A chronic signaling TGFb zebrafish reporter identifies immune response in melanoma
Peer review process
This article was accepted for publication as part of eLife's original publishing model.
History
- Version of Record published
- Accepted
- Preprint posted
- Received
Decision letter
-
Owen J TamplinReviewing Editor; University of Wisconsin–Madison, United States
-
Paivi M OjalaSenior Editor; University of Helsinki, Finland
-
Marina C MioneReviewer; University of Trento, Italy
Our editorial process produces two outputs: (i) public reviews designed to be posted alongside the preprint for the benefit of readers; (ii) feedback on the manuscript for the authors, including requests for revisions, shown below. We also include an acceptance summary that explains what the editors found interesting or important about the work.
Decision letter after peer review:
Thank you for submitting your article "A chronic signaling TGFb zebrafish reporter identifies immune response in melanoma" for consideration by eLife. Your article has been reviewed by 3 peer reviewers, one of whom is a member of our Board of Reviewing Editors, and the evaluation has been overseen by Paivi Ojala as the Senior Editor. The following individual involved in the review of your submission has agreed to reveal their identity: Marina C Mione (Reviewer #3).
The reviewers have discussed their reviews with one another, and the Reviewing Editor has drafted this to help you prepare a revised submission.
Essential revisions (for the authors):
Although the reviewers are very positive about the manuscript, there are several areas that could be explored further. There was a consensus that the role of macrophages in particular needs more data for the model to be supported.
1. There are multiple issues with Figures 1 and Extended Data Figure 1 as seen in the reviewers' comments below. These relate to quality and quantification and should be addressed.
2. The reviewers also suggested that Figure 3 data be improved with better confocal images and time-lapse imaging.
3. In Figure 4, SATB2 data should be compared to Figures 1-3 without SATB2. Does SATB2 facilitate melanoma progression?
Reviewer #1 (Recommendations for the authors):
Data exists of spatially mapped transcriptomes in zebrafish melanoma:
Hunter, et al. Spatially resolved transcriptomics reveals the architecture of the tumor-microenvironment interface. Nat Commun 12, 6278 (2021).
Are the results in this study showing regions of active TGFb signaling in advanced melanoma consistent with existing data of endogenous expression patterns?
The proposed model of preferential phagocytosis of TIE:GFP+ cells vs negative cells is interesting, however, the data is still somewhat preliminary. Higher resolution imaging data and possibly time-lapse would better support the model. Could the authors provide some candidate factors produced by TIE:GFP+ cells that could trigger this response?
In the Extended Data Figure 1C-D experiments, if the mitf:mCherry is transiently injected, then the conclusions may be overstated. Line 90: "TIE:EGFP was also never expressed in mitfa:mCherry high early melanomas (Extended Data Figure 1D)". How many melanomas were observed? What level of GFP expression is required to score as positive in TIE:GFP fish? The conclusions should be reconsidered because transient F0 transgenics are inherently mosaic.
Extended data 1B images are small and difficult to evaluate. Need better images that are quantified for fluorescence intensity.
In the Figure 1C (20 and 22 wpf) panels, why is there a yellow signal in the mitfa:mCherry channel alone in the majority of the tumor but not in the merged image?
Which data does S1B refer to in this line?
Line 115: melanomas were processed for single-cell RNA-sequencing at 23 and 42 weeks post-fertilization (wpf), respectively (Figure 1C and S1B).
Could the authors describe the marco marker and its significance here?
Line: 120 melanoma cells expressing mitfa and/or sox10, but we identified a marco-expressing TIE:EGFP+.
Does the analysis of Figure S2B include macrophages or is it only the melanomas? Is there any independent validation of the GSEA results? This is regarding:
Line: 130 in TIE:EGFPhigh cells were interferon α and γ (Figure S2B).
In the following line, which genes are referred to specifically? Are these genes associated with a specific table or figure?
Line: 293 of melanoma patients exhibit up-regulation of these genes52,53.
Are there experiments that could better support the following statement:
Line: 306 Our work identified macrophage subclusters that contain low levels of mitfa and/or sox10 expression suggestive of phagocytosis of melanoma cells.
Is there a GFP/mCherry merged image to support this statement, as it is not clear from the figure?
311 Figure 3C, there is co-localization of mpeg:mCherry and TIE:EGFP signal.
The following figure legend is insufficient and needs more details (e.g. live or fixed sample? Optical section or z projection? Single frame of time-lapse?):
Line: 943 Supplement Figure 4. Example of a macrophage actively phagocytosing a TIE:EGFP+ cell in a melanoma.
Reviewer #2 (Recommendations for the authors):
1. Figure 1, Extended data 1 and supplemental Figure 1: better characterization of the reporter lines is important at different ages and in melanomas. Only one melanoma is shown in Figure 1C and another one in S1B, and, curiously, the pattern of TIE:EGFP expression is completely different: clusters vs homogenous expression. More pictures and quantitation of TIE:EGFP+ and TIE:EGFP- cells are required.
2. Figure 3 and results about macrophages: Figure 3A: most macrophages do not express mitfa and sox10. Figure 3B is not convincing: I do not see any white cells that would represent macrophages taken up TIE:EGFP+ melanoma cells. I can see several green macrophages that may indicate that macrophages express the TIE:EGFP construct. Additional pictures and video, if possible are needed.
3. Figure 3D: I do not understand why Q1 represent non-melanoma cells that were phagocytosed. I think these are non-phagocytosing macrophages. So, Q2 has to be plotted not Q1+Q2. Anyway, I think that allotransplant experiments of melanoma cells from TIE:EGFP fish need to be performed in mpeg1:cherry recipient, and vice versa, to demonstrate that macrophages are phagocytosing TIE:GFP+ cells rather than expressing the construct. In addition.
4. Figure 4 and SATB2 data: these data should be compared with those obtained in Figure 1-3 w/o SATB2. Does SATB2 facilitate melanoma progression? Is there any difference in the TGFB targets between control and SATB2 TIE:EGFP+ cells?
5. Lines 286-295: it is speculated that the novel chronic TGFB targets are involved in cell migration. I think it would be a nice complement to perform an allotransplant of TIE:EGFP+ and TIE:EGF- melanoma cells and study their aggressiveness.
Reviewer #3 (Recommendations for the authors):
1) The confocal pictures in Figure 3B are not convincing. We cannot see the edge of the cells to make sure that they are not debris. At the same time, how do we expect that melanoma cells that have been phagocytosed by macrophages and that are present in fragments (only puncta of TIE:EGFP signal are seen in macrophages) would maintain intact the mitfa and sox10 mRNAs retrieved in the single cell RNA analysis? This is why we need absolutely perfect confocal images of tumor sections with clearly visible macrophages and tumor cells to support the hypothesis that macrophages preferentially phagocyte TGFbeta-expressing melanoma cells. In addition, this claim should be supported by some in vivo live observation to test the ability of macrophages to recognize TGFbeta-expressing melanoma cells versus other melanoma cells.
The doubt that this reporter labels a subpopulation of macrophages or macrophages in a particular status of activation induced by the presence of the tumor, rather than macrophages that have phagocytosed a rare population of melanoma cells that turn on the TGFb reporter remains.
2) To understand if the overexpression of SATB2 would activate the reporter, and generate melanomas with a larger proportion of TIE:EGFP positive cells, they produced tumors overexpressing SATB2 in the TIE:EGFP background. Then, they searched for differences in the macrophage populations. Here the authors admit that TGFbeta is known to convert macrophages from a pro-inflammatory M1-like to an anti-inflammatory M2-like state, thus implicitly suggesting that the reporter, which is not specific for melanoma but could be expressed in any cell of these transgenic fish, could be activated directly in the macrophages, with no need to justify its expression with the phagocytosis of a rare population of melanoma cells expressing it.
Related to Figure 1C, sections through the melanoma would help to distinguish whether the green fluorescence signal is part of the tumor and not just the residual brain tissue that express the reporter, and that we see clearly also in extended data Figure 1D.
One of the arrowheads in Figure 1C, in the 22 wks image, does not point to gfp signal. What is the yellow halo in the middle panel mitfa:mcherry at 20 and 22 weeks?
In Extended data 1 what is the GFP halo that we see in figures C and D that does not correspond to mitfa:mCherry? Please explain.
TIE:EGFP expressing melanoma cells downregulate interferon signaling
What are the controls here that were used for comparisons, please state in the figure legend (Suppl Figure 2B). It would be interesting to know how the expression of interferon-related genes compares to surrounding non-tumoral cells for the TIE:EGFP negative tumor cells.
In Figure 4 A the mCherry signal is too high and gives a yellow halo which is not due to co-expression with TIE:EGFP (as clearly visible in the TIE:EGFP upper panel). It is necessary to reduce the intensity of the mCherry signal until no spurious yellow is seen. The same applies to Figure 4D.
https://doi.org/10.7554/eLife.83527.sa1Author response
Essential revisions (for the authors):
Although the reviewers are very positive about the manuscript, there are several areas that could be explored further. There was a consensus that the role of macrophages in particular needs more data for the model to be supported.
1. There are multiple issues with Figures 1 and Extended Data Figure 1 as seen in the reviewers' comments below. These relate to quality and quantification and should be addressed.
2. The reviewers also suggested that Figure 3 data be improved with better confocal images and time-lapse imaging.
3. In Figure 4, SATB2 data should be compared to Figures 1-3 without SATB2. Does SATB2 facilitate melanoma progression?
Reviewer #1 (Recommendations for the authors):
Data exists of spatially mapped transcriptomes in zebrafish melanoma:
Hunter, et al. Spatially resolved transcriptomics reveals the architecture of the tumor-microenvironment interface. Nat Commun 12, 6278 (2021).
Are the results in this study showing regions of active TGFb signaling in advanced melanoma consistent with existing data of endogenous expression patterns?
This is an excellent connection that would help to confirm the existence of regional TGFb signaling hubs through transcriptional analyses. In collaboration with Dr. Richard White’s lab, we analyzed this spatially resolved transcriptomics data set for regional expression of a TGFb pathway target gene signature.1 In order to confidently determine cells in which the TGFb pathway was activated, this required a given “gene expression spot” (10X Visium Spatial Gene Expression) that expressed many TGFb target genes. However, Dr. White’s data only identified spots in which two TGFb target genes were simultaneously expressed, lacking the resolution needed to identify regions of active TGFb signaling. This analysis was not performed targeting TGFb target genes with probes, and so the limited identification of simultaneous TGFb target gene expression in a single cell was likely due to low detection sensitivity.
The proposed model of preferential phagocytosis of TIE:GFP+ cells vs negative cells is interesting, however, the data is still somewhat preliminary. Higher resolution imaging data and possibly time-lapse would better support the model. Could the authors provide some candidate factors produced by TIE:GFP+ cells that could trigger this response?
We thank the reviewer for this feedback and agree that additional imaging would provide more solid evidence of phagocytosis. To address this, we have performed additional high-resolution confocal imaging of TIE:EGFP+ cells and macrophages in the tumor. These images reveal a subset of macrophages that express the TIE:EGFP reporter endogenously as well as a subset of macrophages that contain TIE:EGFP+ fragments, often in endosome-like structures Figure 3—figure supplement 1 (previously Figure S4). We have found active phagocytosis to be difficult to capture in snapshots, making it challenging to quantify preferential phagocytosis by imaging. Our FACS experiment presented in Figure 3D provides a robust measurement of preferential phagocytosis of TIE:EGFP+ melanoma cells (vs. TIE:EGFP- melanoma cells) by macrophages.
We have looked into identifying several candidate factors regulated in TIE:EGFP+ cells that could trigger macrophage-mediated phagocytosis. Our single cell RNA-seq data showed differential upregulation of serpine1 mRNA expression in TIE:EGFP+ melanoma cells compared to TIE:EGFP- melanoma cells, and the serpine1 receptor, lrp1ab, is upregulated in TIE:EGFP+ macrophages compared to TIE:EGFP- macrophages. This may indicate that macrophages are recruited to and subsequently phagocytose serpine1-expressing TIE:EGFP+ melanoma cells. Indeed, SERPINE1, which encodes plasminogen activator inhibitor-1 (PAI-1), has been shown to promote cancer cell invasiveness and macrophage recruitment in an esophageal squamous cell carcinoma model.2 Multiple reports indicate a role for PAI-1 in macrophages recruitment and polarization.3,4,5 Furthermore, TIE:EGFPhigh macrophages differentially express mmp14b and the cysteine protease legumain, both of which have been described to promote TGFb bioavailability.6,7,8 These preliminary transcriptional data may indicate a mechanism by which macrophages are recruited to TIE:EGFP+ regions by PAI-1 gradients. Recruited macrophages may subsequently amplify the pool of active, bioavailable TGFb through the activity of enzymes such as mmp14b and lgmn. Such a model would explain the local clustering of macrophages in TGFb-responding regions of the tumor, and their subsequent endogenous activation of TGFb signaling and phagocytic behavior in these regions. (Figure 3B and C). In the paper, we have discussed this hypothesis in the Discussion, lines 350-363.
Chronic exposure to TGFb may affect macrophage polarization, resulting in the induction of M2-like markers, including mrc1b and vegfaa. Notably, in addition to M2-like markers, TIE:EGFPhigh macrophages differentially express a number of genes involved in cholesterol efflux, lipid metabolism, and apoptosis including abca1a, abca1b, lipf, npc2, adipor2, caspa, and casp3b. Recent reports indicate that TGFb signaling can drive lipid droplet formation and ABCA1 expression in macrophages.9,10 Thus, chronic zonal TGFb exposure and uptake may skew macrophages toward a stressed, anti-inflammatory phenotype. In this model, macrophages are continuously recruited to TGFb-high zones, where they facilitate TGFb amplification, phagocytose TGFb-responsive cells, and are gradually rendered incapable of tumoricidal activity. This effect on macrophages is discussed in the Discussion section, lines 364-376.
In the Extended Data Figure 1C-D experiments, if the mitf:mCherry is transiently injected, then the conclusions may be overstated. Line 90: "TIE:EGFP was also never expressed in mitfa:mCherry high early melanomas (Extended Data Figure 1D)". How many melanomas were observed? What level of GFP expression is required to score as positive in TIE:GFP fish? The conclusions should be reconsidered because transient F0 transgenics are inherently mosaic.
It is true that mitfa:mCherry is transiently expressed. We have changed the word “never” to “has yet to be observed” to address this overstatement in line 92 (previously line 90). 56 tumors were observed, as stated in line 101. While FACS of each tumor may be able to give a quantitative threshold of EGFP expression by which to score TIE:EGFP+ tumors, we used stereomicroscope imaging to identify TIE:EGFP+ tumors due to feasibility and to track changes during further tumor progression. Using imaging, it is not easy to give a quantitative threshold of EGFP expression due to differences of EGFP brightness resulting from several factors including the depth at which the cells are located in the tumor volume. To identify TIE:EGFP+ tumors, we looked for tumors in which there were several visible EGFP+ cells or an EGFP+ patch of cells. We also tracked the expansion of these EGFP+ zones over several months, confirming that they were not imaging artifacts.
Extended data 1B images are small and difficult to evaluate. Need better images that are quantified for fluorescence intensity.
For previous Extended Data 1B (current Figure 1—figure supplement 1B) images depicting EGFP signal, we have clarified using white arrows which regions containing EGFP fluorescence are referenced, making it easier to evaluate. In addition, we have added in situ hybridization (ISH) data probing for GFP, which has been quantified for staining intensity. These changes are now reflected in Figure 1—figure supplement 1B and C.
In the Figure 1C (20 and 22 wpf) panels, why is there a yellow signal in the mitfa:mCherry channel alone in the majority of the tumor but not in the merged image?
In the mitfa:mCherry channel, yellow signal is due to saturated high mCherry signal. In our melanoma model, high mitfa expression is characteristic of zebrafish tumors. Using a standard imaging protocol (with defined zoom and exposure) to define melanoma lesions, oversaturation of mitfa:mCherry is displayed by the imaging software in yellow. Using Image J, we have removed the yellow oversaturation signal in these images by rendering each mCherry channel image as an 8-bit image and re-coloring the pixels in red. This way, all pixels are treated the same and the pixel intensity is retained and represented by red color intensity in the new images. We then re-merged the EGFP and new mCherry channels for all tumors in all figures.
Which data does S1B refer to in this line?
Line 115: melanomas were processed for single-cell RNA-sequencing at 23 and 42 weeks post-fertilization (wpf), respectively (Figure 1C and S1B).
The two tumors that were processed for single-cell RNA-sequencing and referred to in previous Line 115 are (1) the tumor shown in manuscript Figure 1C (dissected at 23 wpf) and (2) the tumor shown in manuscript Figure 1—figure supplement 2B (dissected at 42 wpf).
Could the authors describe the marco marker and its significance here?
Line: 120 melanoma cells expressing mitfa and/or sox10, but we identified a marco-expressing TIE:EGFP+.
Marco is a known marker of macrophages, as well as a marker of M2-polarized macrophages. To be more straightforward in identifying the macrophage population in Figures 3 and 4 (and previous line 120), we defined the macrophage cluster using mpeg1.1 expression instead of marco expression.
Does the analysis of Figure S2B include macrophages or is it only the melanomas? Is there any independent validation of the GSEA results? This is regarding:
Line: 130 in TIE:EGFPhigh cells were interferon α and γ (Figure S2B).
Manuscript Figure 2—figure supplement 1B (previously Figure S2B) is labeled “Melanoma Cells,” it does not include macrophages. To provide an independent validation of the GSEA results showing down-regulation of IFNa and IFNγ pathway activation in TIE:EGFPhigh melanoma cells, we have isolated TIE:EGFPhigh and TIE:EGFP- melanoma cells from two additional tumors by FACS and performed qPCR for several IFN-response genes. This data confirms that IFN target genes go down in TIE:EGFPhigh tumor cells.
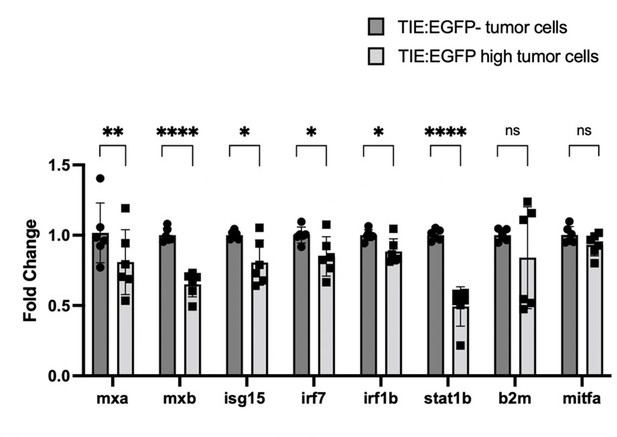
qPCR shows that IFN target genes are down-regulated in TIE:EGFPhigh melanoma cells compared to TIE:EGFP- melanoma cells.
Two Tg(mitfa:BRAFV600E);p53-/-;mitfa-/- tumors expressing TIE:EGFP, mitfa:BFP and mpeg:mCherry were dissociated for FACS. We sorted two populations of cells: TIE:EGFPhigh mitfa:BFP+ mpeg:mCherry-, and TIE:EGFP- mitfa:BFP+ mpeg:mCherry-. Here, we gated for mitfa:BFP+ melanoma cells, gating out the macrophage population which may contain mitfa:BFP due to tumor cell phagocytosis. We isolated RNA from these sorted populations, performed cDNA synthesis, and performed qPCR. We used housekeeping gene b-actin to normalize values according to loading differences. Here, we confirmed that 4 major IFN genes discussed in the manuscript are down-regulated: irf7, irf1b, stat1b, and b2m. We included 3 additional IFN genes here to further validate the IFN pathway trend: mxa, mxb, and isg15. Fold change of mitfa is included here as a negative control, as we do not expect its expression to change between the two conditions. N=2 tumors, 3 technical replicates each. 2-tailed, unpaired t-tests were performed.
In the following line, which genes are referred to specifically? Are these genes associated with a specific table or figure?
Line: 293 of melanoma patients exhibit up-regulation of these genes52,53.
This sentence refers to the 29 upregulated chronic TGFb genes shown in manuscript Figure 2C (first 29 genes listed on the dot plot).
Are there experiments that could better support the following statement:
Line: 306 Our work identified macrophage subclusters that contain low levels of mitfa and/or sox10 expression suggestive of phagocytosis of melanoma cells.
To further support the claim that mitfa and sox10 are expressed in macrophages due to melanoma cell phagocytosis, we have performed additional high-resolution imaging of TIE:EGFP+ cells and macrophages in zebrafish tumors (Figure 3—figure supplement 1). This imaging reveals fragments of TIE:EGFP within macrophages, occasionally contained in endosome-like structures. It also reveals co-localization of mitfa:BFP, mpeg:mCherry, and TIE:EGFP, suggesting phagocytosis of TGFb responsive melanoma cells.
Additionally, we isolated TIE:EGFP+ and TIE:EGFP- macrophages (using the mpeg:mCherry reporter) by FACS and performed qPCR for mitfa and sox10. This data (shown in manuscript Figure 3—figure supplement 2) shows that mitfa and sox10 expression is enriched in TIE:EGFP+ macrophages compared to TIE:EGFP- macrophages, further supporting that macrophages preferentially phagocytose TIE:EGFP+ melanoma cells.
Is there a GFP/mCherry merged image to support this statement, as it is not clear from the figure?
311 Figure 3C, there is co-localization of mpeg:mCherry and TIE:EGFP signal.
Thank you for requesting this clarification. To address this, we have included a merged image of the EGFP and mCherry channels in manuscript Figure 3C and have indicated the region of co-localization of mpeg:mCherry and TIE:EGFP with a white arrow.
The following figure legend is insufficient and needs more details (e.g. live or fixed sample? Optical section or z projection? Single frame of time-lapse?):
Line: 943 Supplement Figure 4. Example of a macrophage actively phagocytosing a TIE:EGFP+ cell in a melanoma.
To address this, we have added additional imaging information to the legend for manuscript Figure 3—figure supplement 1 (previously S4), such as methods for live imaging of zebrafish tumors, and optical sectioning vs. Z projection.
Reviewer #2 (Recommendations for the authors):
1. Figure 1, Extended data 1 and supplemental Figure 1: better characterization of the reporter lines is important at different ages and in melanomas. Only one melanoma is shown in Figure 1C and another one in S1B, and, curiously, the pattern of TIE:EGFP expression is completely different: clusters vs homogenous expression. More pictures and quantitation of TIE:EGFP+ and TIE:EGFP- cells are required.
To show more characterization of the TIE:EGFP reporter in melanoma, we have included an additional supplemental figure (Figure 1—figure supplement 3) showing its expression in 5 additional melanomas. Almost all tumors have only a small cluster of cells in which the TIE:EGFP reporter is activated, however an occasional tumor activates the reporter as highly as that represented in manuscript Figure 1—figure supplement 2B (previously Figure S1B). Although imaging was ideal for our purposes of tracking changes of EGFP expression over time during tumor progression, FACS would be able to more accurately quantify TIE:EGFP+ and TIE:EGFP- cells in the tumor, since imaging does not capture internal EGFP expression. As written in previous line 192 (current line 202), FACS determined that less than 1% of total cells from a tumor are EGFP+ (n=3).
2. Figure 3 and results about macrophages: Figure 3A: most macrophages do not express mitfa and sox10. Figure 3B is not convincing: I do not see any white cells that would represent macrophages taken up TIE:EGFP+ melanoma cells. I can see several green macrophages that may indicate that macrophages express the TIE:EGFP construct. Additional pictures and video, if possible are needed.
We thank the reviewers for this comment and agree this should be clarified. We do not expect all/most macrophages to express mitfa and sox10. Because only a subset of macrophages expresses these genes, this supports the hypothesis that some of them have phagocytosed melanoma cells, rather than the possibility that all macrophages express these genes endogenously. We have clarified in the legend for manuscript Figure 3B that if a macrophage expresses the TIE:EGFP reporter endogenously, we would expect to see the entire volume of the macrophage to express EGFP, rather than fragments. In lines 183-198, we have clarified that the majority of TIE:EGFP+ macrophages contain EGFP+ fragments, suggesting that they phagocytosed TIE:EGFP+ cells.
In three out of thirteen tumors imaged by confocal microscopy, we did capture a cluster of TIE:EGFP+ mitfa:BFP+ melanoma cells closely co-localized with macrophages, indicating that macrophages are engulfing these cells (Figure 3B, Figure 3—figure supplement 1B). To clarify which macrophages have engulfed TIE:EGFP+ melanoma cells in manuscript Figure 3B and Figure 3—figure supplement 1, we have added boxes around areas including white cells (which represent mitfa:BFP+ TIE:EGFP+ mpeg:mCherry+ cells). It is rare to capture active phagocytosis in snapshots (as seen in Figure 3—figure supplement 1C). However, the FACS experiment shown in manuscript Figure 3D and qPCR in manuscript Figure 3—figure supplement 2 confirms that macrophages preferentially phagocytose TIE:EGFP+ melanoma cells over TIE:EGFP- melanoma cells.
In the majority of tumors, however, most TIE:EGFP+ fragments within macrophages appear mitfa:BFP- via imaging. We have mentioned in the manuscript, particularly in lines 183-198 and in the Figure 3 legend, that macrophages likely phagocytose TIE:EGFP+ non-melanoma cells (in addition to TIE:EGFP+ melanoma cells). These non-melanoma cells may not have been identified in our SORT-seq data because the tissue dissociation protocol used is ideal for tumor cell viability, but not for that of various immune cell populations. Indeed, by confocal imaging, the majority of TIE:EGFP+ signal appears to be within clusters of macrophages, while FACS analysis and SORT-seq analysis shows that a large proportion of TIE:EGFP+ cells are tumor cells. This may be attributed to the technicality that tumor dissociation using Liberase (used for FACS and SORT-seq) favors tumor cell survival/dissociation over immune cell populations.
One alternative reason we may not often observe (via imaging) three-way co-localization between TIE:EGFP, mpeg:mCherry, and mitfa:BFP may be attributed to timing of observation. We most often observe macrophages with TIE:EGFP fragments, which may be the remnants of previously engulfed TIE:EGFP+;mitfa:BFP+ tumor cells, and the BFP expression is no longer strong enough to see at the time of imaging. Mitfa:BFP is expressed mosaically in these tumor cells and may not be expressed in every tumor cell. Because we do occasionally observe macrophages engulfing clusters of TIE:EGFP+ melanoma cells, and that FACS confirms that macrophages contain TIE:EGFP+ mitfa:BFP+ fragments, we are confident that melanoma cells are one TIE:EGFP+ population being phagocytosed by macrophages. We focused our studies on this particular interaction to show that macrophages preferentially phagocytose TIE:EGFP+ melanoma cells compared to TIE:EGFP- melanoma cells.
3. Figure 3D: I do not understand why Q1 represent non-melanoma cells that were phagocytosed. I think these are non-phagocytosing macrophages. So, Q2 has to be plotted not Q1+Q2. Anyway, I think that allotransplant experiments of melanoma cells from TIE:EGFP fish need to be performed in mpeg1:cherry recipient, and vice versa, to demonstrate that macrophages are phagocytosing TIE:GFP+ cells rather than expressing the construct. In addition.
We appreciate this being brought to our attention, and we agree that this alternative hypothesis is likely. Although imaging shows that the majority of TIE:EGFP+ macrophages contain only fragments of EGFP, suggesting phagocytosis, it is possible that Q1 can represent a subset of non-phagocytosing macrophages that express the TIE:EGFP construct endogenously, or macrophages that phagocytosed TIE:EGFP+ non-melanoma cells. We have clarified this in the figure legend and re-plotted Q2 only in manuscript Figure 3D.
While an allotransplant would help answer this question of phagocytosis, this experiment would have caveats in our system. Because we do not have isogenic lines expressing these reporters, macrophages from the recipient fish would phagocytose transplanted TIE:EGFP+ cells due to foreign recognition in a “graft vs. host” response, confounding our results.
4. Figure 4 and SATB2 data: these data should be compared with those obtained in Figure 1-3 w/o SATB2. Does SATB2 facilitate melanoma progression? Is there any difference in the TGFB targets between control and SATB2 TIE:EGFP+ cells?
SATB2 overexpression in our zebrafish model does facilitate melanoma progression, as stated in previous lines 204-206 (currently line 215-217), in which we referenced a paper previously published in our lab showing this data. We re-worded this sentence to include that SATB2 overexpression accelerates melanoma onset. We have also revised manuscript Figure 4 to include images of tumor development and TIE:EGFP activation in a control MCR:MCS tumor for reference.
There are differences in TGFb target genes in TIE:EGFP+ tumor cells between MCR:MCS and MCR:SATB2 tumors, although some genes are shared. Shown below is a table listing target genes that are upregulated in TIE:EGFP+ melanoma cells in a control MCR:MCS tumor and a MCR:SATB2 tumor, including those that are shared between the two genotypes (Author response table 1). In manuscript Figure 4C, we have indicated the shared genes in red text.
List of top up-regulated genes in TIE:EGFP+ melanoma cells of MCR:MCS and MCR:SATB2 tumors, as defined by SORT-seq.
These genes are also listed in manuscript Figures 2C and 4C.
| MCS | Shared | SATB2 |
|---|---|---|
| arhgdia | bsg | mmp14b |
| lama4 | col6a1 | igf2r |
| lrp1ab | itgb2 | igfbp3 |
| rhoq | marcksl1a | igfbp5b |
| rnd3a | nid1a | pak1 |
| ddr1 | rhoab | pxnb |
| fgd1 | sparc | nme4 |
| itga4 | lamb1a | ephb2b |
| itga5 | tnc | cdh2 |
| col1a2 | smad7 | |
| dv1b | sox2 | |
| cdkn2a | vim | |
| cdkn2b | rac1a | |
| serpine1 | lrrc17 | |
| fn1a | sema3fb | |
| snai1b | ctnnb1 | |
| twist1b | ||
| snai2 |
5. Lines 286-295: it is speculated that the novel chronic TGFB targets are involved in cell migration. I think it would be a nice complement to perform an allotransplant of TIE:EGFP+ and TIE:EGF- melanoma cells and study their aggressiveness.
We agree that an allotransplant of TIE:EGFP+ melanoma cells would be a nice experiment to determine if they are more migratory than TIE:EGFP- melanoma cells. However, this is beyond the scope of this paper. In this manuscript, we merely speculate that TGFb signaling may promote migration of melanoma cells, and we plan to directly determine this in future experiments.
Reviewer #3 (Recommendations for the authors):
1) The confocal pictures in Figure 3B are not convincing. We cannot see the edge of the cells to make sure that they are not debris. At the same time, how do we expect that melanoma cells that have been phagocytosed by macrophages and that are present in fragments (only puncta of TIE:EGFP signal are seen in macrophages) would maintain intact the mitfa and sox10 mRNAs retrieved in the single cell RNA analysis? This is why we need absolutely perfect confocal images of tumor sections with clearly visible macrophages and tumor cells to support the hypothesis that macrophages preferentially phagocyte TGFbeta-expressing melanoma cells. In addition, this claim should be supported by some in vivo live observation to test the ability of macrophages to recognize TGFbeta-expressing melanoma cells versus other melanoma cells.
The doubt that this reporter labels a subpopulation of macrophages or macrophages in a particular status of activation induced by the presence of the tumor, rather than macrophages that have phagocytosed a rare population of melanoma cells that turn on the TGFb reporter remains.
We thank the reviewer for this comment and have made revisions to address this concern. We have performed additional confocal imaging of zebrafish tumors, for which we have included images in manuscript Figure 3—figure supplement 1. This consistently shows TIE:EGFP+ fragments contained in mpeg:mCherry+ macrophages, representing macrophages that have phagocytosed TIE:EGFP+ cells. These EGFP+ fragments are not debris/imaging artifacts, since they are observed within the bounds of mpeg:mCherry+ cells, they are not fluorescent in every channel (as debris usually is), and they are contained in endosome-like structures in macrophages (manuscript Figure 3—figure supplement 1A, right). We have clarified in lines 183-198 and Figure 3 legend that macrophages likely phagocytose TIE:EGFP+ non-melanoma cells (in addition to TIE:EGFP+ melanoma cells), which may be why we observe a subset of macrophages containing TIE:EGFP fragments without mitfa:BFP fragments. One alternative reason we may not often observe (via imaging) three-way co-localization between TIE:EGFP, mpeg:mCherry, and mitfa:BFP may be attributed to timing of observation. We most often observe macrophages with TIE:EGFP fragments, which may be the remnants of previously engulfed TIE:EGFP+;mitfa:BFP+ tumor cells, and the BFP expression is no longer strong enough to see at the time of imaging. Mitfa:BFP is expressed mosaically in these tumor cells and may not be expressed in every tumor cell.
We have included additional images of tumors in which macrophages do contain TIE:EGFP and mitfa:BFP fragments (represented by white cells contained in boxes in manuscript Figure 3B and Figure 3—figure supplement 1B). Together with the FACS experiment depicted in Figure 3D, this data confirms that macrophages phagocytose TIE:EGFP+ melanoma cells. While our imaging data suggests that macrophages likely phagocytose TIE:EGFP+ non-melanoma cells as well, we decided to focus our studies on the melanoma cell population to show that macrophages preferentially phagocytose TIE:EGFP+ melanoma cells compared to TIE:EGFP- melanoma cells. It is possible for macrophages that have recently phagocytosed TIE:EGFP+ melanoma cells to maintain mitfa and sox10 mRNAs, if these cells are contained in phagosomes that have not yet fused with lysosomes and undergone degradation. This is likely why mitfa and sox10 mRNAs are retrieved in single cell RNA analysis in a subset of macrophages. The retainment of mitfa and sox10 transcripts was validated in an additional qPCR we performed of TIE:EGFP+ and TIE:EGFP- macrophages, as described in Figure 3—figure supplement 2.
In addition to clarifying the phagocytic behavior of TIE:EGFP+ cell populations, our additional imaging revealed a subset of macrophages that express TIE:EGFP endogenously (manuscript Figure 3—figure supplement 1A, left). A description of these two subsets of TIE:EGFP+ macrophages is included in lines 183-198.
2) To understand if the overexpression of SATB2 would activate the reporter, and generate melanomas with a larger proportion of TIE:EGFP positive cells, they produced tumors overexpressing SATB2 in the TIE:EGFP background. Then, they searched for differences in the macrophage populations. Here the authors admit that TGFbeta is known to convert macrophages from a pro-inflammatory M1-like to an anti-inflammatory M2-like state, thus implicitly suggesting that the reporter, which is not specific for melanoma but could be expressed in any cell of these transgenic fish, could be activated directly in the macrophages, with no need to justify its expression with the phagocytosis of a rare population of melanoma cells expressing it.
Indeed, additional confocal imaging reveals a subset of macrophages that express TIE:EGFP endogenously, which populations will be interesting to focus on going forward (manuscript Figure 3—figure supplement 1A, left). This imaging also reveals a subset of macrophages that phagocytose TIE:EGFP+ cells (manuscript Figure 3B and Figure 3—figure supplement B), which is a separate interesting phenomenon discussed in this manuscript. We have included a description of these two TIE:EGFP+ macrophage populations in lines 183-198.
Related to Figure 1C, sections through the melanoma would help to distinguish whether the green fluorescence signal is part of the tumor and not just the residual brain tissue that express the reporter, and that we see clearly also in extended data Figure 1D.
We are confident that the patch of EGFP fluorescence indicated by an arrowhead near the brain in manuscript Figure 1C is not residual brain fluorescence because it is expressed superficially and is expressed in individual cells, rather than the typical hazy, internal brain fluorescence pattern we see. We also observed this EGFP+ patch grow over the course of a few weeks. However, to further show that tumors do indeed contain clusters of TIE:EGFP activation, we have included an additional supplemental figure (Figure 1—figure supplement 3) displaying images of five additional advanced tumors, including those at different anatomical locations.
One of the arrowheads in Figure 1C, in the 22 wks image, does not point to gfp signal. What is the yellow halo in the middle panel mitfa:mcherry at 20 and 22 weeks?
This arrowhead in manuscript Figure 1C does point to EGFP+ cells, but they are hard to see at this magnification, so we deleted the arrowhead to minimize confusion.
In the mitfa:mCherry channel, yellow signal is due to saturated high mCherry signal. In our melanoma model, high mitfa expression is characteristic of zebrafish tumors. Using a standard imaging protocol (with defined zoom and exposure) to define melanoma lesions, oversaturation of mitfa:mCherry is displayed by the imaging software in yellow. Using Image J, we have removed the yellow oversaturation signal in these images by rendering each mCherry channel image as an 8-bit image and re-coloring the pixels in red. This way, all pixels are treated the same and the pixel intensity is retained and represented by red color intensity in the new images. We then re-merged the EGFP and new mCherry channels for all tumors in all figures.
In Extended data 1 what is the GFP halo that we see in figures C and D that does not correspond to mitfa:mCherry? Please explain.
The EGFP halo in previous Extended Data 1 (current Figure 1—figure supplement 1) indicates endogenous TIE:EGFP reporter expression in the spine (Figure 1—figure supplement 1D) and brain (Figure 1—figure supplement 1E). We have now clarified this in the legend for this figure.
TIE:EGFP expressing melanoma cells downregulate interferon signaling
What are the controls here that were used for comparisons, please state in the figure legend (Suppl Figure 2B). It would be interesting to know how the expression of interferon-related genes compares to surrounding non-tumoral cells for the TIE:EGFP negative tumor cells.
The internal control for this experiment was TIE:EGFP- melanoma cells, to which we compared interferon stimulated gene (ISG) expression in TIE:EGFPlow and TIE:EGFPhigh melanoma cells from the same tumor. We have clarified this in the legend of manuscript Figure 2—figure supplement 1 (previously S2B).
Using a scRNA-seq dataset we had available in the lab that contained melanocytes from zebrafish normal skin, early melanoma, and an advanced tumor, we analyzed relative interferon-related genes at these different stages (Author response image 2). Generally, it appears that interferon-related genes often increase in tumors compared to normal skin and precursor lesions. While IFN genes are generally up-regulated in melanoma cells within a tumor, the data in this manuscript describes a rare subset of tumor cells that up-regulate TGFb and thus down-regulate IFN.
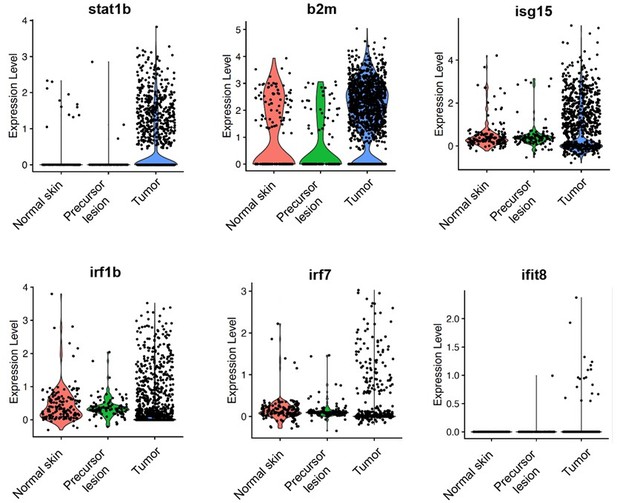
ScRNA-seq shows that expression of IFN target genes increases in tumors compared to normal skin and precursor lesions.
Mitfa:BFP+ melanocytes were sorted via FACS from normal skin, an early melanoma, and an advanced melanoma and 10X single-cell RNA-seq was performed. Data was analyzed using Seurat and cell types were called based on gene expression signatures. Depicted here are violin plots of several IFN target genes in melanocytes across the three stages.
In Figure 4 A the mCherry signal is too high and gives a yellow halo which is not due to co-expression with TIE:EGFP (as clearly visible in the TIE:EGFP upper panel). It is necessary to reduce the intensity of the mCherry signal until no spurious yellow is seen. The same applies to Figure 4D.
In manuscript Figure 4A, the yellow signal in the mitfa:mCherry channel represents saturated mCherry signal, indicative of characteristic mitfa high state of our zebrafish tumors. Using Image J, we have removed the yellow oversaturation signal in these images by rendering each mCherry channel image as an 8-bit image and re-coloring the pixels in red. This way, all pixels are treated the same and the pixel intensity is retained and represented by red color intensity in the new images. We have excluded the merged images in this panel to minimize confusion about the yellow signal, and to make space for a panel including development of a reference MCR:MCS tumor.
References:
Hunter, M.V., Moncada, R., Weiss, J.M. et al. Spatially resolved transcriptomics reveals the architecture of the tumor-microenvironment interface. Nat Commun 12, 6278 (2021).
Sakamoto, H., Koma, Yi., Higashino, N. et al. PAI-1 derived from cancer-associated fibroblasts in esophageal squamous cell carcinoma promotes the invasion of cancer cells and the migration of macrophages. Lab Invest 101, 353–368 (2021).
Kubala, M. H., Punj, V., Placencio-Hickok, V. R., Fang, H., Fernandez, G. E., Sposto, R., & DeClerck, Y. A. Plasminogen Activator Inhibitor-1 Promotes the Recruitment and Polarization of Macrophages in Cancer. Cell reports, 25(8), 2177–2191 (2018).
Honjo, K., Munakata, S., Tashiro, Y., Salama, Y., Shimazu, H., Eiamboonsert, S., Dhahri, D., Ichimura, A., Dan, T., Miyata, T., Takeda, K., Sakamoto, K., Hattori, K., & Heissig, B. Plasminogen activator inhibitor-1 regulates macrophage-dependent postoperative adhesion by enhancing EGF-HER1 signaling in mice. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 31(6), 2625–2637 (2017).
Baumeier, C., Escher, F., Aleshcheva, G. et al. Plasminogen activator inhibitor-1 reduces cardiac fibrosis and promotes M2 macrophage polarization in inflammatory cardiomyopathy. Basic Res Cardiol 116, 1 (2021).
Bai, P. et al. Macrophage-Derived Legumain Promotes Pulmonary Hypertension by Activating the MMP (Matrix Metalloproteinase)-2/TGF (Transforming Growth Factor)-β1 Signaling. Arteriosclerosis, Thrombosis, and Vascular Biology. 39 (4) (2019).
Sounni, N.E., Dehne, K., van Kempen, L., Egeblad, M., Affara, N.I., Cuevas, I., Wiesen, J., Junankar, S., Korets, L., Lee, J., Shen, J., Morrison, C.J., Overall, C.M., Krane, S.M., Werb, Z., Boudreau, N., Coussens, L.M. Stromal regulation of vessel stability by MMP14 and TGFbeta. Dis Model Mech. 3, (2010).
Robertson, I.B., Rifkin, DB. Regulation of the Bioavailability of TGF-β and TGF-β-Related Proteins. Cold Spring Harb Perspect Biol. 8, (2016).
Hu, Y. W., Wang, Q., Ma, X., Li, X. X., Liu, X. H., Xiao, J., Liao, D. F., Xiang, J., & Tang, C. K. TGF-beta1 up-regulates expression of ABCA1, ABCG1 and SR-BI through liver X receptor α signaling pathway in THP-1 macrophage-derived foam cells. Journal of atherosclerosis and thrombosis, 17(5), 493–502 (2010).
Bose, D., Banerjee, S., Chatterjee, N., Das, S., Saha, M., & Saha, K. D. Inhibition of TGF-β induced lipid droplets switches M2 macrophages to M1 phenotype. Toxicology in vitro : an international journal published in association with BIBRA, 58, 207–214 (2019).
https://doi.org/10.7554/eLife.83527.sa2




