched in cells at the apical-most layers; leading edge of the invasive front is indicated by an arrowhead. Nucleus is marked by DAPI (white). Scale bars: 20 µm. (C) Relative enrichment of F-actin (red) and Shg (DE-Cad; green) along the AP axis across the biggest distance between the apical-most and basal-most cells in the multilayer shown in (B) (right panel). Intensities are measured across a 10 µm thickness (Z-axis) and a trendline (Gaussian fit) is shown. The black arrowhead marks the leading edge corresponding to that shown in (B). (D) Box-and-whisker plot showing quantification of the different tjTS>lglRNAi (72 hr in permissive temperature) phenotypes. Data was collected from five replicate trials (color-coded individually), consisting of 1250 intact ovarioles from a total of 165 flies. (E) Principal component analysis (PCA) plot showing the distribution of whole-tissue RNA-seq samples for tjTS experimental controls kept for 24 hr and 96 hr in permissive temperature and their experimental counterparts containing tjTS>lglRNAi. Each uniquely colored sample has two replicates that are grouped. (F) Overview of the single-cell (sc) RNA-seq workflow to isolate follicle cell-specific clusters from 14,537 tjTS>lglRNAi ovarian cells, embedded on lower UMAP dimensions (top). Clusters containing nonepithelial cell types are identified by the enrichment of specific markers (bottom).
-
Figure 1—source data 1
Raw data for (unnormalized) IntDen values of RFP, GFP, and DAPI enrichment.
Related to Figure 1C.
- https://cdn.elifesciences.org/articles/80956/elife-80956-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Quantification of the stage-specific phenotypic characterization of ovarioles containing egg chambers with 96 hr Lgl-KD in their follicle cells.
Related to Figure 1D.
- https://cdn.elifesciences.org/articles/80956/elife-80956-fig1-data2-v2.xlsx
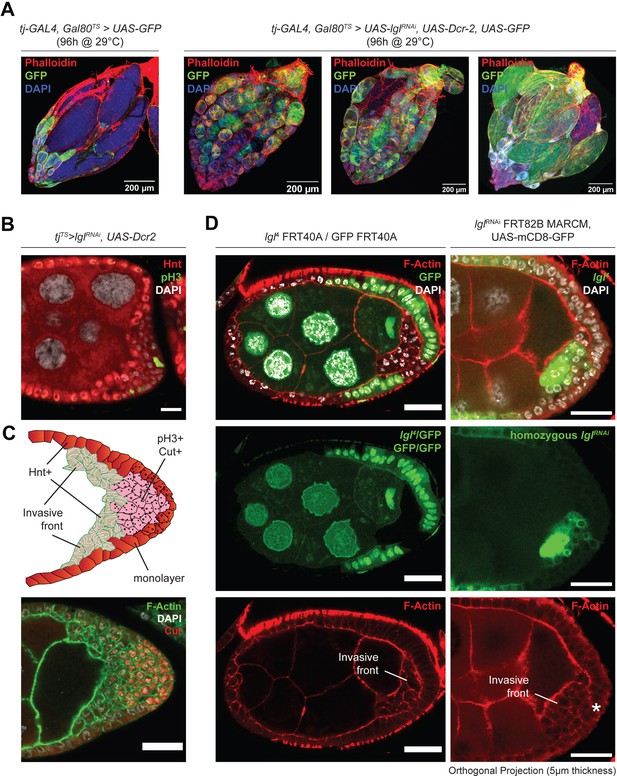
Lgl loss of function in follicle cells causes invasive multilayering and cell fate heterogeneity.
(A) Confocal images of tjTS-driven Lgl-KD ovaries (right) compared to that of a tjTS experimental control (left). Inducing Lgl-KD in follicle cells causes late-stage egg chambers to abort germline development, which leads to an accumulation of follicle cells near the oviduct (top-right part of the image). Some egg chambers also exhibit the accumulation of fully developed eggs posteriorly within the ovarian sheath (right-most column). F-actin is marked by Phalloidin (red), nucleus with DAPI (blue) and GFP (green) marks the follicle cells. Scale bars, 200 µm. (B) Confocal Image of an egg chamber cross-section shows the mutually exclusive enrichment of the endocycling follicle cell marker Hnt and that of the mitotic marker pH3. (C) Top panel: model depicting apically invasive multilayers exhibiting cell fate heterogeneity. Spatially restrictive marker expression and terminologies are indicated. Lower panel: cut expression is observed in the basally enriched cells. Nucleus is marked by DAPI (white) and F-actin is marked by Phalloidin (green). Scale bars: 20 µm. (D) Confocal images of egg chambers containing mitotic clones of homozygous (GFP-) lgl4 follicle cells (left column) and GFP+ MARCM clones of homozygous lglRNAi follicle cells (right column). Panel visualizing the MARCM clones was generated by orthogonally projecting Z-stacks across 5 µm thickness. Nucleus is marked by DAPI (white), and F-actin is marked by Phalloidin (red). Scale bars: 20 µm.
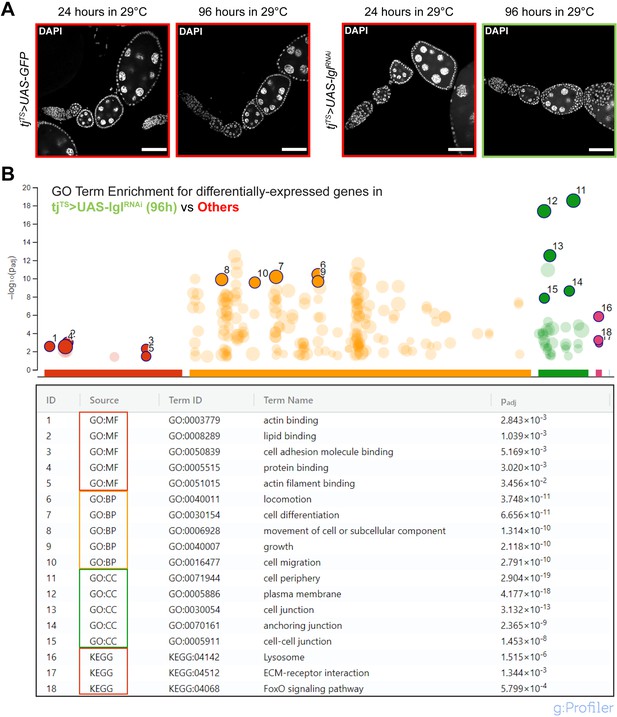
Whole-tissue RNA-seq of samples containing multilayered Lgl-KD follicle cells.
(A) Representative confocal images of representative ovarioles from the four samples used as input for RNA-seq analysis. Nucleus is marked by DAPI (white). Scale bars: 50 µm. (B) Enriched Gene Ontology (GO) Terms in the 477 differentially expressed genes found by comparing 96h-Lgl-KD samples (marked by green border in A) with the rest of the samples (marked by red borders in A). The highlighted GO Terms are mentioned in the table below with their corresponding adjusted p-values.
-
Figure 1—figure supplement 2—source data 1
Differentially expressed genes in 96h-Lgl-KD vs. others comparison.
Related to Figure 1—figure supplement 2B.
- https://cdn.elifesciences.org/articles/80956/elife-80956-fig1-figsupp2-data1-v2.xlsx
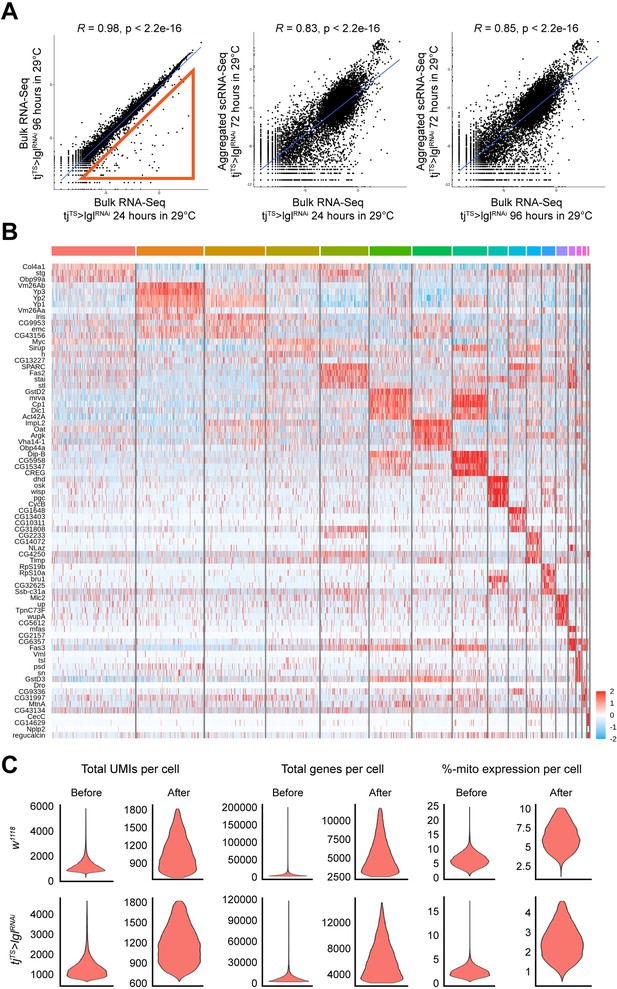
Single-cell RNA-seq of ovaries with 72h-Lgl-KD follicle cells.
(A) Scatter plots depicting the correlation between the different RNA-seq samples. Red triangle in the left plot represents markers from late development stages of oogenesis, representing the late-stage failure in 96h-Lgl-KD follicular development. Middle and right columns depict the similarities between the log-normalized counts of bulk RNA-seq datasets (X-axis) and aggregated single-cell RNA-seq dataset (Y-axis). (B) Heatmap showing the expression of cluster-specific markers in each cluster shown in the UMAP plot in Figure 1F for the 72h-tjTS>lglRNAi dataset before subsetting. (C) Violin plots showing the distribution of total UMI per cell (left column), total genes per cell (middle column), and percentage of cells expressing mitochondrial genes per cell (right column) before and after filtration. Top row represents isolated w1118 follicle cells while bottom row represents tjTS>lglRNAi follicle cells.
-
Figure 1—figure supplement 3—source data 1
Cluster-specific markers of the tjTS>lglRNAi (72 hr) single-cell RNA-seq (scRNA-seq) dataset.
Related to Figure 1—figure supplement 3B.
- https://cdn.elifesciences.org/articles/80956/elife-80956-fig1-figsupp3-data1-v2.xlsx
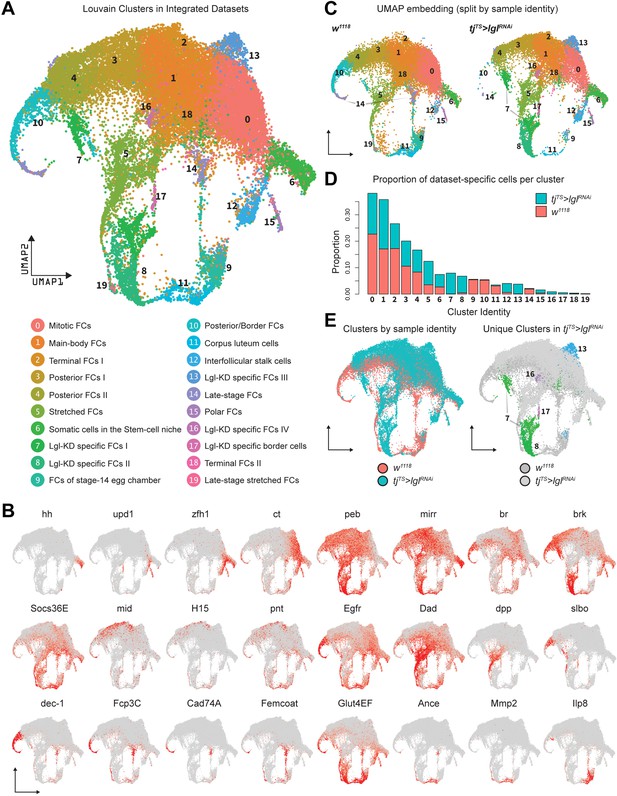
Integration of w1118 and tjTS>lglRNAi single-cell datasets identifies sample-specific clusters.
(A) UMAP plot of 17,874 w1118 follicle cells integrated with 12,923 tjTS>lglRNAi follicle cells, grouped into 20 clusters, with their approximated identities listed below. (B) Canonical marker expression used to annotate the 20 integrated clusters reveals cluster-specific identities of the somatic cells of the stem cell niche (hh), polar follicle cells (upd1), stalk cells (zfh1), immature mitotic follicle cells (ct), mature postmitotic cells (peb), main body follicle cells (mirr and br), terminal follicle cells (brk, Socs36E and Egfr), posterior follicle cells (mid, H15, and pnt), stretched cells (Dad and dpp), border cells (slbo), follicle cells of the vitellogenic (dec-1) and choriogenic stages (Fcp3C, Cad74A, Femcoat, and Glut4EF), and the cells of the terminally fated corpus luteum (Ance, Mmp2, and Ilp8). (C) Distribution of clusters on the UMAP embedding is shown split by sample ID. (D) Bar plot showing the relative proportion of cells in each cluster. Each bar is further divided by the dataset of origin (w1118 is represented by salmon color and tjTS>lglRNAi by teal). (E) UMAP plot showing the overlap between cells from each dataset (left) and clusters unique to the 72h-tjTS>lglRNAi dataset (right).
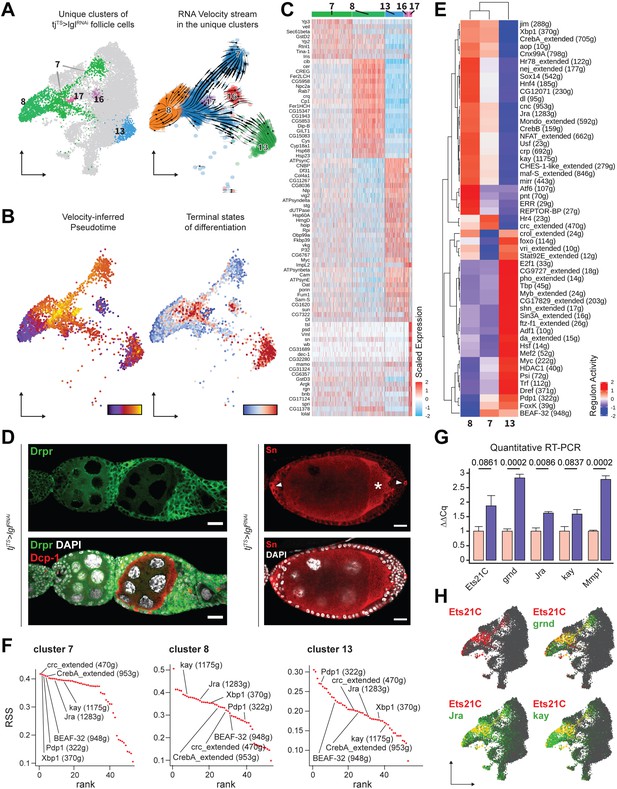
Unique clusters of tjTS>lglRNAi dataset exhibit distinct gene expression and regulon activity.
(A) Left: UMAP plot of the re-embedded tjTS>lglRNAi follicle cells with the unique clusters being highlighted. Right: RNA velocity vectors superimposed on the embedded cells of unique tjTS>lglRNAi clusters revealing the inferred lineage. (B) Left: cells are colored according to their arrangement on the velocity-inferred pseudotime from early (purple) to late (yellow). Right: cells are colored to denote terminal states of differentiation according to their position on the inferred lineage, where stable end points are colored in red and root cells are colored in blue. Observed noncompliance of inferred lineage and terminal states likely represents mixed population of cells. (C) Heatmap of the top 20 cluster-specific markers (if present) in the 72h-tjTS>lglRNAi dataset. Selected genes are expressed in a minimum of 75% cells per cluster. Range of gene expression is scaled within +2 (red) to –2 (blue) log2 fold change. (D) Confocal images showing Drpr (left) in green and Sn staining (right) in red. Sn+ polar cells are indicated by arrowheads, while oocyte is indicated by asterisks (*). Dcp-1 (red) is also shown in left panel to identify dying germline cells within degenerating egg chambers. Nucleus is marked by DAPI (white). Scale bars: 20 µm. (E) Heatmap of the scaled (and centered) activity scores of regulons in clusters 7, 8, and 13. Both columns (clusters) and rows (regulons) are clustered hierarchically and relative similarity between cluster 7 and cluster 8 regulon activity is inferred. (F) Regulon specificity score (RSS) rank plots show the specificity of regulon activity for clusters 7, 8, and 13. (G) Bar plot shows the relative mRNA levels of JNK signaling pathway components Ets21C, grnd, Jra, kay, and Mmp1 in tjTS control (N = 12 pairs of ovaries) and tjTS>lglRNAi sample (N = 15) using quantitative RT-PCR. Bars representing the tjTS experimental control are colored salmon while tjTS>lglRNAi is colored purple. Error bars represent Standard Error (SE) and the p-values obtained for individual t-test comparisons between samples are listed above each pairing. (H) Gene enrichment of Ets21C (red), Jra, kay, and grnd (green) is shown on the UMAP-embedded cells. Overlapping expression is colored in yellow.
-
Figure 3—source data 1
Regulon specificity scores (RSS) of regulons enriched in clusters 7, 8, and 13 in tjTS>lglRNAi (72 hr) single-cell RNA-seq (scRNA-seq) dataset.
Related to Figure 3F.
- https://cdn.elifesciences.org/articles/80956/elife-80956-fig3-data1-v2.xlsx
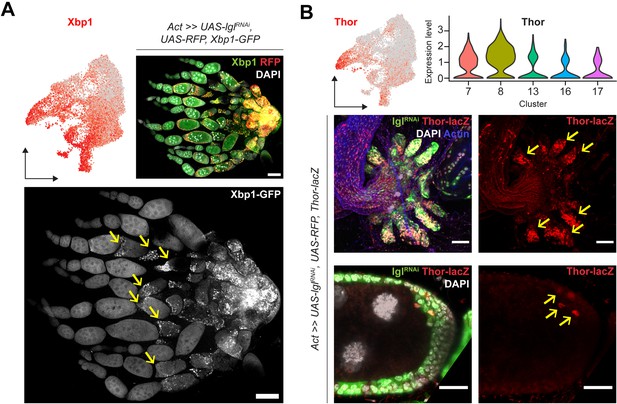
Validation of overlapping markers in Lgl-KD cells.
(A) Gene enrichment plot for Xbp1 expression (red) is shown on top left. Expression of UPR stress sensor Xbp1-GFP is observed in clusters representing early stages of development as well as clusters 7 and 8. Confocal image of the entire ovary shows the enrichment of Xbp1-GFP in the follicle cells accumulating near the oviduct at late stages (represented by cluster 8), as well as in the terminal regions of the egg chambers containing Lgl-KD follicle cells (marked by yellow arrows). Scale bars: 50 µm. (B) Gene enrichment plot for Thor expression (red) is shown on top left. Violin plot showing the relative expression (Y-axis) of Thor in the unique clusters (X-axis) of Lgl-KD cells identifies high expression of Thor in cluster 8 and mid-level expression in cluster 7. Confocal images of the RFP+ (green) Lgl-KD follicle cells during late- (top panels) and mid- (bottom panels) developmental stages. Thor expression (red) is observed in accumulating follicle cells, as well as cells within the multilayers (yellow arrows). Scale bars: 20 µm.
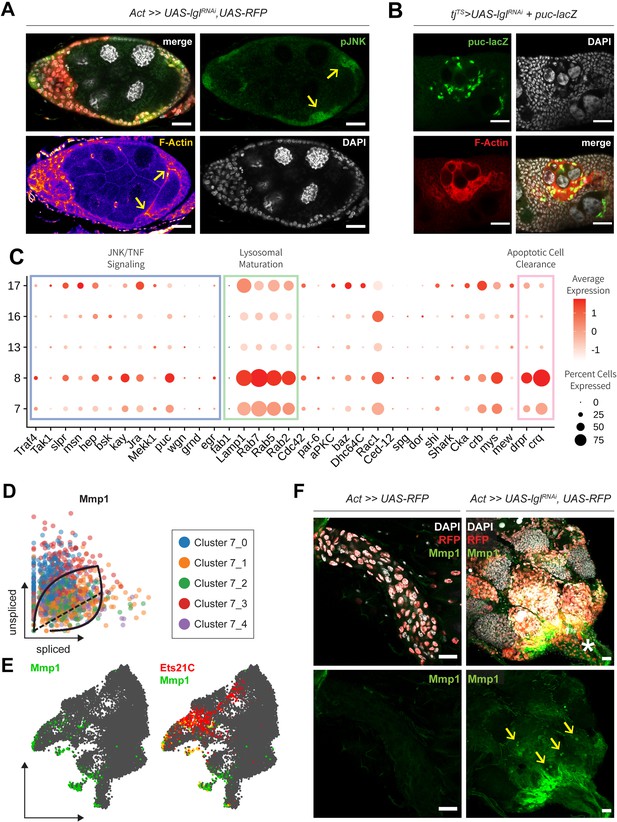
Expression of JNK signaling components in 72h-Lgl-KD cells.
(A) Confocal images showing antibody staining against pJNK (green) in multilayered egg chambers containing RFP+ (red) Lgl-KD follicle cells. Nucleus is marked by DAPI (white). F-actin is marked by Phalloidin staining intensity. Invasive fronts are indicated by yellow arrows. (B) Confocal images showing the expression of puc-lacZ (green), downstream reporter of JNK signaling activity. Puc expression is observed only in cells surrounding pre-apoptotic germline cells. (C) Dot plot showing the relative enrichment of markers related to canonical and noncanonical JNK signaling components in the unique clusters of Lgl-KD dataset. (D) Phase portraits of Mmp1 mRNA distribution in cluster 7 cells. (E) Gene enrichment plot showing the overlap of Mmp1 expression (green) in cluster 7 cells at terminal end points, which also exhibit Ets21C expression (red). Overlap is shown in yellow. (F) Confocal images showing Mmp1 staining in terminal follicle cells of experimental control egg chambers (in corpus luteum; left column) and those containing Lgl-KD (3 days) follicle cells (right column) that accumulate near the oviduct marked by an asterisk (*). Transgene expression is marked by RFP expression in red. Externally secreted Mmp1 proteins are highlighted by yellow arrows. Scale bars, 20 µm.
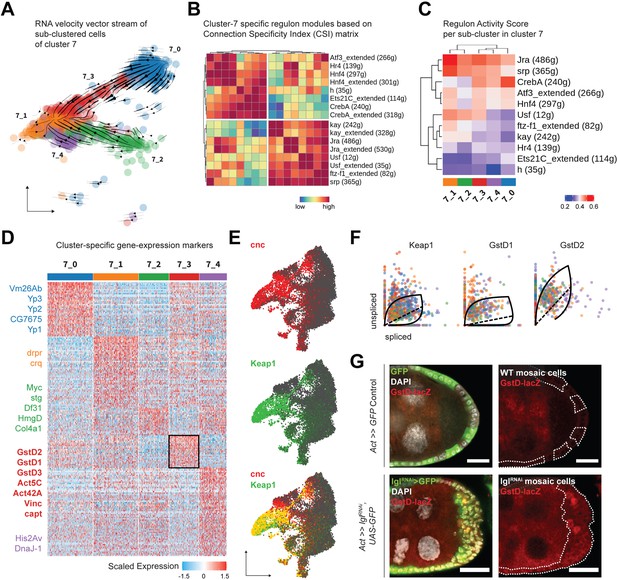
Cluster 7 cells exhibit heterogenous gene expression and regulon activity.
(A) UMAP plot of subdivided cluster 7 cells, superimposed with RNA velocity vectors. (B) Heatmap representing regulon–regulon correlation based on Connection Specificity Index (CSI) of active regulon modules. Both high-confidence and low-confidence (marked by the ‘_extended’ suffix) transcription factor (TF) associations are plotted. (C) Heatmap of the unscaled activity scores of regulons in the subclusters of cluster 7. (D) Heatmap of the (relatively) differentially expressed markers of each subcluster of cluster 7 cells, scaled within +1.5 (red) and –1.5 (blue) log2 fold change. Select markers are mentioned, while those of cluster 7_3 (denoted by black border on the heatmap) are highlighted in bold. (E) Gene enrichment plot for cnc (red), Keap1 (green), and their overlap (bottom). (F) Phase portraits showing dynamic behavior of genes in cluster 7 cells (colored by their subcluster ID as shown in panel A). Solid line represents the learned splicing dynamics while the dotted line represents the inferred gene expression steady state. Keap1, GstD1, and GstD2 exhibit an acute increase in transcription in cluster 7 cells. (G) Reporter expression of GstD-lacZ is detected by β-gal expression (red) within the multilayered cells of lglRNAi follicle cells (green; clonal boundaries are marked by the dotted white lines). Nuclei is marked by DAPI (white). Scale bars: 20 µm.
-
Figure 4—source data 1
Regulon specificity scores (RSS) of regulons enriched in the subclusters of cluster 7 in tjTS>lglRNAi (72 hr) single-cell RNA-seq (scRNA-seq) dataset.
Related to Figure 4C.
- https://cdn.elifesciences.org/articles/80956/elife-80956-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Differentially expressed markers of the subclusters of cluster 7 cells in tjTS>lglRNAi (72 hr) single-cell RNA-seq (scRNA-seq) dataset.
Related to Figure 4D.
- https://cdn.elifesciences.org/articles/80956/elife-80956-fig4-data2-v2.xlsx
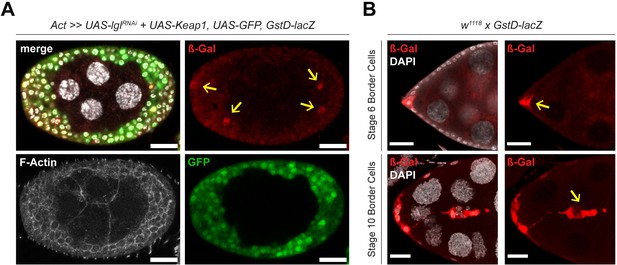
GstD-lacZ expression in Lgl-KD+Keap1-OE follicle cells and w1118 experimental control.
(A) Confocal image of a multilayered egg chamber containing Lgl-KD+Keap1-OE follicle cells (green). GstD-lacZ expression, marked by β-gal staining (red), is observed spuriously in the multilayered cells (yellow arrows) indicating Keap1-Nrf2 signaling activation. (B) GstD-lacZ expression, marked by β-gal staining, is specifically observed in the border cells (yellow arrows) of w1118 egg chambers. Nucleus is marked by DAPI (white). Scale bars, 20 µm.
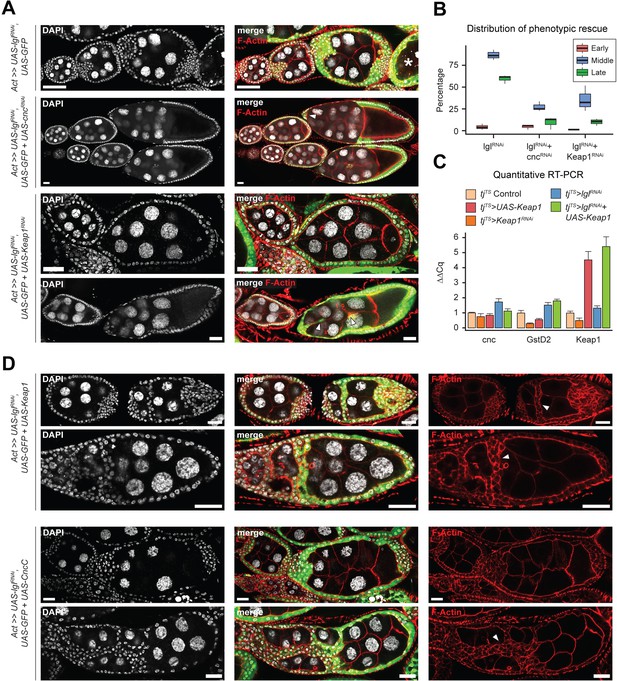
Keap1-Nrf2 signaling drives invasiveness of Lgl-KD multilayers.
(A) Representative confocal images of ovarioles containing egg chambers with transgene expression in follicle cells (green) driving Lgl-KD (top), Lgl-KD+Cnc-KD (middle), and Lgl-KD+Keap1-KD (bottom panels). Degenerating egg chambers are marked by asterisks (*). Border cell migration defects are indicated by arrowheads. Nucleus is marked by DAPI (white). F-actin is marked by Phalloidin (red). Scale bars: 20 µm. (B) Box-and-whisker plot showing the quantification of phenotypic rescue at early (red), mid (blue), and late (green) oogenesis. For the multilayering phenotype at midoogenesis, we have only counted instances of >2 follicular layers. (C) Bar plot showing relative expression levels of genes involved in the Keap1-Nrf2 signaling pathway in relevant genotypes (N = minimum 10 pairs of ovaries). Samples are color-coded as shown in legend and the error bars represent Standard Error (SE). (D) Confocal images of ovarioles with Lgl-KD+Keap1-OE (above) and Lgl-KD+CncC-OE (below) in follicle cells (green). Arrowheads mark the epithelial bridging or fusion phenotype. Nucleus is marked by DAPI (white). F-actin is marked by Phalloidin (red). Scale bars, 20 µm.
-
Figure 5—source data 1
Quantification of the rescue of stage-specific Lgl-KD phenotypes upon Cnc-KD and Keap1-KD.
Related to Figure 5B.
- https://cdn.elifesciences.org/articles/80956/elife-80956-fig5-data1-v2.xlsx
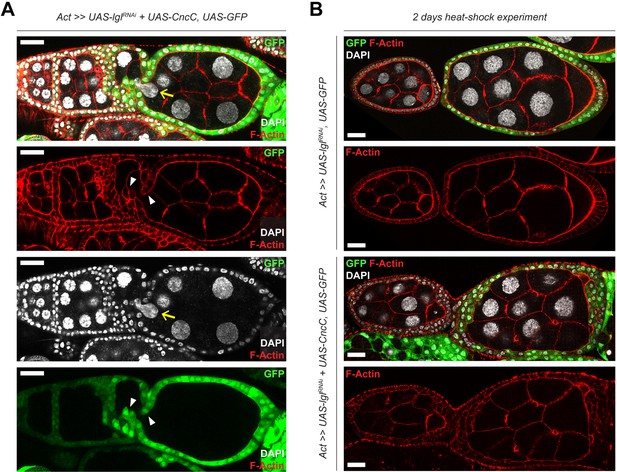
Epithelial multilayering and invasiveness are increased in Lgl-KD+CncC-OE egg chambers.
(A) Confocal images showing increased invasiveness of the epithelia containing Lgl-KD+CncC-OE follicle cells (green). Nurse cell (yellow arrow) is depicted being compressed by invading Lgl-KD+CncC-OE epithelia (marked by white arrowheads). (B) Confocal images comparing mid-stage egg chambers containing Lgl-KD (top panels) and Lgl-KD+CncC-OE (bottom panels) follicle cells, 2 days following heat shock induction of transgene expression. Enhancement of multilayer formation in Lgl-KD+CncC-OE egg chambers at midoogenesis is observed in 22.85% ovarioles (N = 16/70) compared to 2% ovarioles (N = 50) containing Lgl-KD egg chambers. Nucleus is marked by DAPI (white), and F-actin is marked by Phalloidin (red). Scale bars, 20 µm.
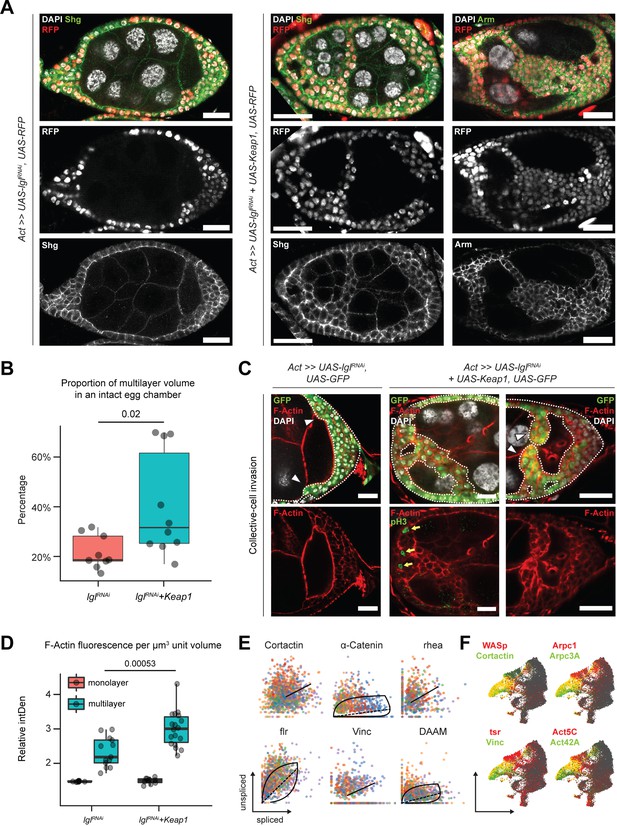
Invading Lgl-KD+Keap1-OE cells exhibit extensive cytoskeletal remodeling.
(A) Confocal images of egg chambers with Lgl-KD (left) and Lgl-KD+Keap1-OE (middle and right panels) in the follicle cells (red; shown in the middle row). Egg chambers are stained with Shg (DE-Cad; left and center columns) and Arm (right column) (green). Nucleus is marked by DAPI (white). F-actin is marked by Phalloidin (red). Follicle cells with transgene expression are separately shown in the middle row, while Shg and Arm staining intensities are shown in the bottom row. Scale bars, 20 µm. (B) Box-and-whisker plot showing the proportion of delaminated epithelial volume of Lgl-KD (N = 9 egg chambers) and Lgl-KD+Keap1-OE (N = 10) follicle cells compared to the volume of the entire egg chamber. The p-value (p=0.02) obtained from t-test comparing the two samples is listed on top. (C) Confocal images of collectively invading follicle cells (green) in Lgl-KD (left) and Lgl-KD+Keap1-OE (middle and right). Nucleus is marked by DAPI (white). F-actin is marked by Phalloidin (red). Invasive fronts are marked by arrowheads. Positive pH3 staining (green in bottom middle panel) shows mitotically dividing cells, marked by yellow arrows. Scale bars, 20 µm. (D) Box-and-whisker plot to show the relative intensities of F-actin in the monolayer (red) and multilayers (teal) of egg chambers with Lgl-KD (N = 10 egg chambers) and Lgl-KD+Keap1-OE (N = 10) follicle cells. Individual egg chambers are shown as distinct dots. The p-value (p=0.00053) obtained from t-test comparing the two samples is listed on top. (E) Phase portraits of cytoskeletal genes in cluster 7. Straight line indicates that the gene exhibits steady-state dynamics, while curved solid line with dashed straight line represents upregulation. Cells are colored according to their subcluster identity. (F) Gene enrichment plot shows overlapping enrichment of actin-remodeling genes in cluster 7 cells.
-
Figure 6—source data 1
Related to Figure 6B.
- https://cdn.elifesciences.org/articles/80956/elife-80956-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Raw data for the IntDen values of F-actin enrichment in the monolayered and multilayered follicle cells expressing Lgl-KD and Lgl-KD+Keap1-OE.
Related to Figure 6D.
- https://cdn.elifesciences.org/articles/80956/elife-80956-fig6-data2-v2.xlsx
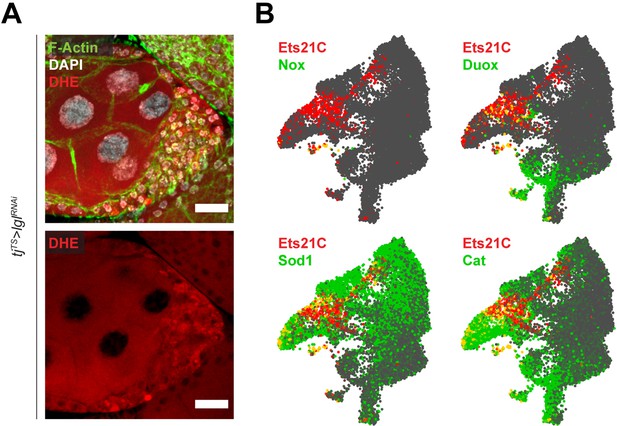
Oxidative stress response is not detected in cluster 7 cells.
(A) Confocal images showing dihydroethidium (DHE) staining (red) in the multilayers. Significant DHE staining is not observed within the multilayered cells. Nucleus is marked by DAPI (white). F-actin is marked by Phalloidin (green). Scale bars, 20 µm. (B) Gene enrichment plots showing the enrichment overlap of common oxidative stress response genes (green) with Ets21C (red) that is specific to cluster 7. Overlap of gene expression is observed as cells colored yellow.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/80956/elife-80956-mdarchecklist1-v2.docx
-
Source data 1
Enriched transcription factor binding motifs detected in genes expressed in the cells of cluster 7.
- https://cdn.elifesciences.org/articles/80956/elife-80956-data1-v2.xlsx
-
Source data 2
Genes showing dynamic behavior of transcription driving the underlying RNA velocity of cluster 7 cells.
- https://cdn.elifesciences.org/articles/80956/elife-80956-data2-v2.xlsx





