Interdependent progression of bidirectional sister replisomes in E. coli
Figures
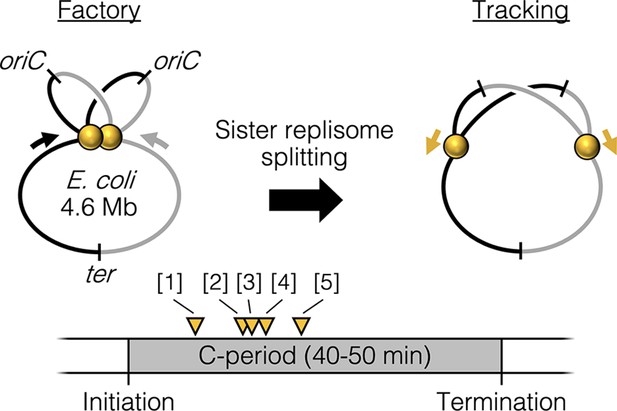
Replisome splitting.
Under slow growth conditions, E. coli bidirectional sister replisomes (yellow spheres) are initially colocalized, then separate part way through the replication period, marking a transition from factory to tracking configurations. Yellow triangles indicate approximate replisome splitting times within the replication phase (C-period) from previous studies: [1] Reyes-Lamothe et al., 2008, [2] Japaridze et al., 2020, [3] Hiraga et al., 2000, [4] Onogi et al., 2000, and [5] Bates and Kleckner, 2005.
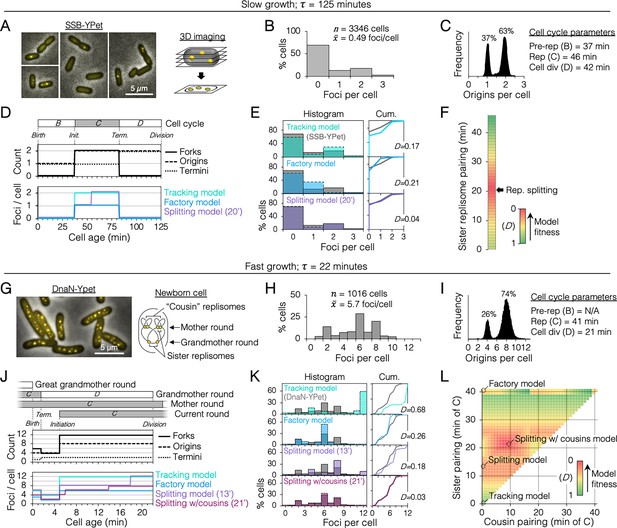
Sister replisome association under slow and fast growth conditions.
(A) Three-dimensional (3D) replisome imaging. E. coli cells with an SSB-YPet tag were grown to mid-exponential phase in M9 minimal media at 37°C then imaged at 0.2 µm z-plane increments, deconvoluted, and combined into a single maximum intensity projection (MIP). (B) Distribution of resolvable SSB-YPet foci. (C) Origins per cell measured by flow cytometry after RIF runout (n = 30,000 cells). Cell cycle parameters are shown. (D) Cell cycle schematic with three models of replisome pairing. Top panel indicates the number of replication forks, origins, and termini per cell as a function of cell age as determined by flow cytometry (Materials and methods). Lower panel indicates the expected number of replisome foci per cell from three models: no replisome pairing (Tracking model), full-time replisome pairing (Factory model), and 20-min transitory replisome pairing (Splitting model). (E) Fitness of replisome modeling. (Left) Overlay of theoretical (colored bars) and SSB-YPet (gray bars) foci per cell histograms. (Right) Kolmogorov–Smirnov (KS) cumulative plots with dissimilarity indexes indicating the difference between modeled and measured data. (F) Heatmap of KS dissimilarity indexes obtained with varying replisome pairing times for the Splitting model. The optimal pairing time (20 min, arrow) is indicated. (G–L) As in (A–F) except cells were grown in Luria-Bertani (LB) media at 37°C and replisomes were tagged with DnaN-YPet. Graphical representation of a newborn cell under fast growth conditions with multiple rounds of replication initiated in prior cell generations (G, right panel). Optimal (lowest D) replisome pairing times under fast growth conditions for Splitting (13 min) and Splitting with cousins (21 min) models are indicated (L).
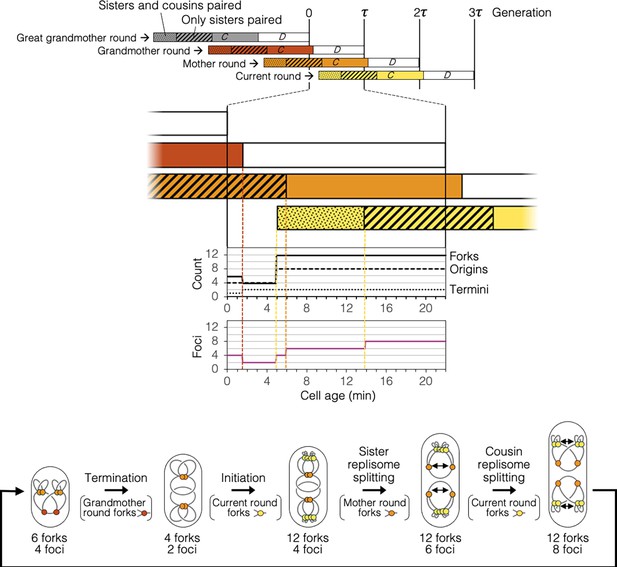
Timeline and spatial dynamics of replication under the Splitting with cousins model.
Arrangement of replisomes and DNA in cell drawings are speculative and are based solely on measurements of genomic content and number of DnaN-YPet foci per cell (Figure 2H, I).
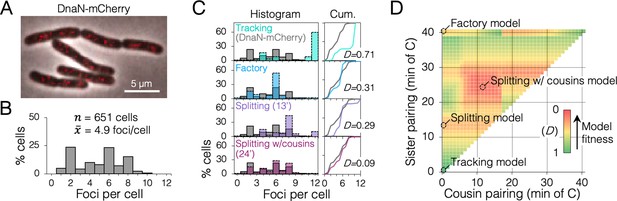
Replisome dynamics under fast growth conditions using a monomeric DnaN-mCherry fusion protein.
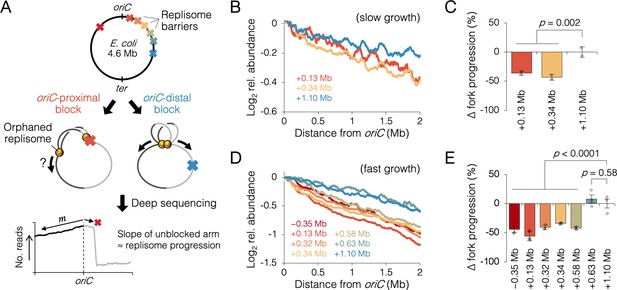
Measuring the effect of replisome pairing on fork progression by whole-genome sequencing.
(A) Experimental approach. Strains carrying an inducible tetO array replication barrier (top) are roadblocked and progression of the unblocked orphaned replisome is quantified by sequencing. Hypothetically, oriC-proximal roadblocks, but not oriC-distal roadblocks, will negatively affect progression of the unblocked replisome. (B) DNA sequencing profiles along the unblocked chromosome arm under slow growth conditions. Values indicate the number of sequencing reads per kilobase relative to oriC, converted to log2 to show fold changes in copy number from oriC. (C) Relative progression of the unblocked replisome under slow growth conditions. Fork progression was quantified as slope−1 of raw sequencing profiles (B), relative to the most oriC-distal roadblock strain (+1.10 Mb). (D, E) As above (B–D) except under fast growth conditions and with additional roadblocks as indicated. Profiles are 100 kb moving average of the means of two to five independent experiments. Error bars are ±1 standard deviation (s.d.); two-tailed t-test.
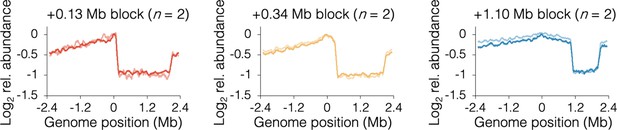
Individual sequencing profiles under slow growth conditions.
Profiles along the blocked chromosome arm are approximately flat between the tetO array and the most distal ter site, which terminates replication emanating from the unblocked arm. Profile noise is a consequence of low copy number under slow growth conditions (see Figure 3—figure supplement 2 for comparison). Strains are DB2185 (+0.13 Mb block), DB2193 (+0.34 Mb block), and DB2146 (+1.10 Mb block).
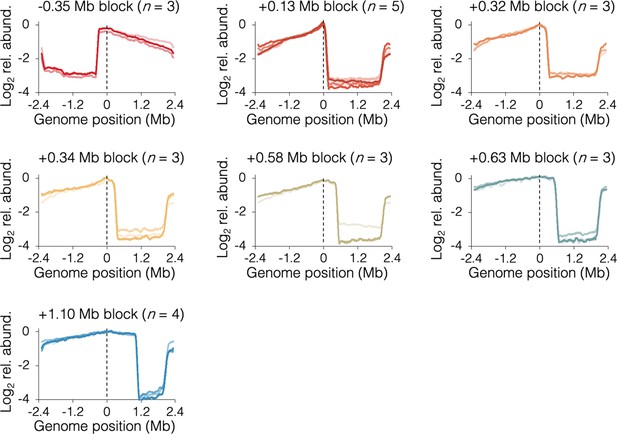
Individual sequencing profiles under fast growth conditions.
Slight slope on the blocked chromosome arm downstream of the tetO array indicates partial ‘read-through’ of opposite arm forks through the terminus region, eventually stopping at the tetO array. Strains are DB3216 (−0.35 Mb block), DB2185 (+0.13 Mb block), DB3198 (+0.32 Mb block), DB2193 (+0.34 Mb block), DB3264 (+0.58 Mb block), DB3195 (+0.63 Mb block), and DB2146 (+1.10 Mb block).
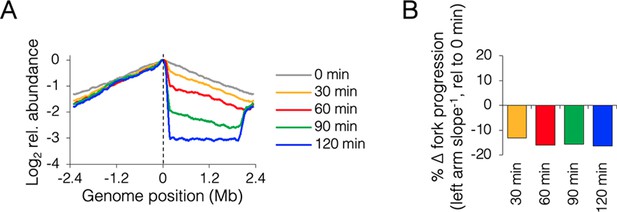
Time course of roadblocking under fast growth conditions.
(A) Sequencing profiles for +0.13 Mb roadblock (DB2185) every 30 min after TetR-YFP induction (n = 1). (B) Change in progression of the unblocked (left) replisome after roadblocking relative to time zero, showing little change in progression after 60 min.
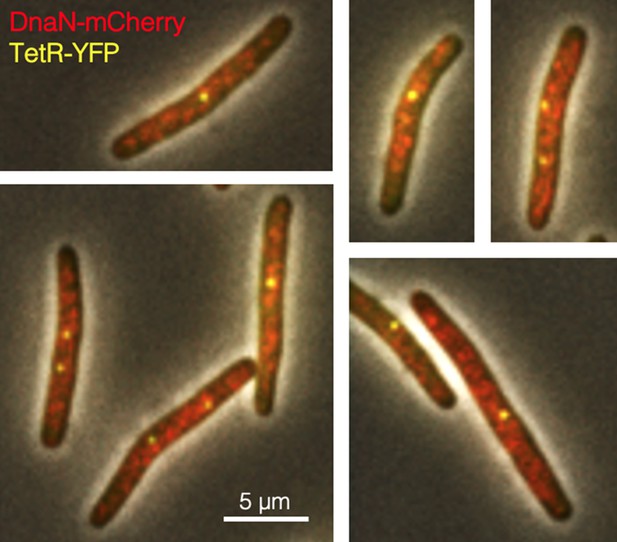
Imaging replisomes in roadblocked cells under fast growth conditions.
DB2972 cells were roadblocked at +0.13 Mb from oriC for 2 hr and imaged for replisomes (red, DnaN-mCherry) and the roadblock (yellow, TetR-YFP). Multiple dispersed replisome foci rarely colocalizing with the roadblock suggests that unblocked replisomes were unattached to any residual replication factory structure.
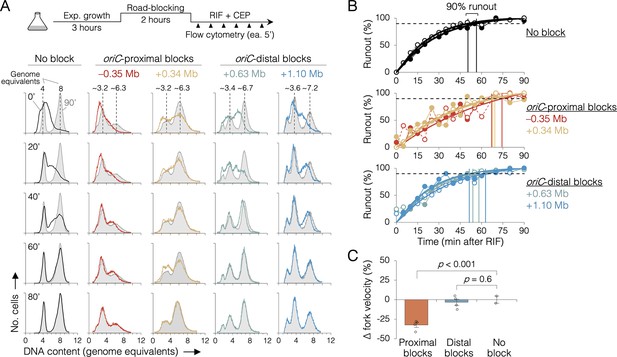
The effect of replisome pairing on replication fork velocity.
(A) Replication elongation measured as a change in cellular DNA content over time after inhibiting replication initiation with rifampicin (RIF) and cell division with cephalexin (CEP). (Top) Experimental procedure. Representative DNA histograms of runout samples in wild-type MG1655 cells (no block) or cells with one replisome blocked proximally or distally to the origin are shown. DNA contents were measured by flow cytometry every 5 min for 90 min after rifampicin treatment (20-min intervals shown) with 90 min time points (light gray) overlaid for comparison. (B) Completion of replication (% runout) quantified by KS dissimilarity analysis between DNA histograms at each time point and at 90 min. Exponential regressions and times at 90% runout (vertical lines) are shown. (C) Change in fork velocity relative to unblocked cells. Values are average time to 90% runout (n = 2 replicates per strain) as determined in (B) relative to unblocked cells. Error bars are ±1 standard deviation (s.d.); two-tailed t-test. All cells were grown under fast growth conditions.
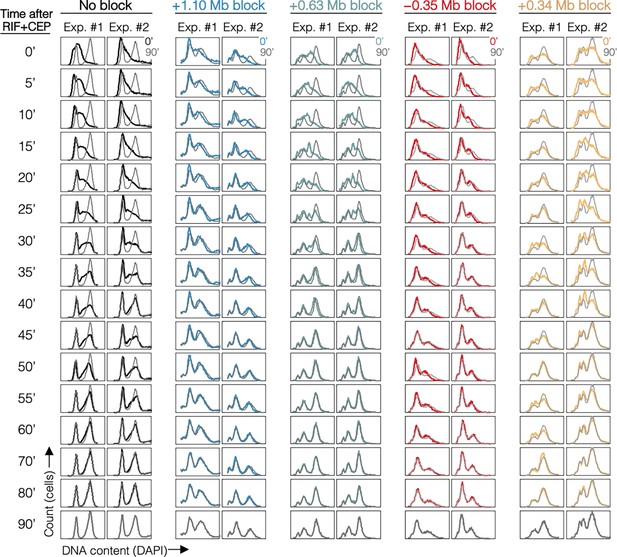
DNA histograms for all samples.
DNA histograms are shown at 5-min intervals with 90-min time point overlayed (gray). Strains are MG1655 (No block), DB2146 (+1.10 Mb block), DB3195 (+0.63 Mb block), DB3216 (−0.35 Mb block), and DB2193 (+0.34 Mb block).
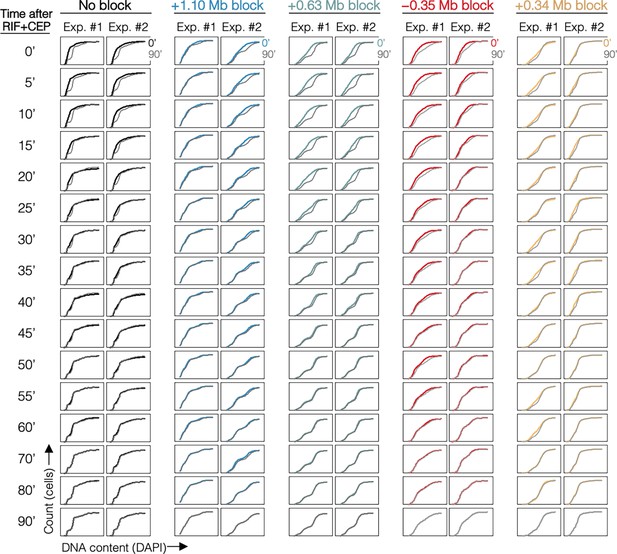
Cumulative curve plots for all samples.
Cumulative curves of DNA histograms (Figure 4—figure supplement 1) with 90-min time point overlayed (gray).
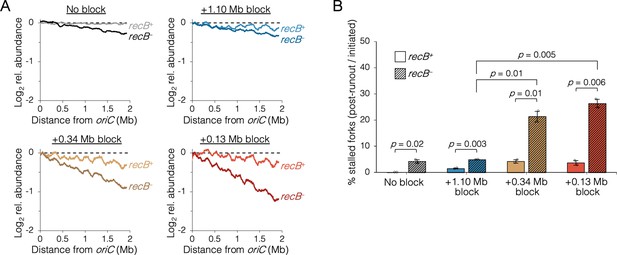
The requirement of RecB-dependent fork restart in uncoupled replisomes.
(A) Raw sequencing profiles of the unblocked chromosome arm in recB+ and recB− cells after rifampicin runout. Strains contained either no block (wild-type MG1655 or its recB− derivative), an oriC-distal block at +1.10 Mb, or an oriC-proximal block at +0.34 Mb or +0.13 Mb as indicated (n = 2 replicates per strain). Dashed lines indicate the theoretical DNA profile after complete (100%) runout. (B) The frequency of stalled forks relative to unblocked recB+ . Stalled forks are quantified as the number of remaining forks on the unblocked arm after rifampicin runout (Materials and methods). Cells were grown under fast growth conditions. Error bars are ±1 standard deviation (s.d.); two-tailed t-test.
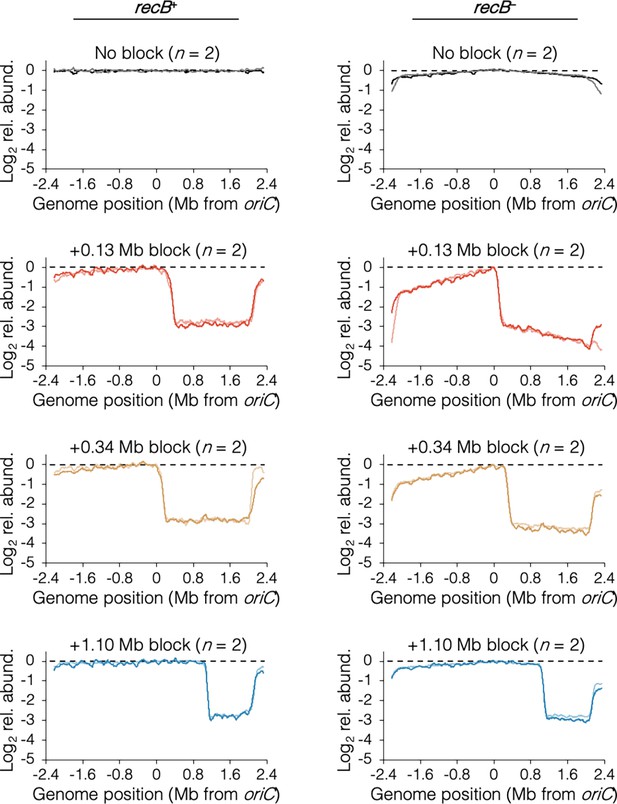
Individual sequencing profiles after rifampicin runout.
Data as described in Figure 5A. Strains are MG1655 (recB+, No block), DB2257 (recB−, No block), DB2185 (recB+, +0.13 Mb block), DB2403 (recB−, +0.13 Mb block), DB2193 (recB+, +0.34 Mb block), DB2722 (recB−, +0.34 Mb block), DB2146 (recB+, +1.10 Mb block), and DB2725 (recB−, +1.10 Mb block).
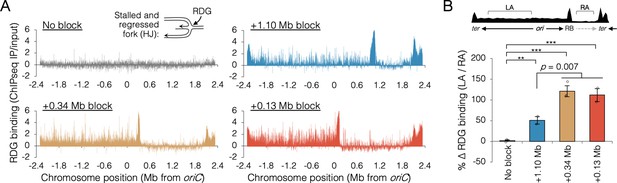
Holiday junction frequency on DNA replicated by uncoupled replisomes.
(A) Genome binding of the Holliday junction protein RuvCDef-GFP (RDG) by chromatin immunoprecipitation and sequencing (ChIP-seq). RDG-expressing cells either contained no roadblock (DB2945) or were roadblocked at position +0.13, +0.34, or +1.10 Mb as indicated. Chromatin immunoprecipitation was performed using RuvC antibody. Log2 ChIP-seq reads (pull down/input) values >0 indicate RDG enrichment. (B) Percent change in RDG binding on the unblocked chromosome arm. Values are average RDG binding on the left arm between −2 Mb and oriC (region LA) divided by average RDG binding on the right arm (region RA). All cells were grown under fast growth conditions (n = 2–3). Error bars are ±1 standard deviation (s.d.); ***P < 0.001; **P < 0.01; two-tailed t-test.
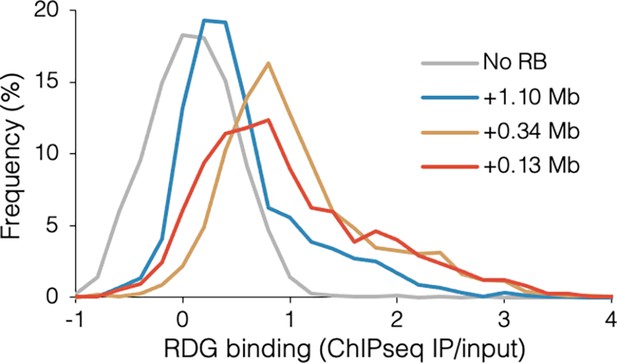
Distribution of RuvCDef-GFP (RDG) binding.
Frequency indicates the percentage of RDG chromatin immunoprecipitation and sequencing (ChIP-seq) IP/input values in 0.2-increment bins for wild-type DB2945 (No RB), DB3124 (+1.10 Mb blocked), DB2956 (+0.34 Mb blocked), and DB2954 (+0.13 Mb blocked).
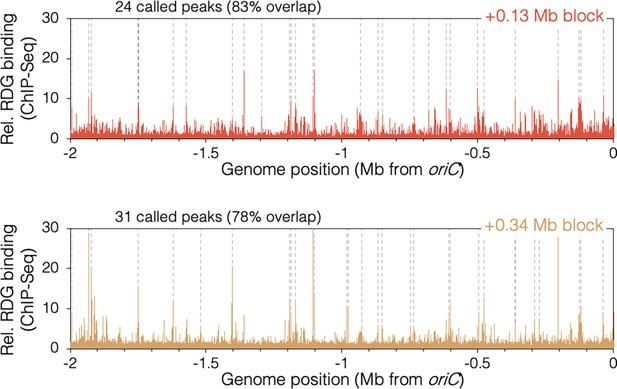
RuvCDef-GFP (RDG)-binding peaks in oriC-proximal roadblocked cells.
Unblocked chromosome arm is shown between −2 Mb and oriC. Peaks (dashed lines) are locations of RDG signal ≥4 standard deviation (s.d.) above the 100 kb local median.
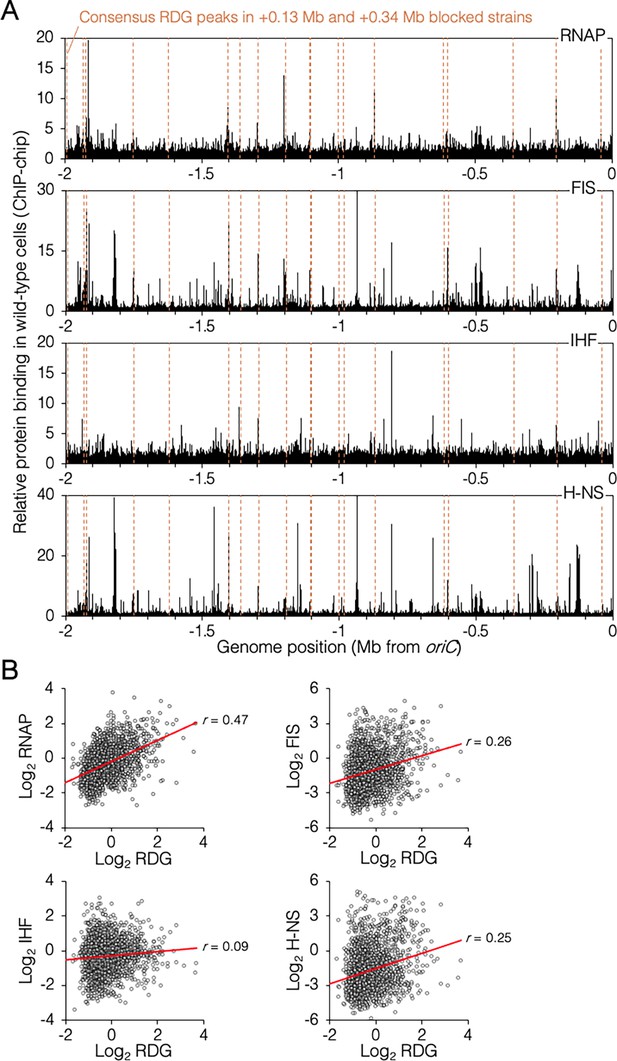
Genomic binding of RuvCDef-GFP (RDG) and major nucleoid proteins.
(A) Nucleoid protein-binding profiles (black) and consensus RDG-binding peaks (dashed lines). (B) Correlation plots of nucleoid protein and RDG binding (average +0.13 Mb, +0.34 Mb blocked cells). All data are along the unblocked chromosome arm only, between oriC and −2 Mb.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | AB1157 dnaN-mCherry FRTKanFRT | Moolman et al., 2014 | BN1682 | |
| Strain, strain background (E. coli) | AB1157 dnaN-ypet FRT | This paper | DB1568 | RRL190 flipped to KmS w/pCP20 |
| Strain, strain background (E. coli) | MG1655 141x tetO array @ lac, pDM15 (+1.10 Mb block) | Joshi et al., 2011 | DB2146 | |
| Strain, strain background (E. coli) | MG1655 141x tetO array @ glnA, pDM15 (+0.13 Mb block) | Joshi et al., 2011 | DB2185 | |
| Strain, strain background (E. coli) | MG1655 141x tetO array @ dnaB, pDM15 (+0.34 Mb block) | Joshi et al., 2011 | DB2193 | |
| Strain, strain background (E. coli) | MG1655 ΔrecB FRTKanFRT | This paper | DB2257 | MG1655 × P1.JW2788 |
| Strain, strain background (E. coli) | MG1655 141x tetO array @ gln, ΔrecB FRTKanFRT, pDM15 (+0.13 Mb block) | This paper | DB2403 | DB2185 × P1.JW2788 |
| Strain, strain background (E. coli) | MG1655 141x tetO array @ dnaB, ΔrecB FRTKanFRT, pDM15 (+0.34 Mb block) | This paper | DB2722 | DB2193 × P1.JW2788 |
| Strain, strain background (E. coli) | MG1655 141x tetO array @ lac, ΔrecB FRTKanFRT, pDM15 (+1.10 Mb block) | This paper | DB2725 | DB2146 × P1.JW2788 |
| Strain, strain background (E. coli) | MG1655 dnaN-mCherry FRTKanFRT | This paper | DB2943 | MG1655 × P1.BN1682 |
| Strain, strain background (E. coli) | MG1655 λcIts857 PR::ruvCDef-gfp FRTKanFRT (42°C inducible RDG cassette) | This paper | DB2945 | SMR19379 recombineered with primers P1/P2; P3/P4 |
| Strain, strain background (E. coli) | MG1655 141x tetO array @ gln, λcIts857 PR::ruvCDef-gfp FRTKanFRT, pDM15 (+0.13 Mb block, RDG) | This paper | DB2954 | DB2185 × P1.DB2945 |
| Strain, strain background (E. coli) | MG1655 141x tetO array @ dnaB, λcIts857 PR::ruvCDef-gfp FRTKanFRT, pDM15 (+0.34 Mb block, RDG) | This paper | DB2956 | DB2193 × P1.DB2945 |
| Strain, strain background (E. coli) | MG1655 dnaN-mCherry FRT | This paper | DB2962 | DB2943 flipped to KmS w/pCP20 |
| Strain, strain background (E. coli) | MG1655 141x tetO array @ glnA, pDM15 (+0.13 Mb block) dnaN-mCherry FRTKanFRT | This paper | DB2970 | DB2185 × P1.BN1682 |
| Strain, strain background (E. coli) | MG1655 141x tetO array @ glnA, pDM15 (+0.13 Mb block) dnaN-mCherry FRT | This paper | DB2972 | DB2970 flipped to KmS w/pCP20 |
| Strain, strain background (E. coli) | BW25113 yjiC::141x tetO array FRT | This paper | DB3094 | JW4288 × pJZ087 & pCP20 |
| Strain, strain background (E. coli) | BW25113 yjbL::141x tetO array FRT | This paper | DB3096 | JW4007 × pJZ087 & pCP20 |
| Strain, strain background (E. coli) | MG1655 141x tetO array @ lac, λcIts857 PR::ruvCDef-gfp FRTKanFRT, pDM15 (+1.10 Mb block, RDG) | This paper | DB3124 | DB2146 × P1.DB2945 |
| Strain, strain background (E. coli) | MG1655 yjiC::141x tetO array FRT | This paper | DB3191 | MG16556 × P1.DB3094 |
| Strain, strain background (E. coli) | MG1655 yjbL::141x tetO array FRT | This paper | DB3193 | MG1655 × P1.DB3096 |
| Strain, strain background (E. coli) | MG1655 yjiC::141x tetO array FRT, pDM15 (+0.63 Mb block) | This paper | DB3195 | DB3191 × pDM15 |
| Strain, strain background (E. coli) | MG1655 yjbL::141x tetO array FRT pDM15 (+0.32 Mb block) | This paper | DB3198 | DB3193 × pDM15 |
| Strain, strain background (E. coli) | BW25113 yhgN::141x tetO array FRT | This paper | DB3212 | JW3397 × pJZ087 & pCP20 |
| Strain, strain background (E. coli) | MG1655 yhgN::141x tetO array FRT | This paper | DB3214 | MG1655 × P1.3212 |
| Strain, strain background (E. coli) | MG1655 yhgN::141x tetO array FRT, pDM15 (−0.35 Mb block) | This paper | DB3216 | DB3214 × pDM15 |
| Strain, strain background (E. coli) | BW25113 yjgZ::141x tetO array FRT | This paper | DB3258 | JW4236 × pJZ087 & pCP20 |
| Strain, strain background (E. coli) | MG1655 yjgZ::141x tetO array FRT | This paper | DB3262 | MG1655 × P1.DB3258 |
| Strain, strain background (E. coli) | MG1655 yjgZ::141x tetO array FRT, pDM15 (+0.58 Mb block) | This paper | DB3264 | DB3262 × pDM15 |
| Strain, strain background (E. coli) | BW25113 ΔrecB FRTKanFRT | Baba et al., 2006 | JW2788 | Keio collection |
| Strain, strain background (E. coli) | BW25113 ∆yhgN FRTKanFRT | Baba et al., 2006 | JW3397 | Keio collection |
| Strain, strain background (E. coli) | BW25113 ∆yjbL FRTKanFRT | Baba et al., 2006 | JW4007 | Keio collection |
| Strain, strain background (E. coli) | BW25113 ∆yjgZ FRTKanFRT | Baba et al., 2006 | JW4236 | Keio collection |
| Strain, strain background (E. coli) | BW25113 ∆yjiC FRTKanFRT | Baba et al., 2006 | JW4288 | Keio collection Baba et al., 2006 |
| Strain, strain background (E. coli) | AB1157dnaN-ypet FRTKanFRT | Reyes-Lamothe et al., 2010 | RRL190 | |
| Strain, strain background (E. coli) | AB1157 ssb-ypet::Kan | Reyes-Lamothe et al., 2008 | RRL32 | |
| Strain, strain background (E. coli) | MG1655 PN25tetO::ruvCDef-gfp FRT | Xia et al., 2016 | SMR19425 | Doxycycline-inducible RDG cassette |
| Antibody | anti-RuvC (mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-53437, RRID:AB_630213 | IP (1:1000) |
| Recombinant DNA reagent | pCP20 | Cherepanov and Wackernagel, 1995 | Flippase FLP ApR recombinase | |
| Recombinant DNA reagent | pJZ087 | Wang et al., 2019 | FRT-141x tetO GmR integration | |
| Recombinant DNA reagent | pKD46 | Datsenko and Wanner, 2000 | FRTKanFRT for RDG cassette | |
| Recombinant DNA reagent | pDM15 | Magnan and Bates, 2015a | PnahG::tetR-yfp CmR expression | |
| Sequence-based reagent | P1 | This paper | PCR primer | GGTATATCTCCTTCTTAAAGTTAAACAAAATTATTTCTAGAAGGGTTATGCGTTGTTCCA |
| Sequence-based reagent | P2 | This paper | PCR primer | GTTTCATGCTATGCCAAACGAGAATGATTATCAAATTCATGTGTAGGCTGGAGCTGCTTC |
| Sequence-based reagent | P3 | This paper | PCR primer | CCCTAATTCGATGAAGATTCTTGCTCAATTGTTATCAGCGTGTAGGCTGGAGCTGCTTC |
| Sequence-based reagent | P4 | This paper | PCR primer | AGACGTTTGGCTGATCGGCAAGGTGTTCTGGTCGGCGATTCCGGGGATCCGTCGACC |
| Commercial assay or kit | Nextera XT DNA Sample Preparation Kit | Illumina | Cat #: FC-131-1024 | |
| Commercial assay or kit | MiSeq Reagent Kit v3 (150-cycle) | Illumina | Cat #: MS-102-3001 | |
| Chemical compound, drug | Anhydrotetracycline hydrochloride | Sigma-Aldrich | Cat #: 37919 | |
| Chemical compound, drug | Sodium salicylate | Sigma-Aldrich | Cat #: S3007 | |
| Software, algorithm | FocusCounter | https://github.com/DavidBatesLab/Matlab-scripts.git; Joshi et al., 2013 | Matlab script | Counting replisome foci |
| Software, algorithm | FlowJo | FlowJo LLC | Quantifying flow cytometry histograms | |
| Software, algorithm | sequencingcompile.m | https://github.com/DavidBatesLab/Matlab-scripts.git | Matlab script | Extracting read coordinates from bam files |
| Software, algorithm | sequencingcompile2.m | https://github.com/DavidBatesLab/Matlab-scripts.git | Matlab script | Binning reads into 1 kb bins |





