Molecular mechanism of active Cas7-11 in processing CRISPR RNA and interfering target RNA
Figures
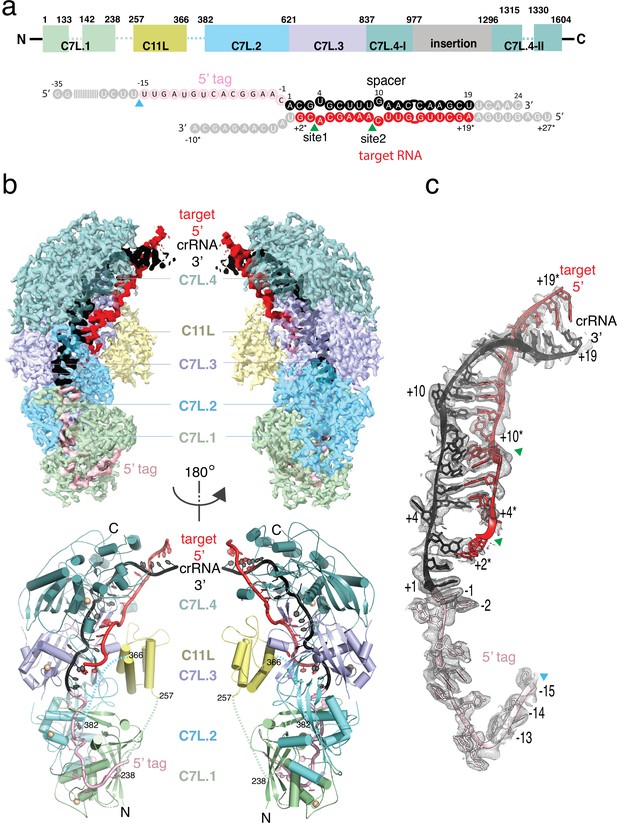
Structure overview of Desulfonema ishimotonii Cas7-11 (DiCas7-11)-crRNA-target RNA ternary complex.
(a) Domain organization of DiCas7-11 and schematic representation of crRNA-target RNA duplexes used in the study. Protein domain and RNA elements are colored differently and labeled. C7L denotes Cas7-like domain and C11L denotes Cas11-like domain. The gray colored nucleotides indicate those included in the constructs but not built due to weak or no density. Dash lines indicate protein regions not built due to weak or no density. The cyan and green colored triangles indicate the precursor crRNA (pre-crRNA) processing and target RNA cleavage sites, respectively. (b) Top, electron potential density map of DiCas7-11-crRNA-target RNA ternary complex shown in two different orientations. Bottom, cartoon representation of DiCas7-11-crRNA-target RNA ternary complex shown the same views as in top panel with corresponding colors representing protein domains and the two RNA strands. The solid spheres in wheat represent Zn atoms. (c) Close-up view of the density for the crRNA (spacer, black; 5’-tag, light pink) and target RNA (red) duplex. RNA nucleotide positions as well as the three cutting sites are indicated as in panel (a).
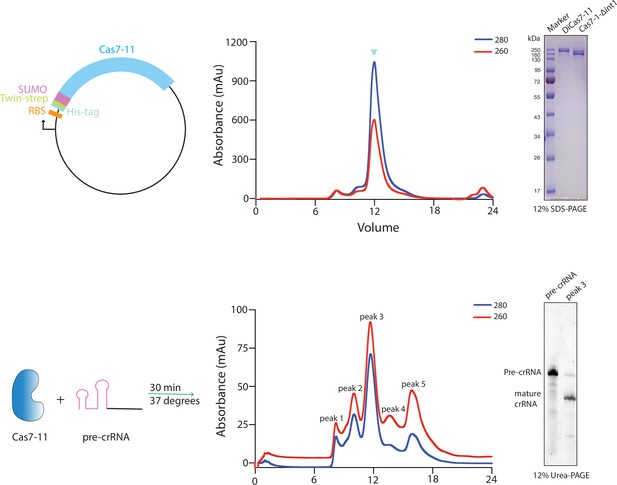
Top, schematic of the plasmid used for over-expression of the wild-type and mutant Desulfonema ishimotonii Cas7-11 (DiCas7-11), the elution profile of full-length DiCas7-11 on size exclusion chromatography, and gel analysis of the purified wild-type (DiCas7-11) and an insertion deleted Δint1-Cas7-11 proteins.
The peak fraction run on gel in lane 2 is marked with inverted blue triangle on the elution profile and was used for subsequent experiments. Proteins are visualized by Coomassie brilliant blue staining. Bottom, schematic of DiCas7-11-precursor crRNA (pre-crRNA) complex formation, elution profile of DiCas7-11-crRNA binary complex, and gel analysis of the crRNA in the peak fraction by SYBR Gold staining.
-
Figure 1—figure supplement 1—source data 1
Polyacrylamide gel images showing Cas7-11 purification and Cas7-11-crRNA complex formation used in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/81678/elife-81678-fig1-figsupp1-data1-v2.zip
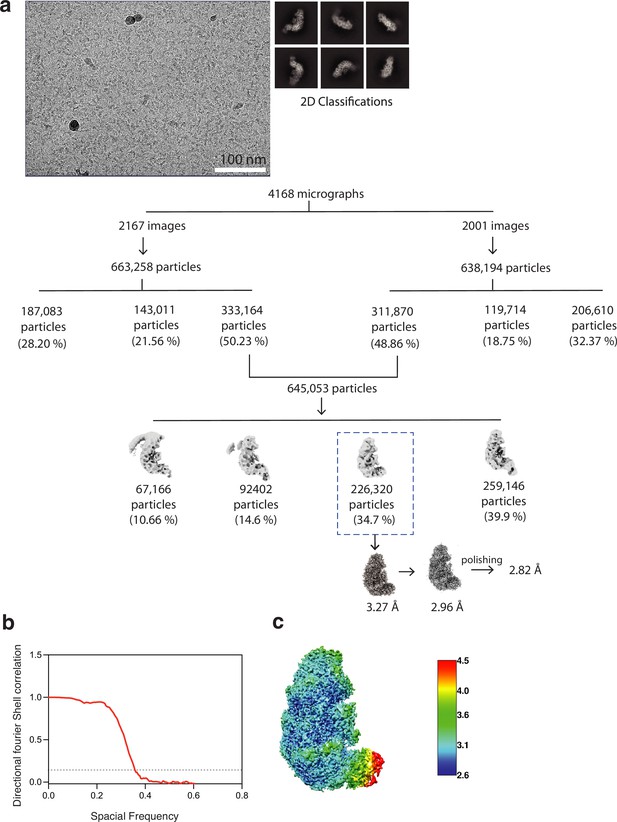
Data collection, processing,and assesment.
(a) Workflow of the cryo-electron microscopy (cryo-EM) image processing and 3D reconstruction of the Desulfonema ishimotonii Cas7-11 (DiCas7-11) ternary complex. The numbers indicate the number of particles while the percent indicates the percent to the total particles for each class. (b) Fourier shell correlations (FSC) of DiCas7-11-crRNA-ternary complex reconstruction, with the FSC cutoff 0.143, marked with a gray line and final resolution. (c) The local resolution of the final density map obtained from ResMap is colored as the color scale bar.
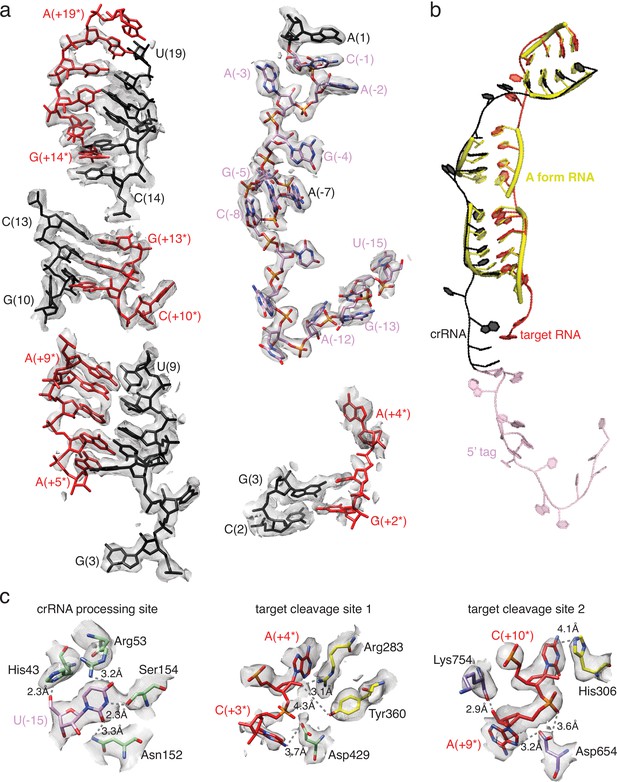
Close-up views of electron potential density maps for selected regions and comparison of the target:crRNA duplex to standard A form helix.
(a) Density for regions of the target and crRNA duplex and the 5’-tag. (b) Superimposed target:crRNA duplex (black and red) with standard A form helices (yellow). (c) Density of the three active sites as indicated. Nucleotide positions are labeled according to the scheme in Figure 1.
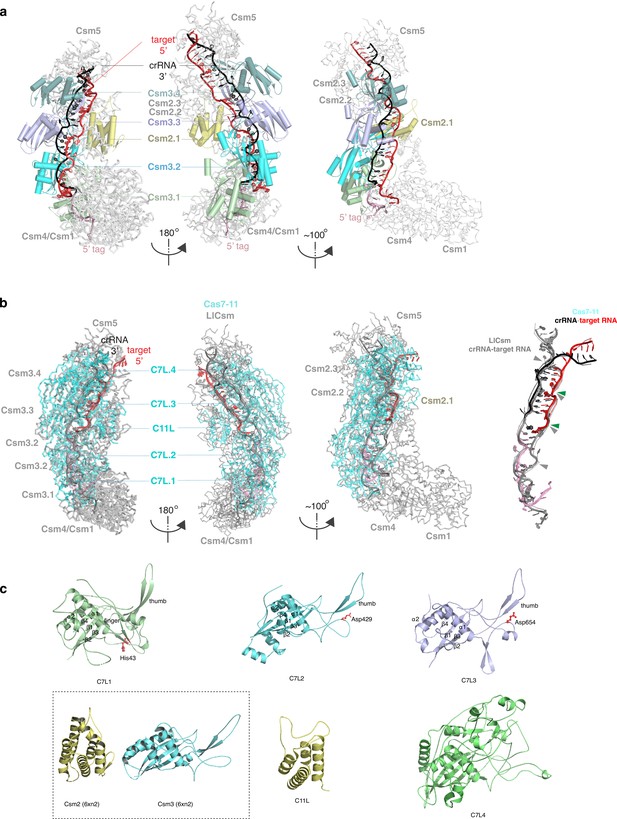
Comparison of Desulfonema ishimotonii Cas7-11 (DiCas7-11) to the homologous multi-subunit Csm complex from Lactococcus lactis (LlCsm) (PDB ID: 6XN2) (a and b) and its individual protein domains (d).
(a) LlCsm overview in three orientations in carton representations. The two left views are oriented exactly the same as those of DiCas7-11 shown in Figure 1. Subunits similar to DiCas7-11 domains and RNA are labeled and colored identically. (b) Superimposed LlCsm (gray) with DiCas7-11 (cyan) in ribbon models in the same three views as in (a). The DiCas7-11 and LlCsm were superimposed by using the C7L.2 domain and Csm2 subunits only that resulted in a root-means-square-difference (r.m.s.d.) of 5.3 Å for 131 Cα atoms. The target RNA that can be superimposed have an r.m.s.d. of 0.897 Å for 60 sugar phosphate backbone atoms. Solid triangles indicate target cleavage sites for LlCsm (gray) and DiCas7-11 (green), respectively. (c) The superimposed crRNA-target RNA duplexes in the DiCas7-11 (black, pink, and red) and LlCsm (gray) complexes, respectively. (d). The four Cas7-like (C7L) domains are superimposed and shown separately in cartoon representations and colored as those in Figure 1. The Cas11-like (C11L) is shown in cartoon representations. Key elements discussed in the text are labels. Boxed structures are the corresponding subunits for C11L and C7L, respectively, from LlCsm (PDB ID: 6XN2).
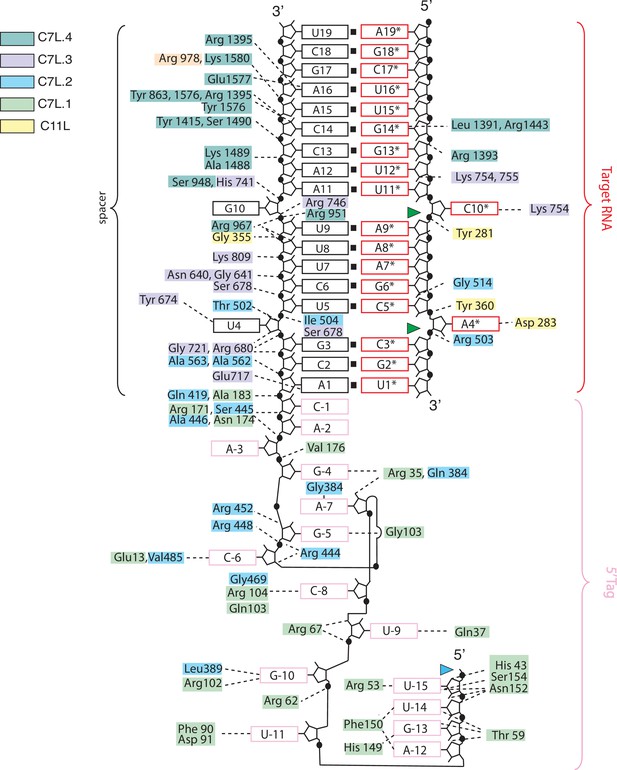
Schematic of RNA-protein interactions observed in the Desulfonema ishimotonii Cas7-11 (DiCas7-11)-crRNA-target RNA ternary complex structure.
Protein residues are colored according to the scheme used in Figure 1 that corresponding to respective domains. The black, red, and light pink color boxes denote spacer, repeat, and target RNA. The green and blue colored triangles indicate target RNA and precursor crRNA (pre-crRNA) processing sites, respectively.
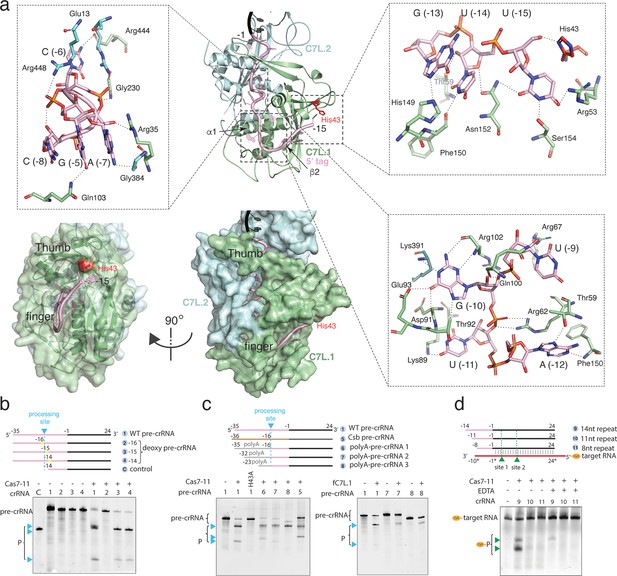
Precursor crRNA (pre-crRNA) processing and recognition.
(a) The mode of Cas7-like domain 1 (C7L.1) and C7L.2 interaction with the processed crRNA nucleotides –15 to –1 in both cartoon (top) and surface (bottom) representations. Key secondary elements involved in crRNA interaction are labeled. Insets indicate close-up views around U(–15)-U(–13)-G(–13), the C(–8)-A(–7)-C(–6)-G(–5) tight RNA turn, and the conserved A(–12)-U(–11)-G(–10)-U(–9) tetranucleotide. The catalytic residue His43 for crRNA processing is colored red. Dash lines indicate close polar contacts. (b–c) Top, various pre-crRNA used in processing reactions. Cyan colored triangles and dash lines indicate the pre-crRNA processing sites. Yellow bars indicate the sites of deoxy modification. The control RNA contains the last 14 nucleotides of the repeat plus the spacer. Spacer and repeat are shown in black and pink, respectively. ‘Csb pre-crRNA’ denotes the pre-crRNA for Candidatus Scalindua broadae Cas7-11. Processed products (P) of pre-crRNA are stained by SYBR Gold and imaged by ChemiDoc MP. Bottom, RNA processing results analyzed on polyacrylamide urea gel for the wild-type (1) and other pre-crRNA (2-8) by the wild-type Desulfonema ishimotonii Cas7-11 (DiCas7-11) (WT), the His43 to alanine mutant (H43A) of DiCas7-11, and the free-standing C7L.1 (fC7L.1). Processing products are indicated by cyan triangles. (d) Target RNA cleavage results analyzed on polyacrylamide urea gel using the wild-type and truncated pre-crRNA in the presence and absence of ethylenediaminetetraacetic acid (EDTA). ‘Cy3’ denotes the target RNA containing a 5’-Cy3 fluorophore. The cleavage products (P) of the Cy3-labeled target RNA are visualized on ChemiDoc MP using 550 nm as the excitation and 564 nm as the emission wavelength, respectively, and are indicated by green triangles.
-
Figure 2—source data 1
Polyacrylamide gel image for deoxy precursor crRNA (pre-crRNA) processing activity shown in Figure 2.
- https://cdn.elifesciences.org/articles/81678/elife-81678-fig2-data1-v2.zip
-
Figure 2—source data 2
Polyacrylamide gel image for precursor crRNA (pre-crRNA) variant processing by DiCas7-11 and free-standing C7L.1 (fC7L.1) shown in Figure 2.
- https://cdn.elifesciences.org/articles/81678/elife-81678-fig2-data2-v2.zip
-
Figure 2—source data 3
Polyacrylamide gel image of target RNA cleavage activities with truncated crRNA and DiCas7-11 shown in Figure 2.
- https://cdn.elifesciences.org/articles/81678/elife-81678-fig2-data3-v2.zip

Sequence comparison among Cas7-11 proteins overlays with the secondary structure of DiCas7.
Regions of interests described in the main text are marked by asterisks. Desulfonema ishimotonii Cas7-11 (DiCas7-11) is from Desulfonema ishimotonii; CjcCas7-11 is from Candidatus Jettenia caeni; CsbCas7-11 is from Candidatus Scalindua broadae; HsmCas7-11 and SstCas7-11 source organism is unknown.
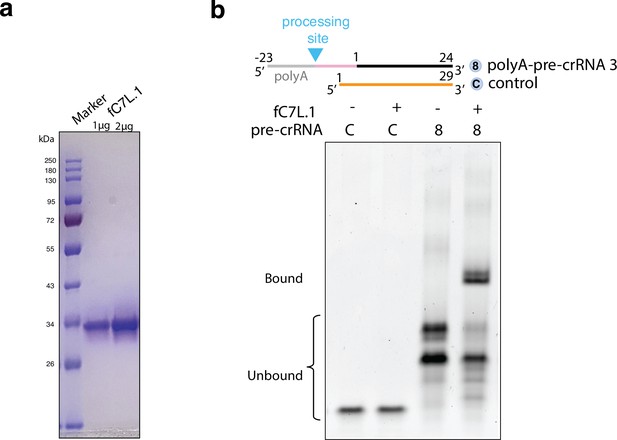
Protein analysis and electrophoretic mobility shift assay of the free-standing C7L.1 (fC7L.1) with precursor crRNA (pre-crRNAs).
(a) SDS-PAGE gel analysis of fC7L.1 in two different concentrations after size exclusion chromatography. Proteins are visualized by Coomassie brilliant blue staining. (b) Native TBE gel analysis of a polyA-containing pre-crRNA (same as substrate 8 in Figure 2c) and a negative control RNA (C) following 30 min incubation with molar excess of fC7L.1. RNA is visualized by SYBR Gold staining.
-
Figure 2—figure supplement 2—source data 1
Native-polyacrylamide gel images for binding activity by free-standing C7L.1 (fC7L.1) used in Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/81678/elife-81678-fig2-figsupp2-data1-v2.zip
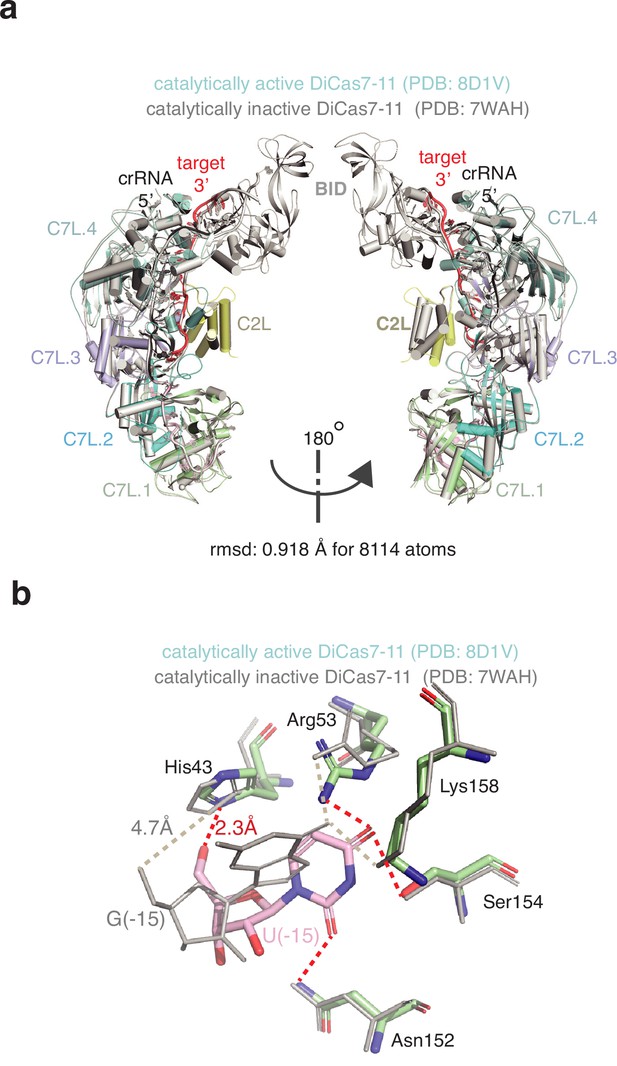
Comparison of the catalytically inactive (PDB: 7WAH) (gray) and the catalytically active (PDB: 8D1V) (color) Desulfonema ishimotonii Cas7-11 (DiCas7-11)-RNA complex structures.
(a) Two views of the superimposed protein-RNA complexes (0.918 Å root-means-square-difference (r.m.s.d.) for 8114 atoms) in cartoon representation. Each protein domain and the RNA are labeled. (b) Comparison of the interactions of the first 5’-tag nucleotide with DiCas7-11. G(–15) is used in the catalytically inactive complex while the native nucleotide U(–15) is used in the catalytically active complex. The close contacts between protein residues and U(–15) or G(–15) are indicated by red or gray dashed lines, respectively. Note that the catalytic residue for processing, His43, is 2.3 and 4.7 Å to the leaving 5’-OH of U(–15) and G(–15), respectively, indicating a cognate interaction in the catalytically active complex at the processing site.
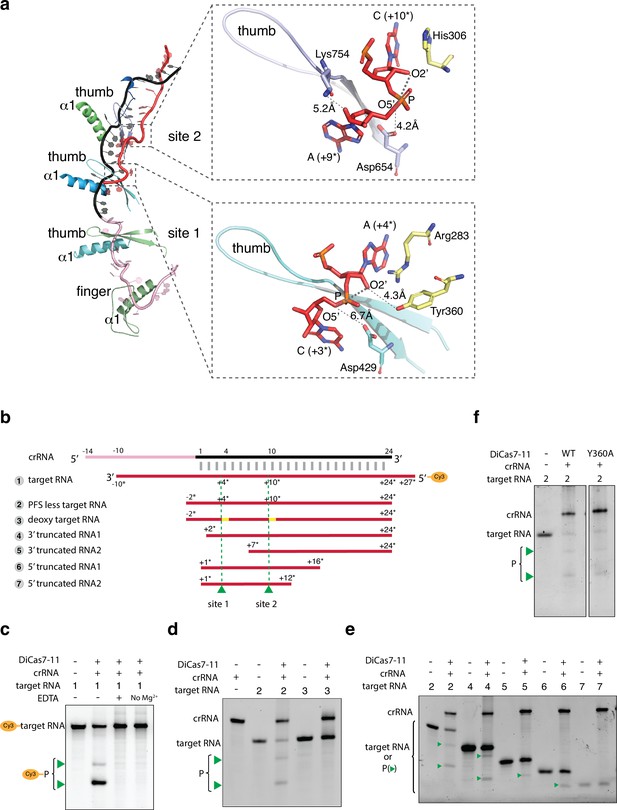
Target RNA cleavage mechanism.
(a) Recognition of target RNA by the crRNA and Desulfonema ishimotonii Cas7-11 (DiCas7-11). The ferredoxin fold α1 and the thumb hairpin for each of the four Cas7-like (C7L) domains are shown as cartoons and colored as in Figure 1. The quoted ‘thumb’ indicates the degenerate thumb motif for the C7L.4 domain. Insets show the two target cleavage sites in close-up views. RNA nucleotides and key amino acids are shown in stick models. The three atoms involved in formation of the ‘in-line’ geometry during phosphodiester bond breakage are labeled and indicated by thick dash lines. The closest of the three atoms to the putative catalytic residues, Asp654 (for site 2) and Asp429 (for site 1), are indicated by a connecting dash line. The close contact between Tyr360 and A(+4*) 2’-hydroxyl oxygen at site 1 and that between Lys754 and A(+9*) 2’-hydroxyl at site 2 are also indicated by dash lines. (b) Schematic of the Cy3-labeled (substrate 1) and other target RNA (substrates 2–7) used in cleavage activity assays. Yellow bars mark the locations of the deoxy modification on the target RNA substrate 3. Green colored triangles and dash lines indicate target RNA cleavage sites. (c–f) Target RNA cleavage by DiCas7-11 and its Tyr360 to alanine mutant (Y360A) are analyzed on polyacrylamide urea gel. The cleavage products (P) of the Cy3-labeled target RNA are visualized on ChemiDoc MP using 550 nm as the excitation and 564 nm as the emission wavelength, respectively. Cleavage products (P) of non-Cy3-labeled target RNA are stained by SYBR Gold and imaged by ChemiDoc MP. All cleavage products are indicated by green triangles.
-
Figure 3—source data 1
Polyacrylamide gel image of target RNA cleavage activity by DiCas7-11 in presence and absence of metal ions shown in Figure 2.
- https://cdn.elifesciences.org/articles/81678/elife-81678-fig3-data1-v2.zip
-
Figure 3—source data 2
Polyacrylamide gel image of deoxy-target RNA cleavage activity by DiCas7-11 shown in Figure 3.
- https://cdn.elifesciences.org/articles/81678/elife-81678-fig3-data2-v2.zip
-
Figure 3—source data 3
Polyacrylamide gel image of target RNA variant cleavage activity by DiCas7-11 shown in Figure 3.
- https://cdn.elifesciences.org/articles/81678/elife-81678-fig3-data3-v2.zip
-
Figure 3—source data 4
Polyacrylamide gel image of target RNA cleavage activity by Desulfonema ishimotonii Cas7-11 (DiCas7-11) Y360A mutant shown in Figure 3.
- https://cdn.elifesciences.org/articles/81678/elife-81678-fig3-data4-v2.zip
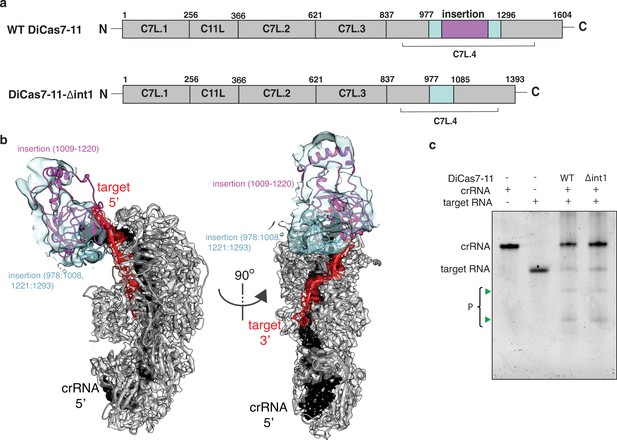
Engineering a compact Desulfonema ishimotonii Cas7-11 (DiCas7-11).
(a) Schematic of domain organization of wild-type and an insertion deletion variant DiCas7-11-Δint1. The region removed is colored in purple and numbered. (b) Cartoon representation of DiCas7-11 overlaying with density map resulted from focused classification using a mask around the insertion domain. The insertion structure model is from AlphaFold prediction. (c) Target RNA cleavage by DiCas7-11 (WT) and DiCas7-11-Δint1(Δint1) are analyzed on a polyacrylamide urea gel. Cleavage products (P) are stained by SYBR Gold and imaged by ChemiDoc MP and are indicated by green triangles.
-
Figure 4—source data 1
Polyacrylamide gel image showing target RNA cleavage activity by DiCas7-11-Δint1.
- https://cdn.elifesciences.org/articles/81678/elife-81678-fig4-data1-v2.zip
Tables
| Reagent type(species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (species) | Escherichia coli NiCo21(DE3) | New England Biolabs | C2529H | Used for proteins expression |
| Recombinant DNA reagent | Sumo-tag-DiCas7-11 expression plasmid | Özcan et al., 2021 | Addgene:172503 | |
| Recombinant DNA reagent | His-tag-DiCas7-11 expression plasmid | This paper | N/A | Constructed in-house. Protein-encoding sequences are inserted into pACYC-duet-1 cloning vector with BamHI and EcoRI cut sites, T7 promoter, p15A origin, and chloramphenicol resistance |
| Recombinant DNA reagent | Cas7-11-Δint-1 expression plasmid | This paper | N/A | Addgene:172503; see Supplementary file 2 for primers |
| Recombinant DNA reagent | fC7L.1 expression plasmid | This paper | N/A | |
| Peptide, recombinant protein | Cas7-11- Δint-1 protein | This paper | N/A | Expressed and purified in house from E. coli NiCo21(DE3) cells |
| Peptide, recombinant protein | fC7L.1 protein | This paper | N/A | |
| Peptide, recombinant protein | ULP1 protease | Protein Expression Facility, FSU | N/A | |
| Software, algorithm | cryoSPARC (v3.3.1) | Punjani et al., 2017 | https://cryosparc.com | |
| Software, algorithm | RELION 4.0 | Kimanius et al., 2021 | https://www2.mrc-lmb.cam.ac.uk/ | |
| Software, algorithm | COOT | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ | |
| Software, algorithm | PHENIX | Afonine et al., 2018 | https://phenix-online.org | |
| Software, algorithm | UCSF ChimeraX UCSF Chimera | Pettersen et al., 2021 Pettersen et al., 2004 | https://www.rbvi.ucsf.edu/chimera/ |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/81678/elife-81678-mdarchecklist1-v2.docx
-
Supplementary file 1
Data acquisition and processing parameters.
- https://cdn.elifesciences.org/articles/81678/elife-81678-supp1-v2.docx
-
Supplementary file 2
Ribonucleic acid sequences used in this study.
- https://cdn.elifesciences.org/articles/81678/elife-81678-supp2-v2.docx





