Parallel evolution of reduced cancer risk and tumor suppressor duplications in Xenarthra
Figures
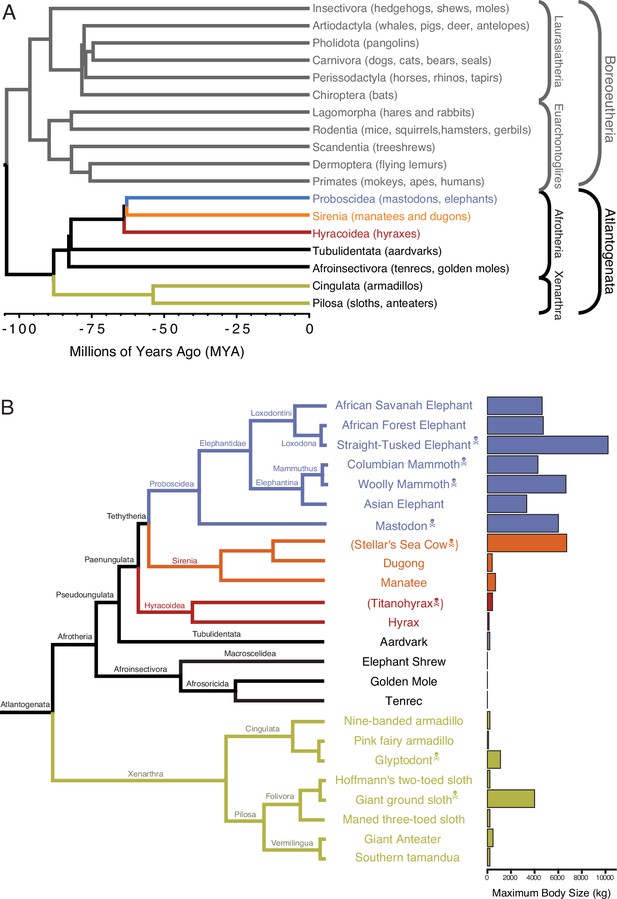
Eutherian phylogenetic relationships.
(A) Phylogenetic relationships between Eutherian orders, examples of each order are given in parenthesis. Horizontal branch lengths are proportional to time since divergence between lineages (see scale, Millions of Ago [MYA]). The clades Atlantogenata and Boreoeutheria are indicated, the order Proboscidea is colored blue, Sirenia is colored orange, and Hyracoidea is colored red. (B) Phylogenetic relationships of extant and recently extinct Atlantogenatans with available genomes are shown along with clade names and maximum body sizes. Note that horizontal branch lengths are arbitrary, species indicated with skull, and crossbones are extinct and do not have available nuclear genomes.
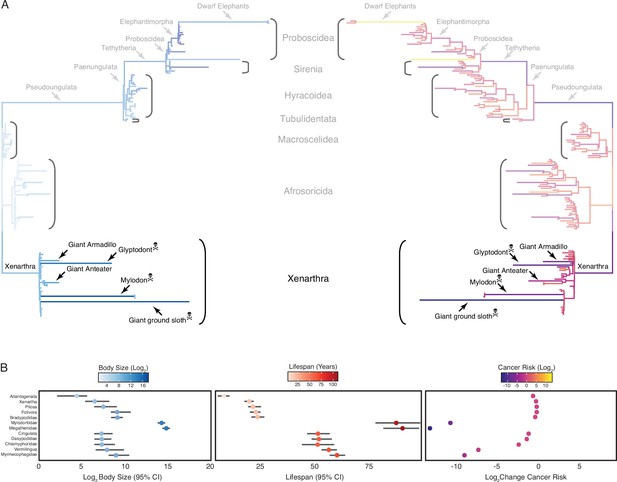
Convergent evolution of large-bodied, cancer-resistant Xenarthrans.
(A) Atlantogenata phylogeny, with branch lengths scaled by log2 change in body size (left) or log2 change in intrinsic cancer risk (right). Branches are colored according to ancestral state reconstruction of body mass or estimated intrinsic cancer risk (scale shown in panel B). The Afrotherian part of the tree is shown opaque because it was analyzed in Vazquez and Lynch, 2021. (B) Ancestral reconstructions of body size (left), lifespan (middle), and change in intrinsic cancer risk (right). Data are shown as mean (dot) along with 95% confidence interval (CI, whiskers) for body size and lifespan (which is estimated from body size data).

Pervasive duplication of tumor suppressors in Xenarthra.
(A) Atlantogenata phylogeny indicating the number of genes duplicated in each Xenarthran lineage inferred by maximum likelihood. Branch lengths are drawn proportional to the per gene duplication rate (see inset scale) and colored according to lineage relationships shown in Figure 1. (B) Word cloud of Reactome pathways in which Xenarthan (green) and Pilsoan (dark green) gene duplicates are enriched (FDR q≤0.25). Pathway term sizes are scaled according to the –log10 hypergeometric p-value of their enrichment (see inset scale).
-
Figure 3—source data 1
Genes duplicated in the stem-lineage of Xenarthra.
- https://cdn.elifesciences.org/articles/82558/elife-82558-fig3-data1-v2.txt
-
Figure 3—source data 2
Genes duplicated in the stem-lineage of Pilosa.
- https://cdn.elifesciences.org/articles/82558/elife-82558-fig3-data2-v2.txt
-
Figure 3—source data 3
Genes duplicated in the stem-lineage of Cingulata.
- https://cdn.elifesciences.org/articles/82558/elife-82558-fig3-data3-v2.txt
-
Figure 3—source data 4
Pathways in which genes duplicated in the stem-lineage of Xenarthra are enriched.
- https://cdn.elifesciences.org/articles/82558/elife-82558-fig3-data4-v2.zip
-
Figure 3—source data 5
Pathways in which genes duplicated in the stem-lineage of Pilosa are enriched.
- https://cdn.elifesciences.org/articles/82558/elife-82558-fig3-data5-v2.zip
-
Figure 3—source data 6
Pathways in which genes duplicated in the stem-lineage of Cingulata are enriched.
- https://cdn.elifesciences.org/articles/82558/elife-82558-fig3-data6-v2.zip
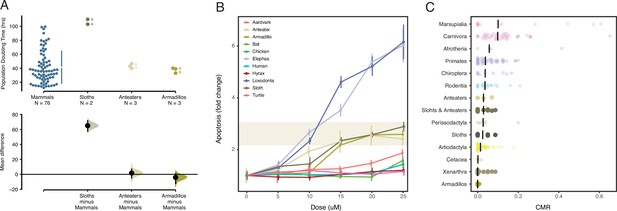
Xenarthrans have anticancer cellular phenotypes and a low prevalence of cancer.
(A) Population doubling time estimates for mammalian cells (upper) and mean differences (lower). Mean differences are depicted as a dot with 95% confidence intervals as vertical error bars; p values are reported from a two-sided permutation t-test with 5000 reshuffles of control and test labels. The unpaired mean difference between mammals and sloths is 65.1 (57.5–71.8), p=4.0×10–4. The unpaired mean difference between mammals and anteaters is 1.71 (5.0–7.16), p=0.905. The unpaired mean difference between mammals and armadillos is –4.1 (12.5–0.86), p=0.781. (B) Induction in apoptosis in vertebrate cells by mitomycin C. Data are shown as fold change in caspase 3/7 activity (apoptosis), mean (dot) and standard deviation (vertical line); n=12 for each data point. (C) Cancer prevalence data for each species (dot) grouped by clade and shown as jittered stripchart. The number of species per clade is: Afrotheria=5, Artiodactyla=68, Carnivora=43, Cetacea=4, Chiroptera=9, Marsupialia=13, Perissodactyla=6, Primates=47, Rodentia=18, anteaters=3, sloths=6, armadillos=5, and Xenarthra=14. Medians for each clade are shown with vertical black bars.
-
Figure 4—source data 1
Data plotted in Figure 4A.
- https://cdn.elifesciences.org/articles/82558/elife-82558-fig4-data1-v2.txt
-
Figure 4—source data 2
Data plotted in Figure 4B.
- https://cdn.elifesciences.org/articles/82558/elife-82558-fig4-data2-v2.txt
-
Figure 4—source data 3
Data plotted in Figure 4C.
- https://cdn.elifesciences.org/articles/82558/elife-82558-fig4-data3-v2.xls
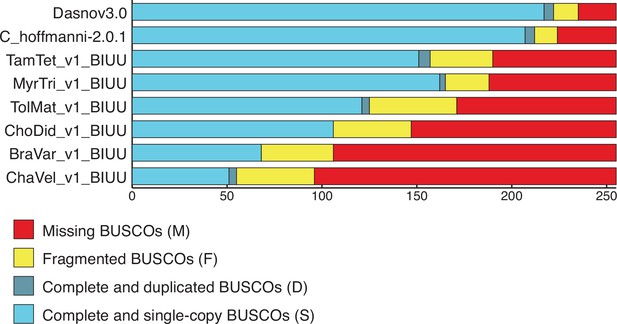
BUSCO scores for Xenarthran genomes used to estimate gene copy number.
BUSCO results from Galaxy (version 4.1.4) using the Eukaryota lineage. The number of complete and single copy (light blue), complete and duplicated (dark blue), fragmented (yellow), and missing (red) genes are shown. We note that the number of duplicated genes identified by BUSCO is inversely proportional to the number of BUSCO genes missing in each genome, suggesting duplicate gene counts in our data may be underrepresented.
Tables
Cancer prevalence in Xenarthrans.
| Common name | Species name | Necropsies | Neoplasia | Prevalence (95% CI) |
|---|---|---|---|---|
| Southern tamandua (combined) | Tamandua tetradactyla | 60 | 1 | 0.07 (0.0004–0.089) |
| Giant anteater (combined)* | Myrmecophaga tridactyla | 139 | 4 | 0.029 (0.008–0.072) |
| Silky anteater | Cyclopes sp. | 3 | 0 | 0 (0–0.708) |
| Maned three-toed sloth | Bradypus torquatus | 25 | 0 | 0 (0–0.137) |
| Pale-throated sloth | Bradypus tridactylus | 2 | 0 | 0 (0–0.841) |
| Brown-throated sloth | Bradypus variegatus | 8 | 0 | 0 (0–0.369) |
| Three-toed sloths (combined) | Bradypus sp. | 69 | 0 | 0 (0–0.050) |
| Linné’s two-toed sloth (combined) | Choloepus didactylus | 23 | 2 | 0.087 (0.011–0.28) |
| Two-toed sloths (combined) | Choloepus sp. | 40 | 2 | 0.050 (0.006–0.169) |
| Nine-banded armadillo (combined)* | Dasypus novemcinctus | 275 | 2 | 0.007 (0.0009–0.026) |
| Six-banded armadillo | Euphractus sexcinctus | 48 | 0 | 0 (0–0.074) |
| Naked-tailed armadillos | Cabassous sp. | 5 | 0 | 0 (0–0.522) |
| Three-banded armadillo | Tolypeutes sp. | 4 | 0 | 0 (0–0.602) |
| Giant armadillo | Priodontes maximus | 1 | 0 | 0 (0–0.975) |
| Vermilingua (anteaters)* | 202 | 5 | 0.023 (0.008–0.057) | |
| Folivora (sloths)* | 109 | 2 | 0.018 (0.002–0.065) | |
| Pilosa (sloths and anteaters)* | 311 | 7 | 0.022 (0.009–0.046) | |
| Cingulata (armadillos)* | 333 | 2 | 0.006 (0.0007–0.021) | |
| Xenarthra (sloths, anteaters, armadillos)* | 644 | 9 | 0.014 (0.006–0.026) |
-
Table 1—source data 1
Vital statistics for armadillos in the NHDP colony 2010–2020.
- https://cdn.elifesciences.org/articles/82558/elife-82558-table1-data1-v2.xlsx





