Ecdysone acts through cortex glia to regulate sleep in Drosophila
Figures
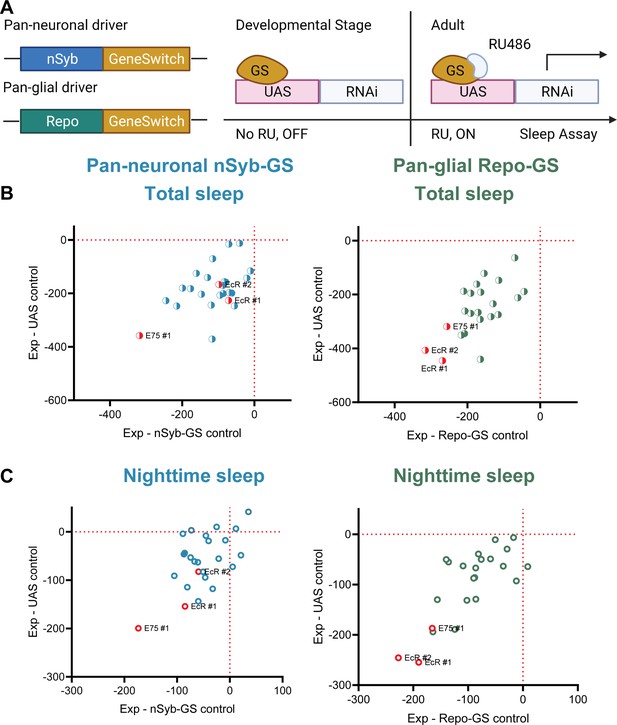
A screen of all nuclear hormone receptors (NHRs) in Drosophila identifies sleep-regulating functions of ecdysone receptor (EcR) and its downstream target, NHR E75.
(A) Flies carrying a pan-neuronal driver nSyb-GS or pan-glial driver Repo-GS were crossed with different UAS lines carrying RNAi constructs against NHRs, and their 5- to 7-day-old F1 female progeny were loaded into DAM monitors to record their activity under 12:12 hr light:dark cycles. GeneSwitch remains inactive during developmental stages and is activated by RU486 in the food in DAM monitors. Behavior data were collected by the DAM system. (B, C) Mean total and nighttime sleep for each group were calculated by Pysolo. Differences between experimental flies and GS and RNAi controls were calculated separately in each independent experiment, and the average values comparing each experimental to its GeneSwitch control (X-axis) and RNAi control (Y-axis) are shown in the plots. The exact numbers of flies used per line are provided in Figure 1—source data 1. While knockdown of most NHRs reduces sleep, effects of EcR RNAi #1, EcR RNAi #2, E75 RNAi #1 predominate, especially in glial knockdown experiments.
-
Figure 1—source data 1
Detailed sleep phenotypes of nuclear hormone receptor (NHR) RNAi screening lines.
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig1-data1-v2.xlsx
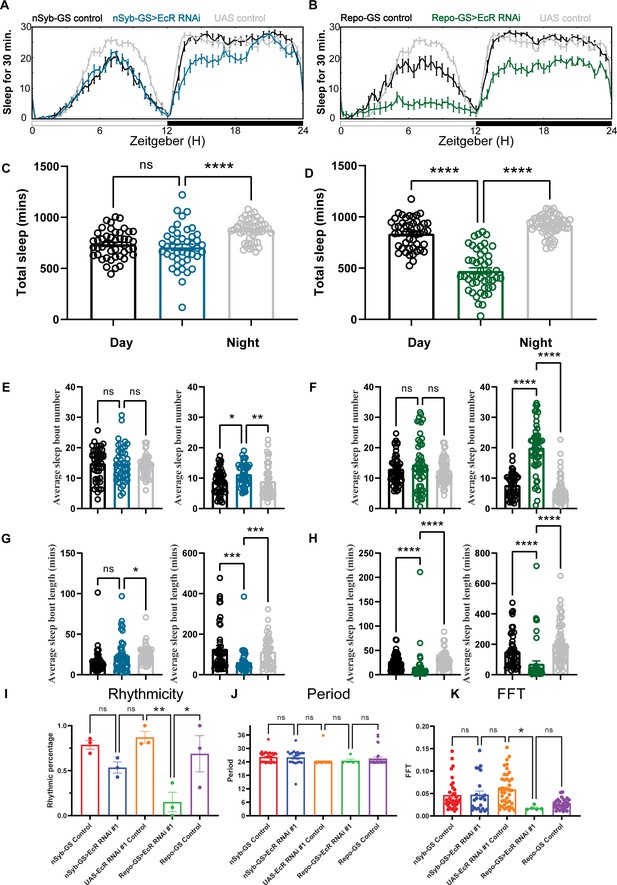
Baseline sleep phenotypes resulting from pan-neuronal or pan-glial knockdown of ecdysone receptor (EcR).
(A, B) Show representative sleep traces of nSyb-GS>EcR RNAi #1 and Repo-GS>EcR RNAi #1. N = 14–16 per genotype. Data are based on at least three independent experiments. Representative sleep traces are showed because Pysolo we used does not allow combining data from repeated experiments. Total sleep of the nSyb-GS>EcR RNAi #1 and Repo-GS>EcR RNAi #1 flies for three replicates, N = 43–63, one-way analysis of variance (ANOVA) with Tukey post hoc test was used for (C), and Kruskal–Wallis test with Dunn’s multiple comparisons test was used for (D). (E–H) The average sleep bout number and average sleep bout length of the nSyb-GS>EcR RNAi #1 and Repo-GS>EcR RNAi #1 flies for all three replicates. Daytime sleep data are quantified in the left panels, and nighttime sleep is quantified in the right panels of each group. One-way ANOVA analysis with Tukey post hoc test was used for (E, F) and Kruskal–Wallis test with Dunn’s multiple comparisons test was used for (G, H). (I–K) The rhythmicity, period, and relative Fast Fourier Transform (FFT) power analysis of nSyb-GS>EcR RNAi #1 and Repo-GS>EcR RNAi #1 flies and controls assayed for locomotor activity rhythms in constant darkness. (I) shows the percentage rhythmic in each genotype from all three independent replicates. Flies used for analysis in (J, K) are rhythmic flies from (I), and experimental files are compared with their GeneSwitch and RNAi control flies. Bar graphs show mean + standard error of the mean (SEM), and p values for each comparison were calculated using the Kruskal–Wallis test with Dunn’s multiple comparisons test. ns = not significant, p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure 2—source data 1.
-
Figure 2—source data 1
Sleep and circadian phenotypes resulting from pan-neuronal or pan-glial knockdown of ecdysone receptor (EcR).
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Sleep phenotypes resulting from pan-neuronal or pan-glial knockdown of E75.
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Sleep phenotypes resulting from pan-neuronal or pan-glial knockdown of Hr51 and sleep phenotypes of adult-specific neuronal and glial knockdown of ecdysone receptor (EcR).
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig2-data3-v2.xlsx
-
Figure 2—source data 4
Sleep phenotypes resulting from pan-glial knockdown of ecdysone receptor (EcR) by temporal and regional gene expression targeting (TARGET) system and different EcR RNAi lines.
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig2-data4-v2.xlsx
-
Figure 2—source data 5
Sleep phenotypes resulting from pan-neuronal or pan-glial knockdown of EcI and overexpression of ecdysone receptor (EcR) common isoforms.
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig2-data5-v2.xlsx
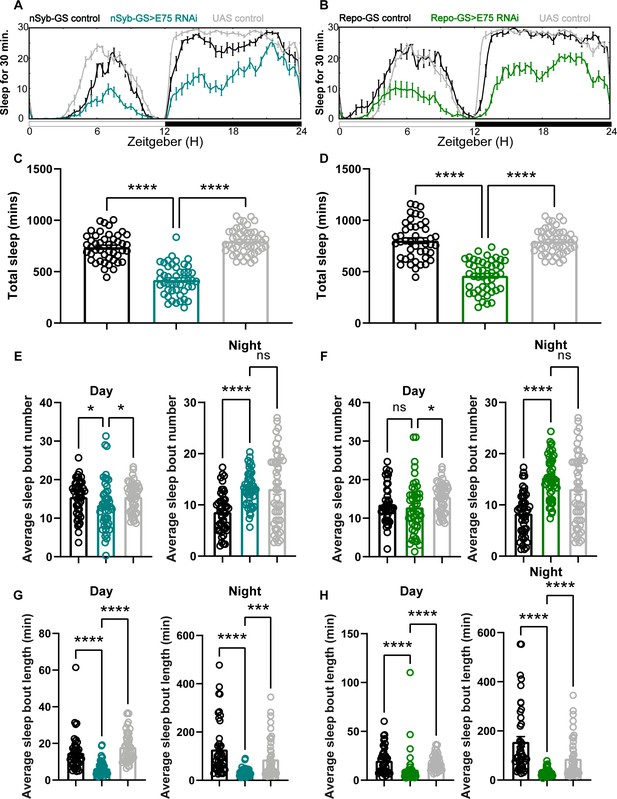
Baseline sleep phenotypes resulting from pan-neuronal and pan-glial knockdown of E75, the direct downstream target of ecdysone receptor (EcR).
(A, B) show representative sleep traces of nSyb-GS>E75 RNAi #1 and Repo-GS>E75 RNAi #1. N = 12–16 per genotype per experiment. Experiments were independently repeated at least two times, and one experiment is shown here. (C, D) Total sleep of the nSyb-GS>EcR RNAi #1 and Repo-GS>EcR RNAi #1 flies shown in (A, B), p values for each comparison were calculated by one-way analysis of variance (ANOVA) with the Tukey post hoc test. (E–H) The average sleep bout number and average sleep bout length of the nSyb-GS>E75 RNAi #1 and Repo-GS>E75 RNAi #1 flies. Daytime sleep data are quantified in the left panels, and nighttime sleep is quantified in the right panels of each group. p values for each comparison in (E–H) were calculated by Kruskal–Wallis test with Dunn’s multiple comparisons test. Bar graphs show mean + standard error of the mean (SEM). ns = not significant, p > 0.05, *p < 0.05, ***p < 0.001, ****p < 0.0001. See also Figure 2—source data 2.
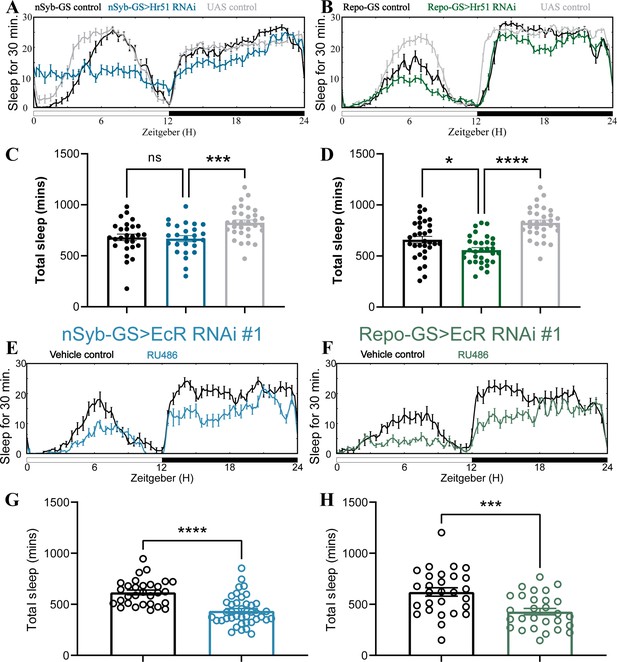
Adult-specific neuronal and glial knockdown of ecdysone receptor (EcR) reduces sleep.
(A, B) show representative sleep traces of nSyb-GS>Hr51 RNAi and Repo-GS>Hr51 RNAi. N = 26–32 per genotype. Experiments were independently repeated two times, and one experiment is shown here. (C, D) Total sleep of the nSyb-GS>Hr51 RNAi and Repo-GS>Hr51 RNAi flies shown in (A, B), p values for each comparison were calculated by one-way analysis of variance (ANOVA) with the post hoc test. (E, F) show representative sleep traces of nSyb-GS>EcR RNAi #1 and Repo-GS>EcR RNAi #1 female flies with vehicle control or RU486; experiments were independently repeated at least two times. (G, H) Total sleep of the nSyb-GS>EcR RNAi #1 RNAi and Repo-GS>EcR RNAi #1 female flies with vehicle control or RU486, N = 28–40 per genotype. Bar graphs show mean + standard error of the mean (SEM), and ns = not significant, p > 0.05, *p < 0.05, ***p < 0.001, ****p < 0.0001. The comparison was calculated with an unpaired parametric Student’s t-test for Repo-GS groups and Mann–Whitney test for nSyb-GS groups. See also Figure 2—source data 3.
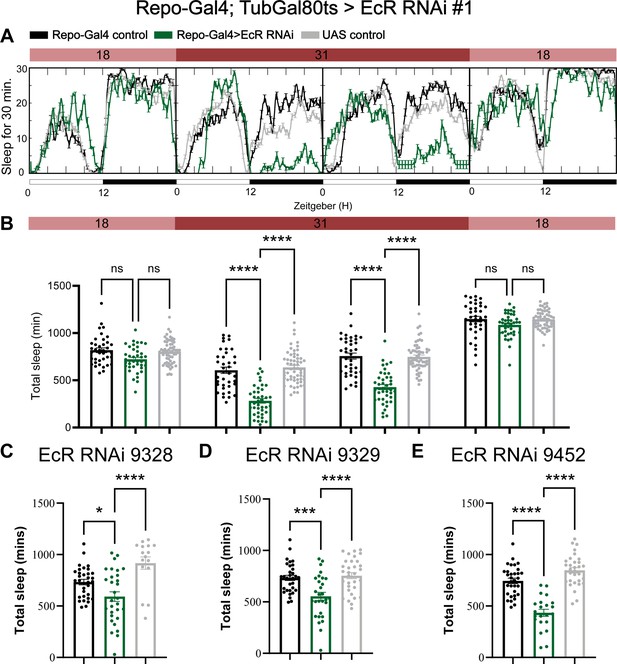
Adult-specific knockdown of ecdysone receptor (EcR) by using the temporal and regional gene expression targeting (TARGET) system reduces sleep.
(A) Sleep traces resulting from pan-glial adult-specific EcR knockdown by using Repo-Gal4; TubulinGal80ts crossed with UAS-EcR RNAi #1. Five-day-old F1 progeny flies were maintained at 18 degrees for 1 day, then switched to 31 degrees to inactivate Gal80ts, resulting in EcR knockdown over the next 2 days. Subsequently, the temperature dropped to 18 degrees in the last day. (B) Quantification of total sleep in pan-glial EcR knockdown flies using Repo-Gal4/TubulinGal80ts, N = 38–59 per genotype and experiments were independently repeated at least two times. Total sleep amount was calculated for each genotype and compared to the control group for four different days. p values for each comparison were calculated by one-way analysis of variance (ANOVA) using Tukey’s post hoc test. (C–E) Total sleep quantification of EcR RNAi knockdown using three different EcR RNAi lines BDSC #9328, #9329, and #9452. N = 16–32 per genotype and experiments were independently repeated two times, p values for each comparison were calculated by one-way analysis of variance (ANOVA) with the Tukey post hoc test. Black bars are Repo-GS control flies, gray bars are UAS-RNAi control flies, and green bars are different Repo-GS>EcR RNAi flies, indicated by their titles. Bar graphs show mean + standard error of the mean (SEM), ns = not significant, p > 0.05, *p < 0.05, ***p < 0.001, ****p < 0.0001. See also Figure 2—source data 4.
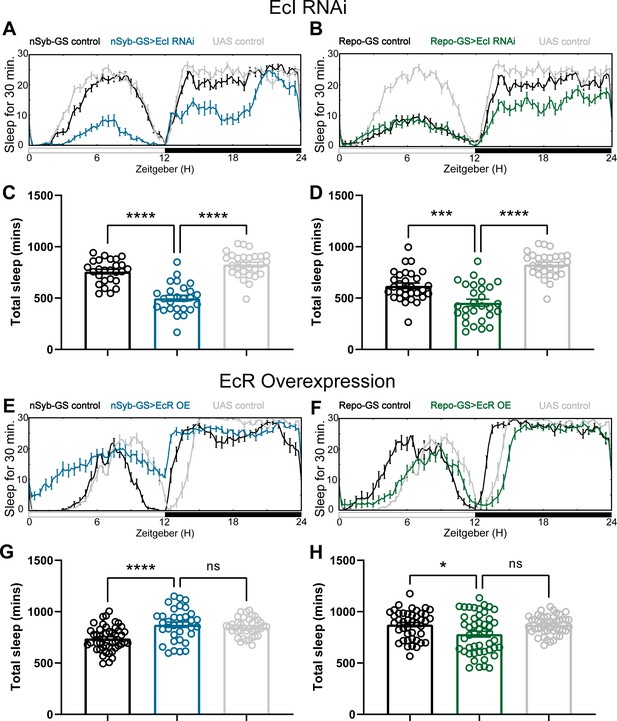
Baseline sleep phenotypes resulting from pan-neuronal and pan-glial overexpression of ecdysone receptor (EcR) common isoforms and knockdown of EcI, the membrane importer of ecdysone.
(A, B) show representative sleep traces of nSyb-GS>EcI RNAi #1 and Repo-GS>EcI RNAi #1 flies, (C, D) Total sleep quantification of the nSyb-GS>EcI RNAi #1 and Repo-GS>EcI RNAi #1 flies and their controls. N = 25–30; data are based on two independent experiments. (E, F) show representative sleep traces of nSyb-GS>EcR_c and Repo-GS>EcR_c flies, N = 35–48 per genotype; data are based on three independent experiments. (G, H) Total sleep of the nSyb-GS>EcR_c and Repo-GS>EcR_c flies. p values for each comparison were calculated by one-way analysis of variance (ANOVA) with Tukey post hoc test. Bar graphs show mean + standard error of the mean (SEM); ns = not significant, p > 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure 2—source data 5.
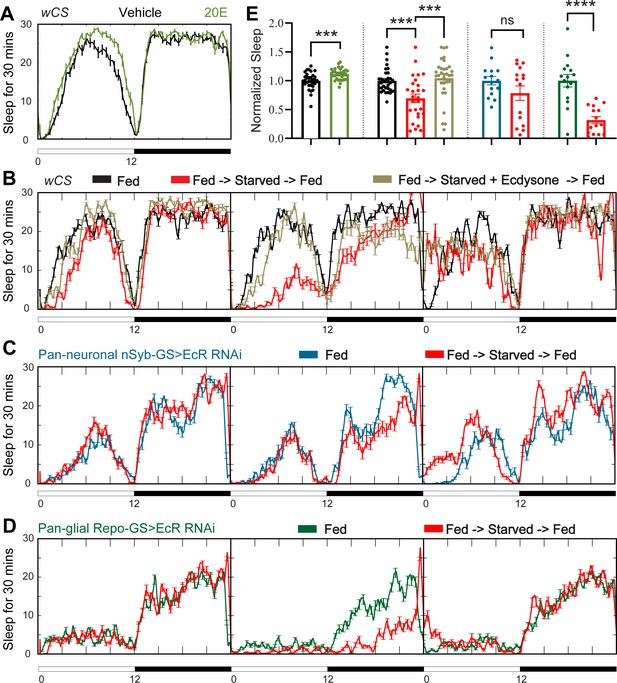
Ecdysone feeding prevents sleep loss in response to starvation, while ecdysone receptor (EcR) KD exacerbates starvation-induced sleep loss.
Sleep was monitored for 3 days either under continuous feeding with and without 0.2 mM ecdysone (A) or under each of the following three conditions—continuous feeding for 3 days or with an intervention on the second day, which consisted of either starvation or starvation accompanied with the feeding of 0.2 mM ecdysone (B). The assay was conducted in (A, B) wild-type flies (wCS) and (C, D) nSyb-GS and Repo-GS EcR RNAi #1 flies. N = 15–32 for each genotype, and the experiment was repeated two times. Quantification of sleep nomalized to vehicle control group for the average daily sleep for (A) and ZT0–ZT23h interval of the second day for (B–D) is shown in (E). ZT0–ZT23 sleep data are shown as flies were flipped back to locomotor tubes with food during the last hour (ZT23–24). Bar graphs show mean ± standard error of the mean (SEM) and ns = not significant, p > 0.05, ***p <0 .001, ****p < 0.0001. p values for comparisons between two groups were based on the Mann–Whitney test, and p values for comparison between three groups were calculated by one-way analysis of variance (ANOVA) with Tukey post hoc test. See also Figure 3—source data 1.
-
Figure 3—source data 1
Sleep phenotypes of starvation on wild-types and ecdysone receptor (EcR) disrupted flies.
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Sleep phenotypes of ecdysone treatment and heat shock when ecdysone receptor (EcR) was disrupted in neurons and glial cells.
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig3-data2-v2.xlsx
-
Figure 3—source data 3
Ecdysone levels in both fly brain and periphery, and the expression levels of ecdysone responsive genes.
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig3-data3-v2.xlsx
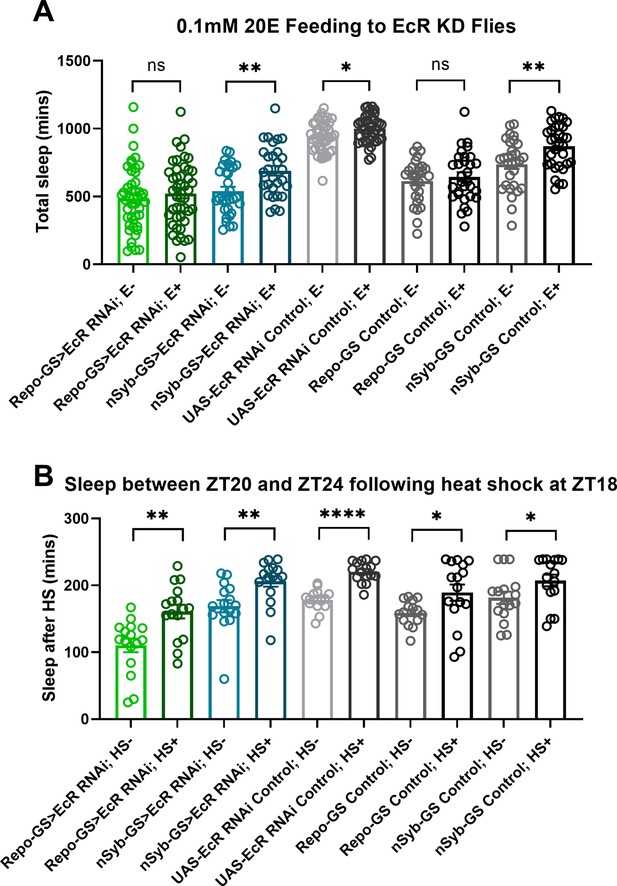
Ecdysone receptor (EcR) disruption in neurons or glia does not affect the response to heat shock.
(A) Total sleep of EcR RNAi #1 knockdown flies in both neurons and glia and their genetic control flies after 0.1 mM 20E feeding, N = 29–46, and experiments were repeated two or three times independently. (B) The last 4 hours’ sleep between ZT20 and ZT24 after heat shock for an hour at ZT18 of EcR RNAi #1 knockdown flies in neurons or glia and their genetic control flies, N = 16. Bar graphs show mean + standard error of the mean (SEM); ns = not significant, p > 0.05, *p < 0.05, **p <0 .01, ****p < 0.0001. p values for each comparison were calculated by one-way analysis of variance (ANOVA) with Tukey post hoc test. See also Figure 3—source data 2.
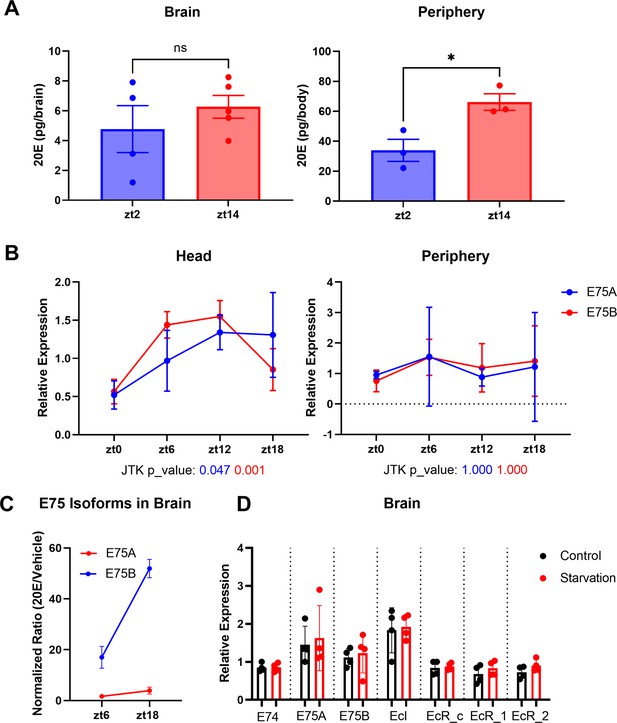
Ecdysone levels are higher in the fly periphery at ZT14 compared to ZT2.
(A) Ecdysone level was measured in the fly’s whole body or brain using an ELISA (enzyme-linked immunosorbent assay) kit. 20E levels are higher at ZT14 than ZT2 in the periphery. (B) A and B isoforms of the direct downstream target gene of ecdysone, E75, were measured by qPCR in fly heads or fly bodies at four different time points. Data indicate a significant rhythm with a mid-afternoon–evening peak in heads (JTK values, which are measures of cycling, are below the x-axis). (C) E75 isoform A and B mRNA levels in the fly brain following 20E/Vehicle injection at ZT6 or ZT18. The mRNA levels were measured 1 hr after flies were injected with 20E or vehicle control and changes seen with 20E (relative to vehicle control) in the brain at different time points were normalized to the changes in the body to control for inconsistency of the injection. (D) mRNA levels of EcR, EcI, E75, and E74 in the fly brains do not respond to 1 day of starvation. All comparisons’ p > 0.05 in (C); thus, these are not shown. Bar graphs show mean + standard error of the mean (SEM), and ns = not significant, p > 0.05, *p < 0.05. p values for each comparison are calculated by unpaired parametric Student’s t-test. See also Figure 3—source data 3.
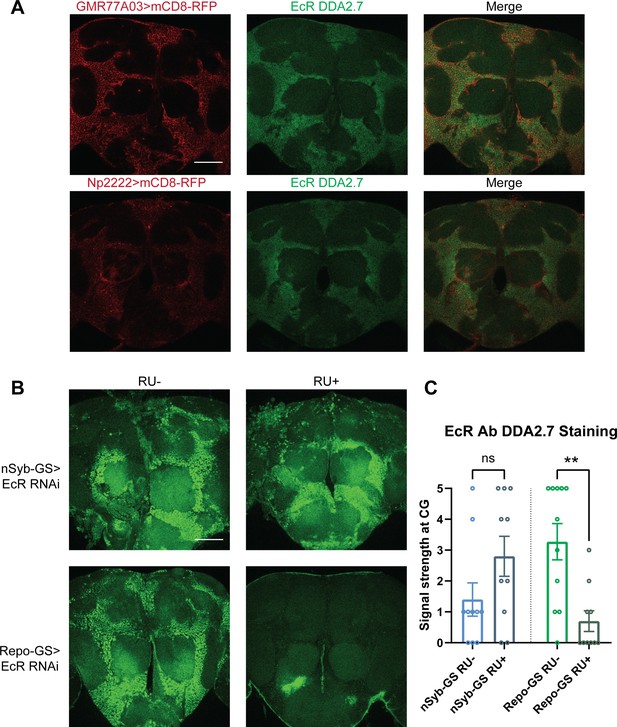
Ecdysone receptor (EcR) is expressed in cortex glia.
(A) EcR antibody DDA2.7 staining overlaps with reporter expression driven by cortex glia drivers (GMR77A03>mCD8 RFP and Np2222>mCD8 RFP), indicating that EcR is expressed in the cortex glia. EcR antibody DDA2.7 staining also shows that EcR can be nuclear or cytoplasmic. (B) EcR antibody DDA2.7 staining is preserved and still observed in the nSyb-GS>EcR RNAi flies but is almost eliminated in the Repo-GS>EcR RNAi flies compared with the vehicle control flies. N = 10 per group. Scale bar: 100 µm. (C) Quantification of antibody staining in the fly brains’ cortex glia layer, see also Figure 4—source data 1. Signal strength was scored manually based on the fluorescence intensity in the cortex glia region. Repo-GS>EcR RNAi #1 flies have significantly reduced staining based on an unpaired parametric Student’s t-test. Bar graphs show mean ± standard error of the mean (SEM) and ns = not significant, p > 0.05, **p < 0.01.
-
Figure 4—source data 1
Quantification of Anti-EcR staining in the cortex glia layer.
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig4-data1-v2.xlsx
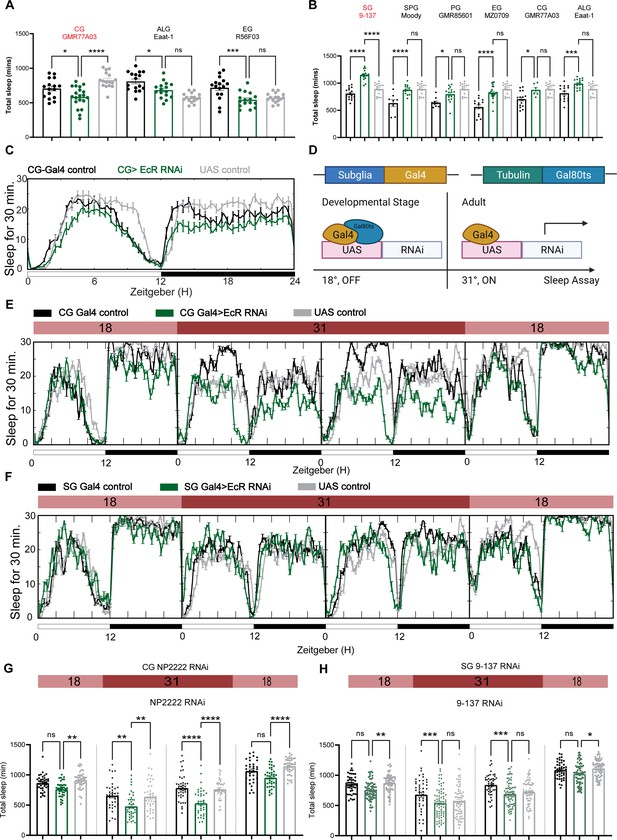
Ecdysone receptor (EcR) functions in cortex glia to affect sleep.
(A) Multiple constitutive Gal4 drivers labeling different subglial populations were crossed to EcR RNAi flies. Only GMR77A03 for cortex glia (CG), Eaat-1 for astrocyte-like glia (ALG), and GMR56F03 for ensheathing glia (EG) drivers produced viable adult progeny flies, and only GMR77A03>EcR RNAi #1 flies showed reduced total sleep compared with control flies. Green chart columns are experimental groups, and neighboring black and gray columns are Gal4 and UAS flies controls, respectively. N = 16–20 per genotype. (B) Overexpression of EcR by subglial Gal4 drivers—9-137 Gal4 to drive expression in the surface glia (SG), Moody-Gal4 for subperineurial glia (SPG), GMR85G01 for perineurial glia (PG), MZ008-Gal4 for ensheathing glia (EG), GMR77A03 for cortex glia (CG), and Eaat-1 for astrocyte-like glia (ALG). Only overexpression of EcR in the surface glia promotes sleep. N = 16–24 per genotype. (C) Representative sleep traces of the cortex glia GMR77A03>EcR RNAi #1 flies. (D) Gal4/Tubulin-gal80ts was used to achieve adult-specific knockdown of EcR in different subglial populations. Under permissive temperature, Gal80ts inhibits Gal4 activation of UAS, but under restrictive temperature, Gal80ts is inactivated, and genes under the regulation of UAS are expressed. (E, F) Sleep traces resulting from EcR knockdown in the cortex glia using NP2222-Gal4/tubulinGal80ts and surface glia using 9-137 Gal4/tubulinGal80ts. F1 progeny flies were kept at 18 degrees for 1 day, and then the temperature was switched to 31 degrees to inactivate the Gal80ts and thus achieve knockdown of EcR over the following 2 days. Subsequently, temperatures were decreased back to 18 degrees. (G, H) show quantification of total sleep of all EcR knockdown flies in the cortex glia using NP2222-Gal4/tubulinGal80ts and surface glia using 9-137 Gal4/tubulinGal80ts, N = 34–77 per genotype. Total sleep of each genotype was calculated and compared to controls for the above 4 days. Bar graphs show mean ± standard error of the mean (SEM), ns = not significant, p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. p values for each comparison were calculated by one-way analysis of variance (ANOVA) with Tukey post hoc test. See also Figure 5—source data 1.
-
Figure 5—source data 1
Sleep phenotypes resulting from subglial knockdown of ecdysone receptor (EcR).
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Sleep phenotypes of adult-specific ecdysone receptor (EcR) disruption in different subglial populations.
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig5-data2-v2.xlsx
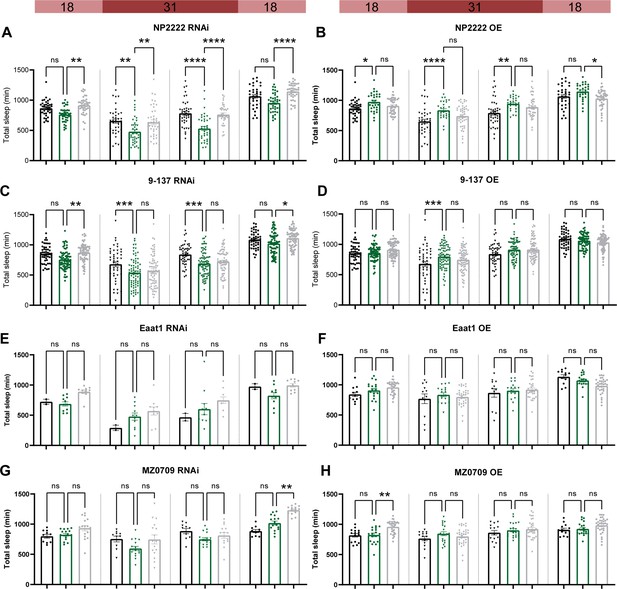
Adult-specific disruption of ecdysone receptor (EcR) in most subglial populations does not affect sleep.
Temperature-sensitive tubulin-Gal80ts were used to restrict the expression of Gal4 to adults. Sleep was monitored for 1 day at the permissive temperature of 18 degrees, then 2 days at the restrictive temperature of 31 degrees data, and then for another day at 18 degrees. EcR knockdown (RNAi) and overexpression (OE) were driven by: (A, B) a cortex glia driver (NP2222>EcR RNAi #1 and NP2222>EcR OE); (C, D) a surface glia driver (9-137>EcR RNAi #1 and 9-137>EcR OE). (E, F) An astrocyte-like glia driver (Eaat-1>EcR RNAi #1 and Eaat-1>EcR OE). (G, H) An ensheathing glia driver (MZ0709>EcR RNAi #1 and MZ0709>EcR OE). Bar graphs show mean + standard error of the mean (SEM); ns = not significant, p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. p values for each comparison were calculated by one-way analysis of variance (ANOVA) with the Tukey post hoc test. See also Figure 5—source data 2.
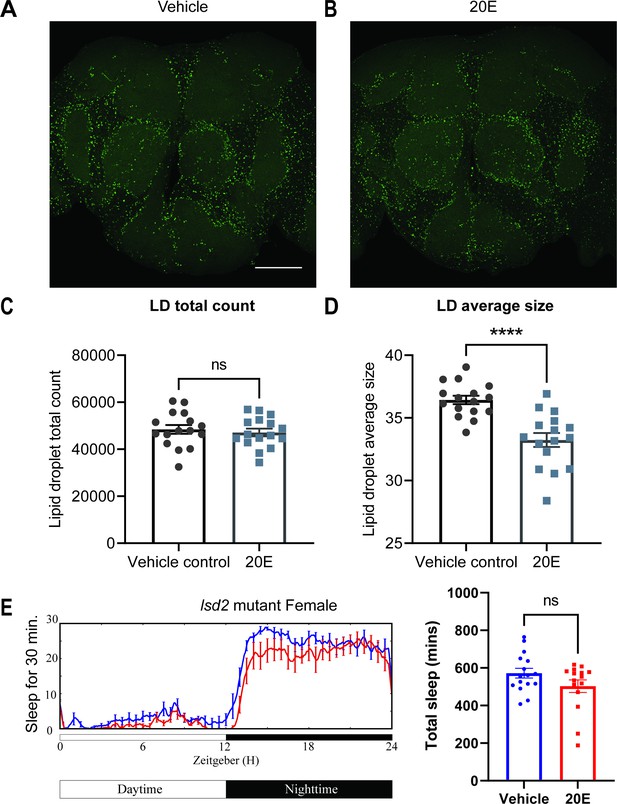
Lipid metabolism mediates the effects of 20E on sleep.
(A, B) Lipid droplet (LD) staining of representative brains from flies treated with vehicle or 0.5 mM 20E. A z-stack slice that shows the maximal structure of the cortex glia was selected, and brightness and contrast were auto-adjusted by ImageJ for better visualization. LDs are stained by the lipophilic dye BODIPY 493. (C, D) 0.5 mM 20E treatment does not significantly affect the total count of LDs but likely mobilizes lipids to lead to smaller LDs. LD count and size were analyzed and calculated using ImageJ as detailed in the Materials and methods. N = 16 per group from two independent repeats. Statistical comparisons used unpaired parametric Student’s t-test. Bar graphs show mean ± standard error of the mean (SEM) and ns = not significant, p > 0.05, ****p < 0.0001. (E) A representative sleep trace of lsd2 mutant flies with vehicle control or 0.2 mM ecdysone. As previously reported, total sleep increases with 20E treatment in wCS,but the sleep-promoting effect of the 20E is lost in the lsd2 mutant flies, N = 16 per group from two separate experiments. Statistical comparisons used unpaired parametric Student’s t-test. See also Figure 6—source data 1.
-
Figure 6—source data 1
The effects of ecdysone on lipid droplets and sleep.
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Lipid droplets changes resulting from glial ecdysone receptor (EcR) knockdown.
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig6-data2-v2.xlsx
-
Figure 6—source data 3
Sleep phenotypes of gaboxadol treatment in lsd-2 mutant flies.
- https://cdn.elifesciences.org/articles/81723/elife-81723-fig6-data3-v2.xlsx
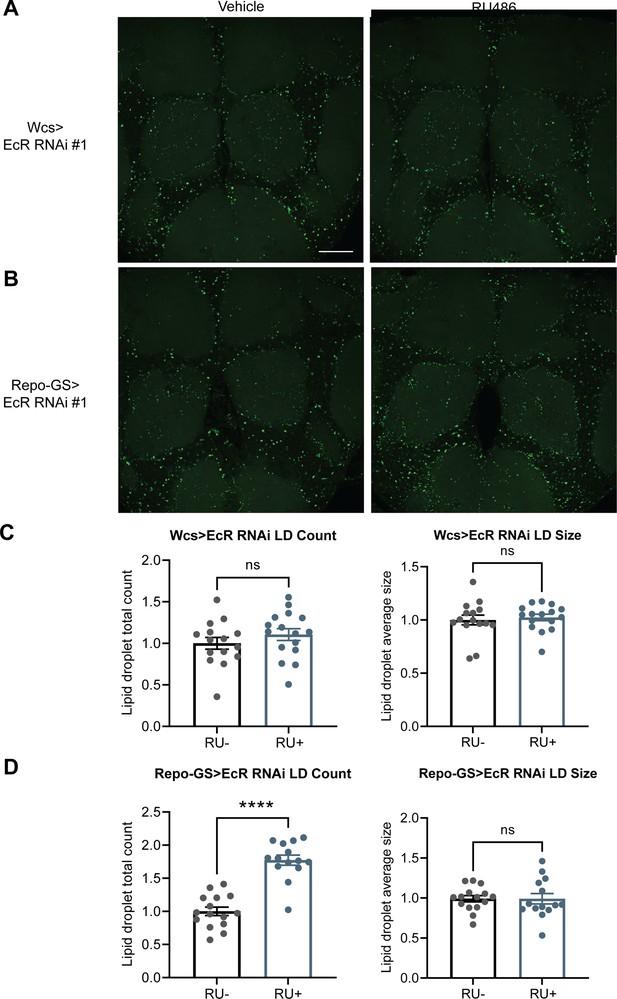
EcR knockdown in glial cells resulted in more LDs.
(A, B) Lipid droplet (LD) staining of representative brains from UAS-EcR RNAi #1 control flies and Repo-GS>EcR RNAi #1 flies treated with vehicle or RU486. A z-stack slice that shows the maximal structure of the cortex glia was selected, and brightness and contrast were auto-adjusted by ImageJ for better visualization. LDs are stained by the lipophilic dye BODIPY 493. (C, D) Both LDs count and size are not significantly different between vehicle- and RU486-treated UAS-EcR RNAi #1 control flies, but LDs counts are significantly higher in the RU+Repo-GS>EcR RNAi #1 flies compared with RU− control group. N = 14–16 per group from two independent repeats. Statistical comparisons used unpaired parametric Student’s t-test. Bar graphs show mean + standard error of the mean (SEM) and ns = not significant, p > 0.05, ****p < 0.0001. See also Figure 6—source data 2.
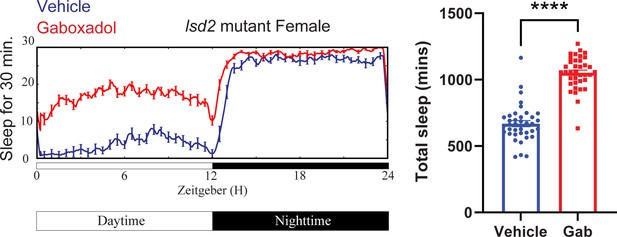
Gaboxadol promotes sleep in lsd-2 mutant flies.
A representative sleep trace of lsd-2 mutant flies with vehicle control or 0.1 mg/ml gaboxadol is shown in the left panel and quantification of all data points are shown on the right. N = 37–38 per group from three separate experiments. Statistical comparisons used unpaired parametric Student’s t-test. Bar graphs show mean + standard error of the mean (SEM) and ns = not significant, p > 0.05, ****p < 0.0001. See also Figure 6—source data 3.
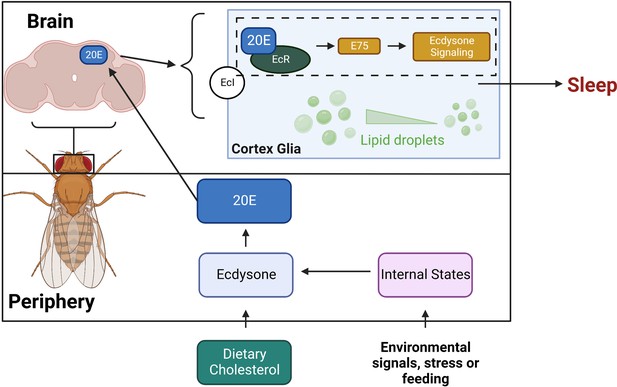
Model for sleep function of ecdysone in cortex glia.
This schematic shows how ecdysone acts as a long-distance signal to affect sleep in cortex glia. Ecdysone synthesis genes are not expressed in the fly brain, suggesting that glial ecdysone comes from the periphery. Glial knockdown of ecdysone receptor (EcR) and its downstream target E75 reduces sleep. Ecdysone promotes sleep by mobilizing lipid droplets stored mainly in glia, and knockdown of glial EcR results in more lipid droplets.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | w[*]; P{w[+mW.hs]=Switch2}GSG2326 | Bloomington Stock Center | BDSC:40990 | |
| Genetic reagent (D. melanogaster) | w[*]; P{y[+t7.7] w[+mC]=UAS-Eip78C.miRNA}attP16/CyO | Bloomington Stock Center | BDSC:44390 | |
| Genetic reagent (D. melanogaster) | w[*]; P{y[+t7.7] w[+mC]=UAS Hr3.miRNA}attP16 | Bloomington Stock Center | BDSC:44399 | |
| Genetic reagent (D. melanogaster) | w[*]; P{y[+t7.7] w[+mC]=UAS Hr96.miRNA}attP16 | Bloomington Stock Center | BDSC:44395 | |
| Genetic reagent (D. melanogaster) | w[*]; P{y[+t7.7] w[+mC]=UAS-Hnf4.miRNA}attP16/CyO | Bloomington Stock Center | BDSC:44398 | |
| Genetic reagent (D. melanogaster) | y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02546}attP2 | Bloomington Stock Center | BDSC:27258 | |
| Genetic reagent (D. melanogaster) | y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMS01620}attP2 | Bloomington Stock Center | BDSC:36729 | |
| Genetic reagent (D. melanogaster) | w[*]; P{y[+t7.7] w[+mC]=UAS Hr78.miRNA}attP16 | Bloomington Stock Center | BDSC:44393 | |
| Genetic reagent (D. melanogaster) | y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02545}attP2 | Bloomington Stock Center | BDSC:27242 | |
| Genetic reagent (D. melanogaster) | y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMS01316}attP2 | Bloomington Stock Center | BDSC:34329 | |
| Genetic reagent (D. melanogaster) | y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMS01951}attP2 | Bloomington Stock Center | BDSC:39032 | |
| Genetic reagent (D. melanogaster) | y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02537}attP2 | Bloomington Stock Center | BDSC:29373 | |
| Genetic reagent (D. melanogaster) | y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMS02272}attP40 | Bloomington Stock Center | BDSC:41707 | |
| Genetic reagent (D. melanogaster) | w[*]; P{y[+t7.7] w[+mC]=UAS Hr83.miRNA}attP16 | Bloomington Stock Center | BDSC:44397 | |
| Genetic reagent (D. melanogaster) | w[*]; P{y[+t7.7] w[+mC]=UAS svp.miRNA}attP16 | Bloomington Stock Center | BDSC:44394 | |
| Genetic reagent (D. melanogaster) | w[*]; P{y[+t7.7] w[+mC]=UAS ERR.miRNA}attP16/CyO | Bloomington Stock Center | BDSC:44391 | |
| Genetic reagent (D. melanogaster) | w[*]; P{y[+t7.7] w[+mC]=UAS Hr38.miRNA}attP16/CyO | Bloomington Stock Center | BDSC:44396 | |
| Genetic reagent (D. melanogaster) | y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02738}attP2 | Bloomington Stock Center | BDSC:27659 | |
| Genetic reagent (D. melanogaster) | y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMS00019}attP2/TM3, Sb[1] | Bloomington Stock Center | BDSC:33625 | |
| Genetic reagent (D. melanogaster) | y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMS00018}attP2/TM3, Sb[1] | Bloomington Stock Center | BDSC:33624 | |
| Genetic reagent (D. melanogaster) | y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02432}attP2 | Bloomington Stock Center | BDSC:27086 | |
| Genetic reagent (D. melanogaster) | w[*]; P{y[+t7.7] w[+mC]=UAS Hr4.miRNA}attP16 | Bloomington Stock Center | BDSC:44392 | |
| Genetic reagent (D. melanogaster) | y[1] w[*]; PBac{y[+mDint2] w[+mC]=I-SceI(FRT.Rab9-GAL4.ATG(loxP.3xP3-RFP))}VK00033/TM3, Sb[1] | Bloomington Stock Center | BDSC:51587 | |
| Genetic reagent (D. melanogaster) | w;; Repo-Gal4, 6 x crossed to Iso | Bloomington Stock Center | BDSC:7415 | |
| Genetic reagent (D. melanogaster) | w[1118]; UAS-EcR.A.dsRNA/TM3 | Bloomington Stock Center | BDSC:9328 | |
| Genetic reagent (D. melanogaster) | w[1118]; P{w[+mC]=UAS EcR.B1.dsRNA}168 | Bloomington Stock Center | BDSC:9329 | |
| Genetic reagent (D. melanogaster) | w[*]; P{w[+mC]=UAS EcR.A.F645A}TP2 | Bloomington Stock Center | BDSC:9452 | |
| Genetic reagent (D. melanogaster) | y1 v1; P{TRiP.JF02257}attP2 | Bloomington Stock Center | BDSC:26717 | |
| Genetic reagent (D. melanogaster) | w[1118]; P{w[+mC]=hs-GAL4-EcR.LBD}SBM | Bloomington Stock Center | BDSC:23656 | |
| Genetic reagent (D. melanogaster) | w; UAS-GFP-Lsd2 | Michael Welte | ||
| Genetic reagent (D. melanogaster) | w;; UAS-EcI RNAi | Naoki Yamanaka | BDSC:37295 | |
| Genetic reagent (D. melanogaster) | w1118; P{GD1434}v44851 | VDRC Stock Center | VDRC:44851 | |
| Genetic reagent (D. melanogaster) | w1118; P{GD1428}v37058 | VDRC Stock Center | VDRC:37058 | |
| Genetic reagent (D. melanogaster) | w1118; P{GD1428}v37059 | VDRC Stock Center | VDRC:37059 | |
| Genetic reagent (D. melanogaster) | UAS-E75 RNAi GD | VDRC Stock Center | VDRC:44851 | |
| Genetic reagent (D. melanogaster) | UAS-E75 RNAi KK | VDRC Stock Center | VDRC:108399 | |
| Genetic reagent (D. melanogaster) | white Canton-S | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | nSyb-GS74 | Laboratory Stocks | PMID: 29590612 | |
| Genetic reagent (D. melanogaster) | Repo-GS11 | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | Actin-GS | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w[*]; P{w[+mW.hs]=Switch2}GSG5961 | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w; 9–137 Gal4; TubGal80ts | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w; MZ0708-Gal4; TubGal80ts | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w; Eaat1-Gal4; TubGal80ts | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w; TubGal80ts; NP222-Gal4 | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w; NP2222-Gal4 | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w; GMR85601-Gal4 | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w; Eaat1-Gal4;;, 6 x crossed to Iso | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w; GMR56F03-Gal4 | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w; NP6293-Gal4, 6 x crossed to Iso | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w; Alarm-Gal4 | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w; 9–137 Gal4; 6 x crossed to Iso | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w; MZ0709-Gal4 | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w; moody-Gal4, 6 x crossed to Iso | Laboratory Stocks | ||
| Genetic reagent (D. melanogaster) | w; TublinGal80ts/CYO; Repo-Gal4/TM6 | Laboratory Stocks | ||
| Antibody | EcR Antibody DDA2.7 (mouse monoclonal) | Developmental Studies Hybridoma Bank (DSHB) | DDA2.7 (EcR common) | 2 ug/ml |
| Sequence-based reagent | E74_FOR | This paper, Integrated DNA Technologies | PCR primers | TGA GAC GCG AGG AAT ACC CTG GAC |
| Sequence-based reagent | E74_REV | This paper, Integrated DNA Technologies | PCR primers | AAC TGC CAG CGT GTA GCC GTT TCC |
| Sequence-based reagent | E75A_FOR | This paper, Integrated DNA Technologies | PCR primers | TCA GCA GGC CAA TCT GCA CCA CTC |
| Sequence-based reagent | E75A_REV | This paper, Integrated DNA Technologies | PCR primers | TGA TGT ACT CGG GAG TCT GGG GAC |
| Sequence-based reagent | E75B_FOR | This paper, Integrated DNA Technologies | PCR primers | AGC AGC ACC AGC ACC AGC AAC AAC |
| Sequence-based reagent | E75B_REV | This paper, Integrated DNA Technologies | PCR primers | ATT GCC CGC ACT GGA GTT GCT CGA |
| Sequence-based reagent | EcI_FOR | This paper, Integrated DNA Technologies | PCR primers | TGC AGT GCC GCT CTC AAC TGT ACC |
| Sequence-based reagent | EcI_REV | This paper, Integrated DNA Technologies | PCR primers | TCA CAG TAA CCG TTG ACC GCC TCC |
| Sequence-based reagent | EcR_C_FOR | This paper, Integrated DNA Technologies | PCR primers | TCA ACC ACA GCC ACA GCT CCT TCC |
| Sequence-based reagent | EcR_C_REV | This paper, Integrated DNA Technologies | PCR primers | TGA TGG GTC CTA TGG CCG CAC TTC |
| Sequence-based reagent | EcR_1_FOR | This paper, Integrated DNA Technologies | PCR primers | GCG GCC AAG ACT TTG TTA AG |
| Sequence-based reagent | EcR_1_REV | This paper, Integrated DNA Technologies | PCR primers | GGC CAA CTG ATT GTA CGT TAA G |
| Sequence-based reagent | EcR_2_FOR | This paper, Integrated DNA Technologies | PCR primers | GCC ATC TGA AGA GGA TCT CAG |
| Sequence-based reagent | EcR_2_REV | This paper, Integrated DNA Technologies | PCR primers | AAC GCT GGT AGA CCT TTA GC |
| Commercial assay or kit | 20-Hydroxyecdysone EIA kit | Cayman | Item No. 501390 | |
| Commercial assay or kit | RNeasy Plus Mini Kit | Qiagen | Item No. 74134 | |
| Chemical compound, drug | BODIPY 493/503 | Fisher | D3922 | 1 ug/ml |
| Chemical compound, drug | 20-Hydroxyecdysone | Sigma | H5142 | 0.5 mM |
| Chemical compound, drug | Gaboxadol | Sigma-Aldrich | T101 | 0.1 mg/ml |
| Software, algorithm | Clocklab | Actimetrics | https://actimetrics.com/ | |
| Software, algorithm | Adobe Illustrator 2020 | Adobe | https://www.adobe.com/ | |
| Software, algorithm | BioRender | BioRender | App.biorender.com | |
| Software, algorithm | Pysolo | Gilestro and Cirelli, 2009 | https://www.pysolo.net/about/ | |
| Software, algorithm | GraphPad Prism v9 | GraphPad Software | https://www.graphpad.com/ | |
| Software, algorithm | JTK_CYCLE | Hughes et al., 2010 | Hughes et al., 2010 | |
| Software, algorithm | DAMFileScan113 | Trikinetics | https://trikinetics.com/ |





