Interaction between Teneurin-2 and microtubules via EB proteins provides a platform for GABAA receptor exocytosis
Figures
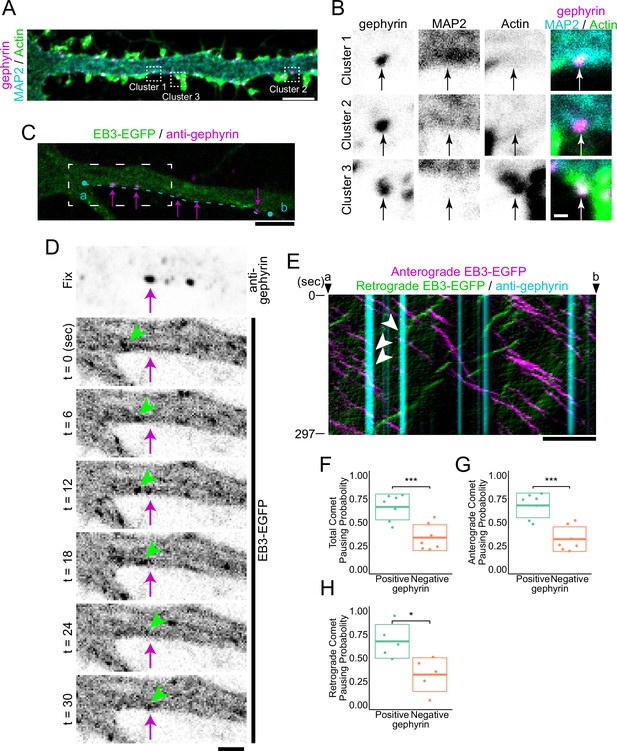
Cluster analysis of inhibitory postsynapses.
(A) Image of immunostaining of gephyrin, MAP2, and actin. Cluster 1 is MT-rich synapses, cluster 2 is synapses with low levels of both MTs and actin, and Cluster 3 is actin-rich synapses. Typical synapses are boxed by dash lines with the cluster number attached to each, and an enlarged view is shown in (B). Scale bar, 5 μm. (B) Enlarged view of the synapses belonging to each cluster. Arrows indicate the position of postsynapses. Scale bar, 500 nm. (C) Overlaid images of live EB3-EGFP with the immunostained image of gephyrin. The timelapse image of the white dash line region is shown in (D). A kymograph of comets passing through an area 6.6 μm wide along the cyan dashed line between points a and b is shown in (E). Arrows indicate representative gephyrin positions. Scale bar, 5 μm. (D) Time-lapse imaging of EB3-EGFP and immunostained image of gephyrin. Arrows indicate the position of gephyrin. Arrowheads indicate tracking of a typical EB3 comet that dissipates at the position of gephyrin. Scale bar, 2 μm. (E) Kymograph of EB3-EGFP and gephyrin, with anterograde comets colored magenta and retrograde comets colored green. Arrowheads indicate typical EB3 comets that dissipate at the position of gephyrin. Scale bar, 5 μm. (F–H) Statistics of comet pausing probability. Total (F), anterograde (G), and retrograde (H) comets all had higher pausing probability at gephyrin-positive positions (p=8.0e-4 in F, p=2.5e-4 in G, p=0.014 in H by Welch’s t-test). n=7 independent experiments. Two of the experiments were excluded from the statistics because a sufficient amount (>4) of retrograde comets were not observed (H). *p<0.05, ***p<0.001.
-
Figure 1—source data 1
4 Excel sheets containing the numerical data used to generate the Figure 1F–H.
- https://cdn.elifesciences.org/articles/83276/elife-83276-fig1-data1-v2.zip
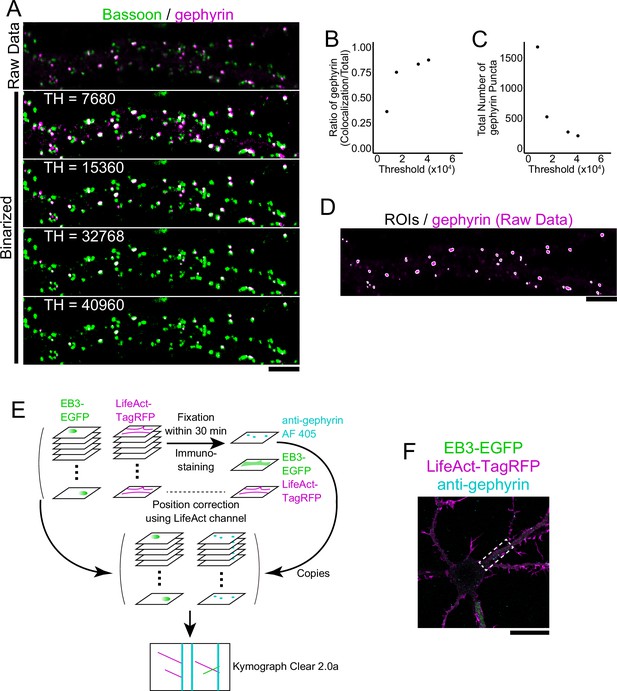
Cluster analysis of inhibitory postsynapses.
(A) Image of immunostaining of bassoon and gephyrin in DIV20 hippocampal cultured neuron. Bassoon is binarized with a threshold value of 32768. Varying the threshold for gephyrin binarization changes the ratio of colocalization. Scale bar, 5 μm. (B) Plots showing the threshold for gephyrin binarization and the colocalization ratio with bassoon. Lowering the threshold lowers the colocalization ratio because more gephyrin is detected. When the threshold is high, over 80% of gephyrin colocalizes with the bassoon. (C) Plot of gephyrin binarization threshold and a number of gephyrin punctures detected. At lower thresholds, more gephyrins are detected. Increasing the threshold decreases the number of gephyrins detected, but the slope of the decrease is slower. (D) ROI (=postsynaptic region) obtained by binarizing the raw data with a gephyrin threshold set to 32768 and overlaying the raw data. Postsynapses that humans can intuitively recognize can be detected almost automatically by ImageJ. Scale bar, 5 µm. (E) Illustration of experimental procedures. Neurons expressing EB3-EGFP and LifeAct-TagRFP were fixed and immunostained within 30 min after live imaging. Since the EB3 comet cannot be observed after fixation, LifeAct-TagRFP was used to correct the position and overlay the images. (F) Overall view of the fixed image. The area boxed by the dashed line is shown in Figure 1C. Scale bar, 20 µm.
-
Figure 1—figure supplement 1—source data 1
An Excel sheet containing the numerical data used to generate the Figure 1—figure supplement 1B and C.
- https://cdn.elifesciences.org/articles/83276/elife-83276-fig1-figsupp1-data1-v2.zip
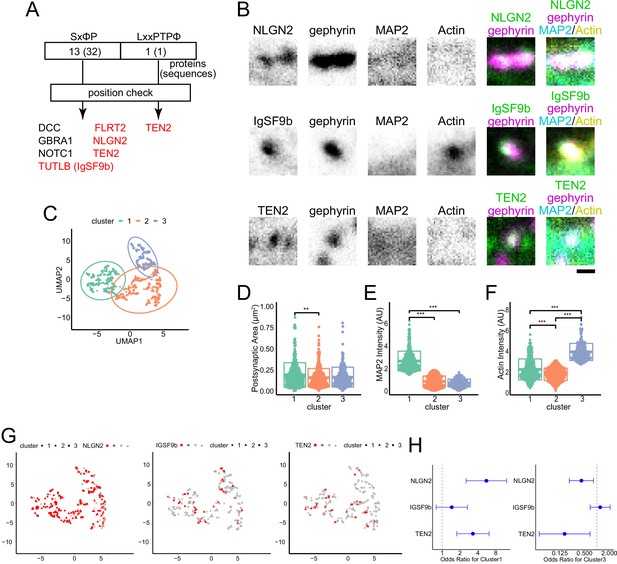
NLGN2 and TEN2 with EB binding motifs localize to MT-rich synapses.
(A) Motif search results: SxφP motifs were found in 32 locations in 13 proteins; LxxPTPφ motifs were found in 1 protein. After checking whether these sequences are intracellular or extracellular, the number of candidate proteins was narrowed down to 7. Of these, those belonging to the adhesion molecule are shown in red. (B) Representative immunostained images of each synaptic organizer and gephyrin, MAP2, and actinin in DIV20 hippocampal cultured neurons. Scale bar, 500 nm. (C) Plots showing the results of cluster analysis. Three-dimensional parameters of synaptic area, MAP2 intensity, and actin intensity evaluated inhibitory postsynapses. After being reduced to two dimensions by UMAP, cluster analysis was performed with the number of clusters pre-specified as 3. The number of synapses belonging to each cluster was 315, 413, and 212 observed by three independent experiments. (D–F) Comparison between clusters for each parameter. (D) Synaptic area: One-way ANOVA showed a significant difference (p=0.0019), and Tukey multiple comparisons showed a significant difference between clusters 1 and 2 (p=0.0016). (E) MAP2 intensity: One-way ANOVA showed a significant difference (p<2e-16), and Tukey multiple comparisons showed significant differences between clusters 1 and 2 (p<1e-07) and between clusters 1 and 3 (p<1e-07). (F) Actin intensity: One-way ANOVA showed a significant difference (p<2e-16), Tukey multiple comparisons showed significant differences between clusters 1 and 2 (p<1e-07), between clusters 2 and 3 (p<1e-07) and between clusters 1 and 3 (p<1e-07). The sample size is the same as (C). **p<0.01, ***p<0.001. (G) Cluster analysis and the relationship between the positivity and negativity of each adhesion molecule. The calculation results by UMAP are the same as in (C). The number of NLGN2 positive and negative synapses are 228 and 65. The number of IgSF9b positive and negative synapses are 53 and 283. The number of TEN2 positive and negative synapses are 49 and 262. TEN2 positive had very little classification to Cluster 3, only 2 synapses. (H) The odds ratio and 95% confidence interval for each adhesion molecule for clusters 1 and 3. For cluster 1: NLGN2, 5.57 (2.54–12.2); IgSF9b, 1.45 (0.80–2.66); TEN2, 3.30 (1.77–6.17). For cluster 3: NLGN2, 0.42 (0.21–0.82); IgSF9b, 1.20 (0.69–2.09); TEN2, 0.16 (0.04–0.68).
-
Figure 2—source data 1
4 Excel sheets containing the numerical data used to generate the Figure 2C–H.
- https://cdn.elifesciences.org/articles/83276/elife-83276-fig2-data1-v2.zip
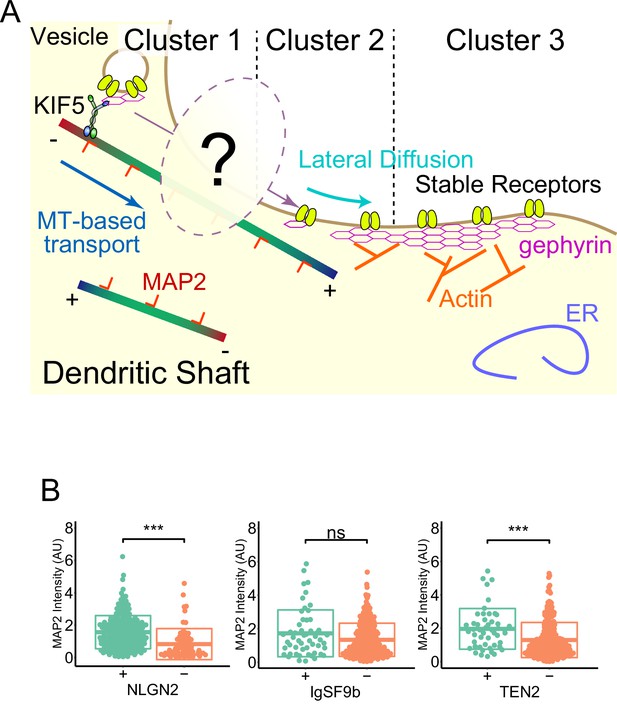
NLGN2 and TEN2 with EB binding motifs localize to MT-rich synapses.
(A) Assumptions of the situation reflected by clustering. Cluster 3 belongs to a stable postsynapse anchored by gephyrin and actin. Cluster 1 belongs to a dynamic postsynapse with receptors being moved in and out by an MT-based transport system. Cluster 2 is an intermediate in which intense lateral diffusion occurs. How the transition between clusters 1 and 2oc curs has been unclear. (B) Classical comparative quantification of MAP2 intensity without reflecting cluster analysis. Welch’s t-test results showed a significant difference between positive and negative synapses for NLGN2 (p=4.08e-07) and TEN2 (p=6.48e-04) but not for IgSF9b (p=0.059). The number of NLGN2 positive and negative synapses are 228 and 65. The number of IGSF9b positive and negative synapses are 53 and 283. The number of TEN2 positive and negative synapses are 49 and 262. ***p<0.001. ns, not significant.
-
Figure 2—figure supplement 1—source data 1
An Excel sheet containing the numerical data used to generate the Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/83276/elife-83276-fig2-figsupp1-data1-v2.zip
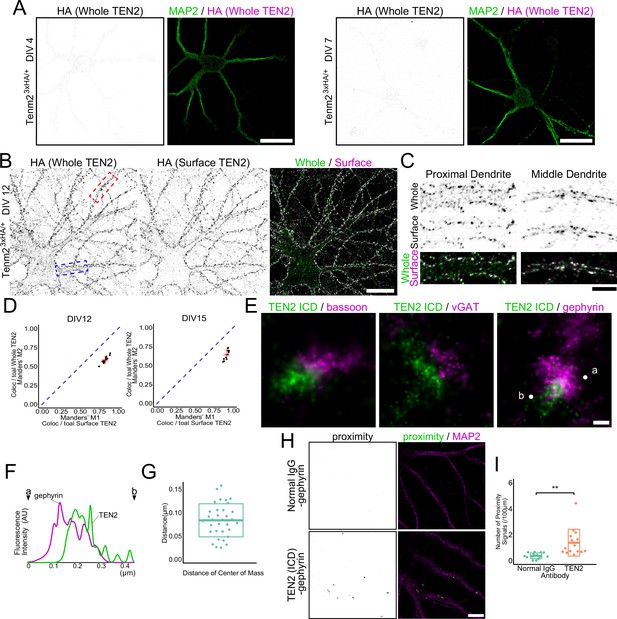
TEN2 is expressed on the surface of the inhibitory postsynapse during early synaptogenesis.
(A) Low expression of TEN2 in early neural development. TEN2 is not expressed in detectable amounts at DIV4 and DIV7. Scale bars, 20 μm. (B) Surface expressed TEN2. TEN2 is expressed, and most TEN2 are surface expressed at DIV12. The area boxed by the dashed line is shown in (C). The blue box indicates the proximal dendrite, and the red box indicates the middle dendrite. Scale bar, 20 μm. (C) Immunostaining of whole TEN2 and surface TEN2 in proximal and middle dendrites. TEN2 is observed intracellularly in the proximal dendrite, while most TEN2 was surface expressed in the middle dendrite. Scale bar, 5 μm. (D) Statistical analysis of whole TEN2 and surface TEN2 measurements. Plots were generated to visualize the values, and red crossbars represent the mean ± SD. The blue dashed line represents M1=M2, which indicates equality according to Manders' overlap coefficient. For DIV12, the Mander’s coefficient values were M1, 0.83±0.037; M2, 0.58±0.044. n=15 neurons. For DIV15, the Mander’s coefficient values were M1, 0.92±0.026; M2, 0.64±0.065. n=12 neurons. (E) dSTORM images. Two-color staining of each presynaptic and postsynaptic molecule suggests that TEN2 is more abundant in the postsynapses. Scale bar, 100 nm. (F) Line graph showing the signal intensity of TEN2 and gephyrin. The horizontal axis shows the length, and the vertical axis shows the fluorescence intensity. Points indicated by letters and arrowheads represent the positions of ‘a’ and ‘b’ in (E). (G) Distance between the centers of mass of TEN2 and gephyrin when observed in dSTORM. The mean ± SD was 83.3±35.3. n=33 synapses. (H) Images showing the results of the proximity ligation assay. When the proximity ligation assay was performed using antibodies against TEN2 and gephyrin, a signal indicating the proximity of less than 20 nm could be detected. On the other hand, no signal was obtained in the negative control. Scale bar, 10 μm. (I) The number of proximity signals per 100 μm. mean ± SD was 0.37±0.23 and 1.38±1.04, respectively. Welch’s t-test showed a significant difference between negative control and TEN2 in proximity to gephyrin (p=0.0021). n=14 and 15 from three independent experiments. **p<0.01.
-
Figure 3—source data 1
4 Excel sheets containing the numerical data used to generate the Figure 3D, F, G and I.
- https://cdn.elifesciences.org/articles/83276/elife-83276-fig3-data1-v2.zip
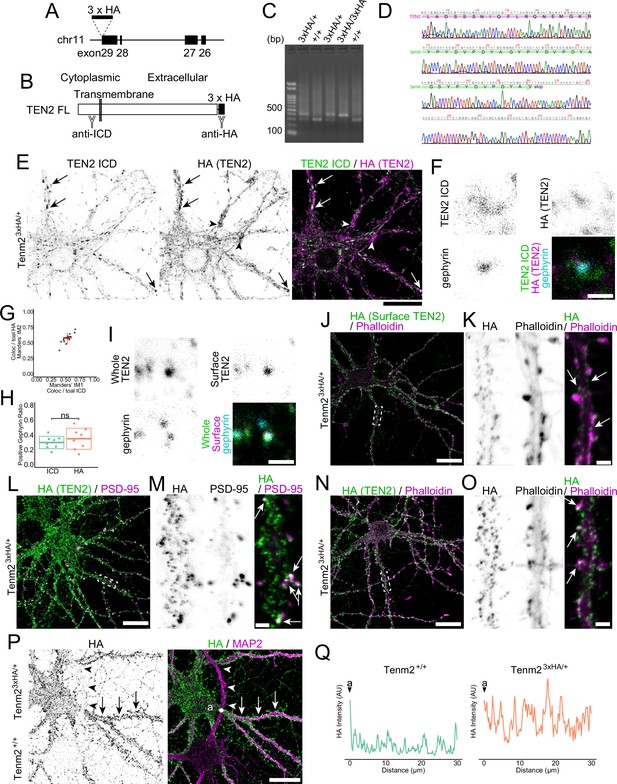
TEN2 is expressed on the surface of the inhibitory postsynapse during early synaptogenesis.
(A) Overview of knock-in mice. TEN2 is encoded on the minus strand of chromosome 11. Knock-in mice were generated by inserting a 3×HA sequence before the stop codon in Exon29. (B) Overview of the TEN2 full-length protein and antibody recognition sites. TEN2 is a type II transmembrane protein that is intracellular at its N-terminus and extracellular at its C-terminus. Therefore, 3×HA, inserted just before the stop codon, is extracellular when translated. (C) Typical genotyping results. A 300 bp band is seen in wild-type mice, while a+100 bp band is seen in knock-in mice; if both bands are seen, the mouse is heterozygous with only one allele being knock-in. (D) Sequence confirmation by Sanger sequencing. Bands amplified by genotyping were purified and Sanger sequenced to confirm the knock-in sequence. (E) Images of double staining for anti-TEN2ICD and anti-HA in knock-in neurons. As shown in (F), overlap is observed in many areas, including inhibitory synapses (arrows). On the other hand, there are also areas where only HA is stained (arrowheads). Scale bar, 20 µm. (F) Images of double-staining of anti-TEN2ICD and anti-HA at inhibitory synapses. (G) Plot and cross bars (mean ± SD) showing the degree of overlap of double staining of anti-TEN2ICD and anti-HA. Statistical analysis shows moderate overlap. Mean ± SD: Manders'tM1, 0.54±0.07; Manders'tM2, 0.57±0.08. n=17 neurons. Scale bar, 1 µm. (H) No effect of HA knock-in on localization to inhibitory synapses. ICR-delivered wild-type neurons at DIV15 were co-stained with ICD and gephyrin antibodies, and HA knock-in neurons at DIV15 were co-stained with HA and gephyrin antibodies. Mean ± SD were 0.30±0.01 and 0.34±0.02, respectively. Since there was no significant difference in the ratio of colocalization (p=0.40), we concluded that HA knock-in did not affect localization to inhibitory synapses. (I) Images of double staining of whole TEN2 and surface TEN2 at inhibitory synapses. Scale bar, 1 µm. (J) Images of immunostaining of HA tag and actin exposed on the cell membrane surface in the knock-in neuron. The dashed box is magnified in (K). (K) Confirmation that the HA tag is exposed at the plasma membrane surface, suggesting that TEN2 functions at the plasma membrane surface along the dendritic shaft and spine-like structures (arrows). (L) Images of immunostaining of HA tag and PSD-95 in the knock-in neuron. The red dashed box is magnified in (M). (M) The strong signal of the HA tag at the site where PSD-95 is localized suggestsx that the molecule is abundant at excitatory synapses (arrows). (N) Images of immunostaining of HA tag and actin in the knock-in neuron. The dashed box is magnified in (O). (O) HA tag signals are present in the dendritic shaft and spine-like structures (arrows), suggesting that the molecule is abundant at excitatory and inhibitory synapses. Scale bars indicate 50 μm in (J), (L), and (N) and 2 μm in (K), (M), and (O). (P) Immunofluorescence images of mixed cultures of knock-in and wild-type neurons. In knock-in neurons with a strong HA signal in the cell body, the HA signal is similarly strong in the dendrites (arrows). In wild-type neurons, the HA signal is weak in the cell body and dendrites (arrowheads). Scale bar, 20 μm. (Q) Line graph of HA signal intensity along the dendritic axis for knock-in and wild-type neurons. From the point with ‘a’ in (P), the signal intensity of HA for 30 µm is shown. Signal intensity is higher in knock-in neurons.
-
Figure 3—figure supplement 1—source data 1
Unprocessed full-size gel photograph showing genotyping of knock-in mice and photograph showing the region used in Figure 3—figure supplement 1C with dashed lines.
- https://cdn.elifesciences.org/articles/83276/elife-83276-fig3-figsupp1-data1-v2.zip
-
Figure 3—figure supplement 1—source data 2
3 Excel sheets containing the numerical data used to generate the Figure 3—figure supplement 1G, H and Q.
- https://cdn.elifesciences.org/articles/83276/elife-83276-fig3-figsupp1-data2-v2.zip
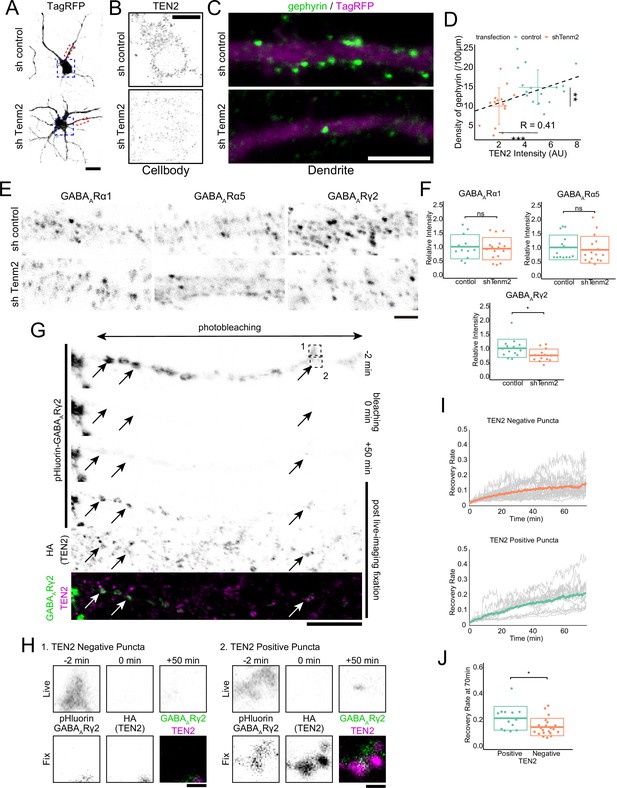
TEN2 provides a platform for the exocytosis of GABAA receptors at inhibitory postsynapses to mature synapses.
(A) Images of neurons transfected with control or knockdown vector. The area boxed by the blue dash line is shown in (B), and the area boxed by the red dash line is shown enlarged in (C). Scale bar, 20 μm. (B) Magnified images of knockdown neurons immunostained with TEN2. Scale bar, 10 μm. (C) Magnified images of knockdown neurons immunostained with gephyrin. Gephyrin accumulation was reduced in TEN2 knockdown neurons. Scale bar, 5 μm. (D) A plot with crossbars (mean ± SD) of the relationship between TEN2 fluorescence intensity in the cell bodies and the density of gephyrin puncta per 100 μm dendrite. The black dashed line represents a linear approximation of the correlation between TEN2 intensity and gefillin density without distinguishing between control and knockdown neurons (R=0.42). It should be noted that transfection with a knockdown vector significantly reduced TEN2 intensity (p=9.9e-8) and gephyrin density (p=0.0058). n=17 for control neurons and n=15 for knockdown neurons. **p<0.01, ***p<0.001 by Welch’s t-test. (E) Magnified images of knockdown neurons immunostained with GABAA receptors subunit α1, α5, and γ2. Only the γ2 receptor is downregulated in TEN2 knockdown neurons of these subunits. Scale bar, 2 μm. (F) Plots and cross bars (mean ± SD) quantifying the relative intensity of GABAA receptor subunits. The fluorescence intensities of receptors present in dendrites within 100 µm from the cell body were quantified comparatively. Mean ± SD were 1±0.43 and 0.93±0.40 for α1, 1±0.40 and 0.92±0.48 for α5, and 1±0.32 and 0.75±0.22 for γ2. Welch’s t-test showed that α1 (p=0.67) and α5 (p=0.62) were not significantly different between control and TEN2 knockdown neurons. γ2 (p=0.027) was predominantly reduced in TEN2 knockdown neurons. n=12, 16, 14, 16, 14, and 13 neurons from three independent experiments. *p<0.05. (G) Time-lapse images showing FRAP assay and immunostaining of TEN2 in post-live-imaging fixation. Arrows indicate exocytosed GABAA receptors puncta in typical TEN2-positive positions. The area boxed by the dashed line is shown in (H). Scale bar, 10 μm. (H) Magnified images of FRAP assay. The pHluorin signal indicating surface expression of GABAARγ2 was observed 50 min after photobleaching in the TEN2 positive position, whereas the signal in the TEN2-negative position was very slight. Scale bar, 1 μm. (I) Statistical analysis showing signal recovery. Gray lines indicate the ratio of pHluorin-GABAARγ2 signal intensity after photobleaching to the intensity before photobleaching in individual puncta. Colored lines indicate mean values. (J) Plot and crossbars (mean ± SD) of recovery rate at 70 min after photobleaching. The recovery rate was significantly higher in TEN2 positive puncta (p=0.032). n=13 positive puncta and 21 negative puncta. *p<0.05 by Welch’s t-test.
-
Figure 4—source data 1
4 Excel sheets containing the numerical data used to generate the Figure 4D, F,I and J.
- https://cdn.elifesciences.org/articles/83276/elife-83276-fig4-data1-v2.zip
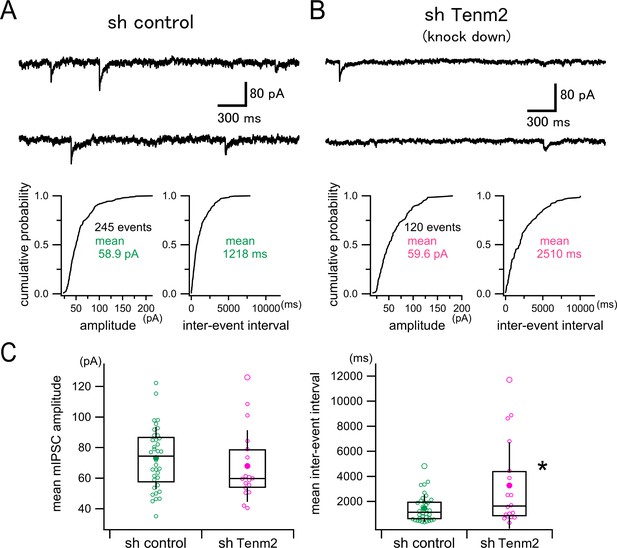
Effect of TEN2 knockdown on miniature inhibitory synaptic currents (mIPSCs) in cultured hippocampal neurons.
(A) Upper panel shows a representative continuous 6 s trace (3 s traces in a row) of mIPSC recording in a control neuron (sh control). The lower panel shows the representative cumulative probability distributions of mIPSC amplitude (left) and inter-event interval (right) measured from a 300 s recording in this neuron. The single mean values of the amplitude and the interval were used to represent each neuron. (B) A representative example of a TEN2 knockdown neuron (sh Tenm2) is shown similarly to (A). (C) Box and whisker plots of mean mIPSC amplitudes (left) and mean inter-event intervals (right). Open circles correspond to individual data points, and the central horizontal lines and the boxes represent the median values and the interquartile ranges, respectively. Filled circles indicate the averaged values, and the error bars indicate one standard deviation above and below the values. TEN2 knockdown had no effect on mIPSC amplitude (sh control, 73.2±20.4 pA, n=36 neurons; sh Tenm2, 67.9±23.5 pA, n=18 neurons, Welch’s t-test, p=0.424), but prolonged inter-event interval (i.e. reduced mIPSC frequency) significantly (sh control, 1455±1077ms, n=36 neurons; sh Tenm2, 3272±3444ms, n=18 neurons, Welch’s t-test, p<0.0418). *p<0.05.
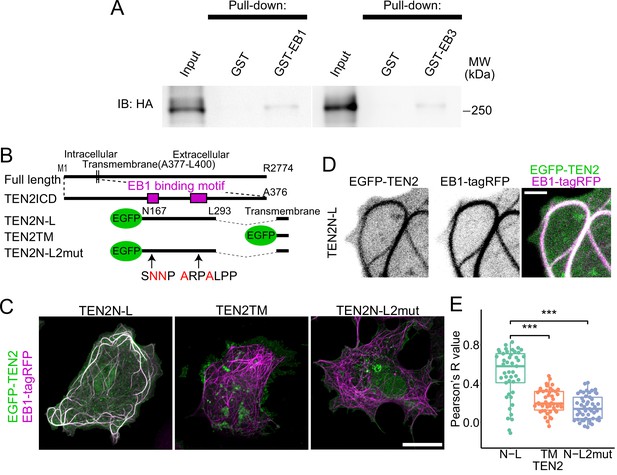
Interaction with MTs via EB1 by two motifs in TEN2.
(A) Interaction between EB and TEN2 by pull-down assay. Pull-down assay was performed on brain lysate of TEN2-HA knock-in mice using GST-EB1/3 as bait, and both assays were positive for HA (TEN2) by Western blot. (B) Overview of the partial domain of TEN2N-L. TEN2N-L was designed to contain the two EB1 binding motifs detected by motif search. TEN2N-L2mut has amino acid mutations in two binding motifs. All proteins have transmembrane domains with predictable topogenic sequences. (C) Co-expression of each truncated mutant with EB1 in COS-7 cells. Cells with MTs patterns of over-expression of EB1 were observed. TEN2N-L colocalized well with EB1 compared to other partial domains, suggesting that TEN2 N-L interacts with EB1. Scale bar, 20 μm. (D) Highly magnified image of COS-7 cells expressing TEN2N-L. Scale bar, 2 μm. (E) Based on correlation coefficients, individual plots, and box plots show the quantitative analysis results of the colocalization index between each TEN2 and EB1. The median Pearson’s correlation coefficient between TEN2N-L and EB1 was 0.58, which was significantly different from that of TEN2TM (0.195; p=1.3e-7), and TEN2N-L 2mut (0.14; p=2.9e-9) by Pairwise comparisons using Wilcoxon rank sum test after Kruskal-Wallis rank sum test (p=5.0e-11). The total number of cells observed was 46, 46, and 49, respectively. ***p<0.001.
-
Figure 6—source data 1
2 unprocessed full-size blot photographs showing western blotting of HA, as well as 2 photographs showing the region used in Figure 6A with dashed lines.
- https://cdn.elifesciences.org/articles/83276/elife-83276-fig6-data1-v2.zip
-
Figure 6—source data 2
An Excel sheet containing the numerical data used to generate the Figure 6E.
- https://cdn.elifesciences.org/articles/83276/elife-83276-fig6-data2-v2.zip
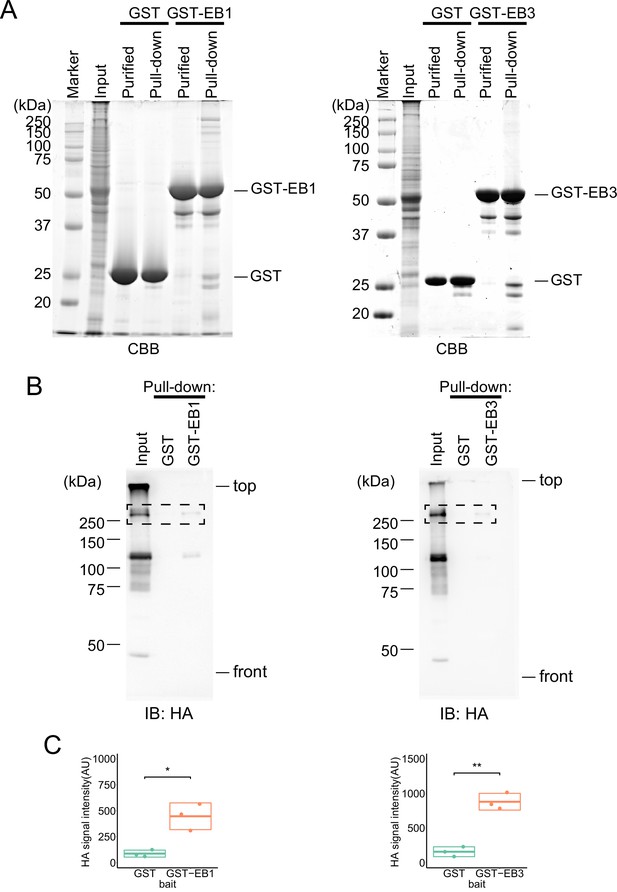
Interaction with MTs via EB1 by two motifs in TEN2.
(A) Images of CBB stained gel showing the results of GST-EBs purification and GST-pulldown assays. The bait of equal amounts is applied, and specific protein bands are seen in the rightmost lane of each gel when pull-down was performed with GST-EBs, (B) Western blot images of GST-pulldown results, showing a band around the expected molecular weight (307 kDa) when pulled down with GST-EBs. (C) Statistical analysis of the GST-pulldown results, showing that the amount of TEN2 pulled down by GST-EBs was significantly higher than that of GST used as a control (GST-EB1, p=0.033; GST-EB3, p=0.0022) in all three trials.
-
Figure 6—figure supplement 1—source data 1
2 Unprocessed full-size gel photographs showing the results of GST pull-down used in Figure 6—figure supplement 1A, 2 unprocessed full-size blot photographs showing western blotting of HA used in Figure 6—figure supplement 1B, and 4 photographs showing the regions used in each figure with dashed lines.
- https://cdn.elifesciences.org/articles/83276/elife-83276-fig6-figsupp1-data1-v2.zip
-
Figure 6—figure supplement 1—source data 2
An Excel sheet containing the numerical data used to generate the Figure 6—figure supplement 1C.
- https://cdn.elifesciences.org/articles/83276/elife-83276-fig6-figsupp1-data2-v2.zip
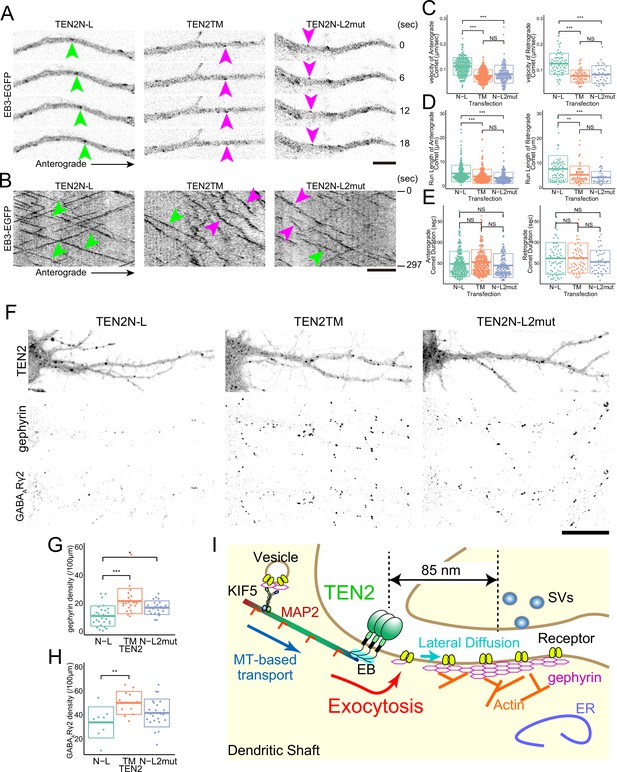
MTs need to be recruited near the cell membrane by TEN2 for inhibitory synapse formation.
(A) Live imaging of EB3-EGFP in neurons expressing each partial domain. Due to the dominant-negative effect, the fast comet was observed in neurons expressing TEN2N-L (green arrowheads). In neurons expressing the other two domains, relatively slow comets (magenta arrowheads) were observed. Scale bar, 5 μm. (B) EB3-EGFP kymographs in the neurons expressing each partial domain, and a linear kymograph was observed in the TEN2N-L-expressing neurons due to the dominant-negative effect (green arrowheads). In neurons expressing the other two, undulation was observed in addition to linear kymograph (magenta arrowheads). Scale bar, 5 μm. (C–E) Statistical analysis of EB3-EGFP separately for anterograde and retrograde motion. The analysis revealed no significant difference in comet duration among the three partial domains (E). However, significant differences were observed in velocity (p<2e-7 for anterograde with TEN2TM, p<2e-7 for anterograde with TEN2N-L2mut, p<2e-7 for retrograde with TEN2TM, and p<2e-7 for retrograde with TEN2N-L2mut) and run length (p=1.5e-4 for anterograde with TEN2TM, p=3e-7 for anterograde with TEN2N-L2mut, p=2.7e-3 for retrograde with TEN2TM, and p=1.9e-4 for retrograde with TEN2N-L2mut), indicating a significant increase in TEN2N-L compared to the other two domains (C and D). For the anterograde motion, the statistical tests were based on a one-way ANOVA (p<2e-16 in C, p=1.4e-7 in D, and p=0.043 in E) followed by post hoc Tukey analysis. For the retrograde motion, the statistical tests were based on a one-way ANOVA (p=1.0e-13 in D, p=7.5e-5 in D, and p=0.147 in E) followed by post hoc Tukey analysis. The number of comets analyzed for the anterograde motion was as follows: TEN2N-L (n=208), TEN2TM (n=235), and TEN2N-L2mut (n=129). For the retrograde motion, the number of comets analyzed was as follows: TEN2N-L (n=66), TEN2TM (n=59), and TEN2N-L2mut (n=39). (F) Immuno-staining of gephyrin and GABAARγ2 subunits in neurons expressing each partial domain. Scale bar, 20 μm. (G) The density of gephyrin puncta in neurons expressing each partial domain. The density of gephyrin puncta was found to be significantly lower in neurons expressing TEN2N-L compared to those expressing TEN2TM (P=5.3e-6) and TEN2N-L2mut (P=0.013). The statistical tests were based on a one-way ANOVA (p=8.8e-6) followed by post hoc Tukey analysis. The sample sizes were as follows: TEN2N-L (n=28), TEN2TM (n=23), and TEN2N-L2mut (n=23). *p<0.05, ***p<0.001. (H) The density of GABAARγ2 puncta in neurons expressing each partial domain. The density of GABAARγ2 puncta was found to be significantly lower in neurons expressing TEN2N-L compared to those expressing TEN2TM (p=0.009). The statistical tests were based on a one-way ANOVA (p=0.011) followed by post hoc Tukey analysis. The sample sizes were as follows: TEN2N-L (n=8), TEN2TM (n=12), and TEN2N-L2mut (n=23). **p<0.01. (I) A working model derived from this study. The interaction of TEN2 and dynamic MTs provides a platform for exocytosis and allows proper transport of components of the inhibitory postsynapse.
-
Figure 7—source data 1
5 Excel sheets containing the numerical data used to generate the Figure 7C–E, G and H.
- https://cdn.elifesciences.org/articles/83276/elife-83276-fig7-data1-v2.zip

MTs need to be recruited near the cell membrane by TEN2 for inhibitory synapse formation (A) Confocal imaging of gephyrin accumulation and MAP2 in neurons expressing each TEN2.
Higher gephyrin accumulation was observed in TEN2TM neurons, whereas it was reduced in dominant-negative TEN2N-L. In addition, biased MAP2 was observed in the TEN2N-L. Scale bar, 20 μm. (B) Line graph showing the signal intensity of MAP2. The horizontal axis shows the length, and the vertical axis shows the fluorescence intensity. Points indicated by letters and arrowheads represent positions of a-d in (A). In TEN2TM neurons, the MAP2 signal is strongly observed around the dendrite axis. In TEN2N-L, on the other hand, the MAP2 peak is biased to be located just below the membrane in the direction parallel to the axis and is sparse near the axis. This suggests that TEN2N-L on the membrane recruits MTs. (C) Ratio of neurons with membrane-biased MTs. Mean ± SD were 0.056±0.096 and 0.48±0.12, which were significantly different (p=0.01) by Welch’s t-test. Observations were based on three independent trials. **p<0.01.
-
Figure 7—figure supplement 1—source data 1
2 Excel sheets containing the numerical data used to generate the Figure 7—figure supplement 1B and C.
- https://cdn.elifesciences.org/articles/83276/elife-83276-fig7-figsupp1-data1-v2.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL/6 J JAX mice | Charles River | Cat# JAX:000664, RRID:IMSR_JAX:000664 | |
| Strain, strain background (Mm) | ICR | Japan SLC | Cat# 5462094, RRID:MGI:5462094 | |
| Strain, strain background (Mm) | ICR | Charles River | Cat# CRL:022, RRID:IMSR_CRL:022 | |
| Strain, strain background (Escherichia coli) | 5-alpha Competent | New England Biolabs | Cat# C2987 | |
| Strain, strain background (Ec) | BL21(DE3) Competent Cells | Agilent | Cat# 200131 | |
| Cell line (Chlorocebus sabaeus) | COS-7 cells | RIKEN Cell Bank | Cat# RCB0539, RRID:CVCL_0224 | |
| Antibody | Rabbit polyclonal anti-TEN2 Cytoplasmic | This study | N/A | 0.2–0.8 µg/mL |
| Antibody | Rabbit polyclonal GABRA1 antibody | Proteintech | Cat# 12410–1-AP, RRID:AB_2108692 | 1:1000 |
| Antibody | Rabbit polyclonal Anti-GABA-A receptor alpha5 | Synaptic Systems | Cat# 224 503, RRID:AB_2619944 | 1:5000 |
| Antibody | Rabbit polyclonal Anti-GABA-A receptor gamma2 | Synaptic Systems | Cat# 224 003, RRID:AB_2263066 | 1:2000 |
| Antibody | Mouse monoclonal anti-gephyrin (mAb7a) | Synaptic Systems | Cat# 147011, RRID:AB_887717 | 1:2000 |
| Antibody | Chicken polyclonal Anti-Bassoon | Synaptic Systems | Cat# 141 016, RRID:AB_2661779 | 1:2000 |
| Antibody | Mouse monoclonal Anti-VGAT(117G4) | Synaptic Systems | Cat# 131 011, RRID:AB_887872 | 1:5000 |
| Antibody | Rabbit polyclonal Anti-Neuroligin 2 | Synaptic Systems | Cat# 129 203, RRID:AB_993014 | 1:2000 |
| Antibody | Mouse monoclonal anti-PSD95 (7E3) | Cell Signaling Technology | Cat# 36233, RRID:AB_2721262 | 1:1000 |
| Antibody | Rabbit monoclonal anti-HA-tag (C29F4) | Cell Signaling Technology | Cat# 3724, RRID:AB_1549585 | 1:1000 (IF, WB) |
| Antibody | Rabbit polyclonal Anti-IGSF9B | Merck | Cat# HPA010802, RRID:AB_1079194 | 1:1000 |
| Antibody | Chicken polyclonal anti-MAP2 | Novus | Cat# NB300-213, RRID:AB_2138178 | 1:50000 |
| Antibody | Donkey polyclonal Anti-Mouse IgG (Alexa Fluor 405) | abcam | Cat# ab175658, RRID:AB_2687445 | 1:1000 |
| Antibody | Donkey polyclonal Anti-Mouse IgG (H+L), Alexa Fluor 488 | Jackson ImmunoResearch Labs | Cat# 715-546-151, RRID:AB_2340850 | 1:2000 |
| Antibody | Donkey polyclonal Anti-Mouse IgG (H+L), Rhodamine Red-X | Jackson ImmunoResearch Labs | Cat# 715-296-151, RRID:AB_2340835 | 1:2000 |
| Antibody | Donkey polyclonal Anti-Mouse IgG (H+L), Alexa Fluor 647 | Jackson ImmunoResearch Labs | Cat# 715-606-151, RRID:AB_2340866 | 1:2000 |
| Antibody | Donkey polyclonal Anti-Rabbit IgG (H+L), DyLight 405 | Jackson ImmunoResearch Labs | Cat# 711-475-152, RRID:AB_2340616 | 1:1000 |
| Antibody | Donkey polyclonal Anti-Rabbit IgG (H+L), Alexa Fluor 488 | Jackson ImmunoResearch Labs | Cat# 711-546-152, RRID:AB_2340619 | 1:2000 |
| Antibody | Donkey polyclonal Anti-Rabbit IgG (H+L), CF568 | Biotium | Cat# 20098–1, RRID:AB_10853318 | 1:2000 |
| Antibody | Donkey polyclonal Anti-Chicken IgY (IgG) (H+L), Alexa Fluor 647 | Jackson ImmunoResearch Labs | Cat# 703-605-155, RRID:AB_2340379 | 1:2000 |
| Antibody | Donkey polyclonal Anti- Rabbit IgG (H+L), HRP | Jackson ImmunoResearch Labs | Cat# 711-036-152, RRID:AB_2340590 | 1:20000 (WB) |
| Recombinant DNA reagent | guide RNA for knock-in | IDT | 5’- GACAGAATGAGATGGGAAAG-3’ | |
| Recombinant DNA reagent | ssODN for knock-in | IDT | 5’-ACAGTAGCAGCAACATCCAGTTCTTAAGACAGAATGAGATGGGAAAGAGATACCCATACGATGTACCTGACTATGCGGGCTATCCCTATGACGTCCCGGACTATGCAGGATCCTATCCTTATGACGTTCCAGATTACGCTGTTTAACAAAATAACCTGCTGCCACCTCTTCTCTGGGTGGCTCAGCAGGAGCAACT-3’ | |
| Recombinant DNA reagent | Homo sapiens TENM2 cDNA | KAZUSA | NCBI AB032953 | TEN2 |
| Recombinant DNA reagent | Mm Tenm2 cDNA | RIKEN | NCBI AK031198 | TEN2 |
| Recombinant DNA reagent | Hs MAPRE1 cDNA | KAZUSA | NCBI AB463888 | EB1 |
| Recombinant DNA reagent | Hs MAPRE3 cDNA | Eurofins Genomics | EB3 gene synthesis | |
| Recombinant DNA reagent | pHluorin-GABAARγ2 | Addgene | plasmid # 49170 RRID:Addgene_49170 | Jacob et al., 2005 |
| Recombinant DNA reagent | pBAsi-mU6 DNA | Takara Bio | Cat# 3222 | |
| Recombinant DNA reagent | Top strand of oligonucleotide cassette for control shRNA | Eurofins Genomics | 5’-GATCCGGCCTAAGGTTAAGTCGC CCTCGCTCGAGCGAGGGCGACT TAACCTTAGGTTTTTGA –3’ | |
| Recombinant DNA reagent | Bottom strand of oligonucleotide cassette for control shRNA | Eurofins Genomics | 5’-AGCTTCAAAAACCTAAGGTTAA GTCGCCCTCGCTCGAGCGAGGG CGACTTAACCTTAGGCCG –3’ | |
| Recombinant DNA reagent | Top strand of oligonucleotide cassette for Tenm2 shRNA | Eurofins Genomics | 5’-GATCCGGGCCAGGTTTG ATTATACCTATCTCGAGATA GGTATAATCAAACCTGGCTTTTTGA –3’ | |
| Recombinant DNA reagent | Bottom strand of oligonucleotide cassette for Tenm2 shRNA | Eurofins Genomics | 5’-AGCTTCAAAAAGCCAGGTTT GATTATACCTATCTCGAGATAGG TATAATCAAACCTGGCCCG –3’ | |
| Recombinant DNA reagent | Top strand of oligonucleotide cassette for LifeAct | Eurofins Genomics | 5’-CTAGCATGGGCGTGGCCGACCTGATCAAGAAGTTCGAATCGATAAGCAAGGAAGAGGGC –3’ | |
| Recombinant DNA reagent | Bottom strand of oligonucleotide cassette for LifeAct | Eurofins Genomics | 5’-GATCGCCCTCTTCCTTGCTTATCGATTCGAACTTCTTGATCAGGTCGGCCACGCCCATG –3’ | |
| Peptide, recombinant protein | synthetic peptide | Eurofins Genomics | CSNTSHQIMDTNPDE | |
| Peptide, recombinant protein | synthetic peptide | GenScript | CQMPLLDSNTSHQIMD TNPDEEFSPNS | |
| Commercial assay or kit | FlexAble CoraLite 488 Antibody Labeling Kit for Rabbit IgG | Proteintech | Cat# KFA001 | |
| Commercial assay or kit | FlexAble CoraLite Plus 555 Antibody Labeling Kit for Rabbit IgG | Proteintech | Cat# KFA002 | |
| Commercial assay or kit | FlexAble CoraLite Plus 647 Antibody Labeling Kit for Rabbit IgG | Proteintech | Cat# KFA003 | |
| Commercial assay or kit | Zenon Mouse IgG1 Labeling Kits Alexa Fluor 405 | Thermo Fisher Scientific | Cat# Z25013 | |
| Commercial assay or kit | Zenon Mouse IgG1 Labeling Kits Alexa Fluor 594 | Thermo Fisher Scientific | Cat# Z25007 | |
| Commercial assay or kit | Duolink In Situ PLA Probe Anti-Mouse PLUS | Merck | Cat# DUO92001 | |
| Commercial assay or kit | Duolink In Situ PLA Probe Anti-Rabbit MINUS | Merck | Cat# DUO92005 | |
| Commercial assay or kit | Duolink In Situ Detection Reagents Green | Merck | Cat# DUO92014 | |
| Commercial assay or kit | High-Efficiency Ca2+ Phosphate Transfection Kit | Takara Bio | Cat# 631312 | |
| Chemical compound, drug | Lipofectamine 2000 Transfection Reagent | Thermo Fisher Scientific | Cat# 11668030 | |
| Chemical compound, drug | Alexa Fluor 555 Phalloidin | Thermo Fisher Scientific | Cat# A34055 | |
| Chemical compound, drug | Can Get Signal Solution | Toyobo | Cat# NKB-101 | |
| Chemical compound, drug | Immunostar Zeta | FUJIFILM Wako | Cat# 291–72401 | |
| Chemical compound, drug | polyethylenimine solution | Merck | Cat# P3143 | |
| Chemical compound, drug | BioCoat poly-D-lysine | Corning | Cat# 354210 | |
| Chemical compound, drug | MEM, no glutamine | Thermo Fisher Scientific | Cat# 11090081 | |
| Chemical compound, drug | GlutaMAX | Thermo Fisher Scientific | Cat# 35050061 | |
| Chemical compound, drug | B27 Plus Plus Supplement (50 X) | Thermo Fisher Scientific | Cat# A3582801 | |
| Chemical compound, drug | SulfoLink Coupling Resin | Thermo Fisher Scientific | Cat# 20401 | |
| Chemical compound, drug | Glutathione Sepharose 4B | Cytiva | Cat# 17075601 | |
| Software, algorithm | Fiji | NIH | https://fiji.sc | |
| Software, algorithm | KymoResliceWide | Eugene Katrukha | https://imagej.net/KymoResliceWide | |
| Software, algorithm | KymographClear 2.0 a | Erwin Peterman’s group | https://sites.google.com/site/kymographanalysis/ | Mangeol et al., 2016 |
| Software, algorithm | R | R Core Team | https://www.r-project.org | |
| Software, algorithm | pCLAMP | Molecular Devices | https://www.moleculardevices.com/ | |
| Software, algorithm | Igor Pro 8 | Wavemetrics | https://www.wavemetrics.com/software/igor-pro-8 | |
| Software, algorithm | NeuroMatic | ThinkRandom | http://www.neuromatic.thinkrandom.com/ | Rothman and Silver, 2018 |
Additional files
-
Supplementary file 1
The results of motif search.
In order to narrow down the candidates for MT recruiter, a motif search was conducted to investigate the potential binding with EB. Based on previous proteomics studies (Loh et al., 2016), proteins containing the motifs SxφP and LxxPTPφ were searched. The cellular localization (extracellular or intracellular) of each motif was manually checked.
- https://cdn.elifesciences.org/articles/83276/elife-83276-supp1-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83276/elife-83276-mdarchecklist1-v2.pdf





