Context-dependent requirement of G protein coupling for Latrophilin-2 in target selection of hippocampal axons
Figures
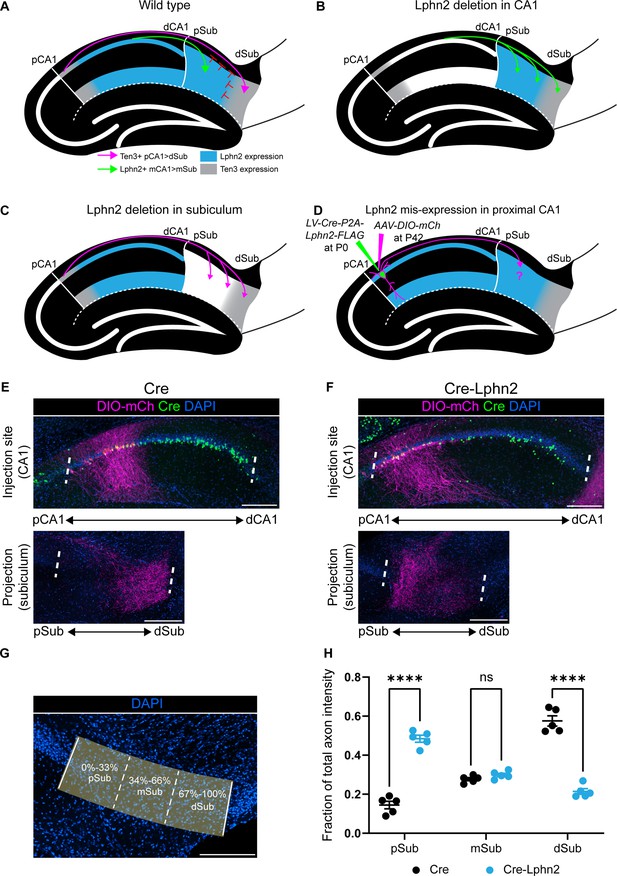
Misexpression of latrophilin-2 (Lphn2) in proximal CA1 axons causes axon mistargeting to the proximal subiculum.
(A) Cartoon depicting the topographic connections from proximal CA1 (pCA1) to distal subiculum (dSub) and distal CA1 (dCA1) to proximal subiculum (pSub). Ten3+ proximal CA1 axons are repelled from Lphn2 expressing (Lphn2+) proximal subiculum and Lphn2+ axons are repelled from Ten3+ distal subiculum. Red symbols indicate the repulsive cues experienced by CA1 axons, previously described in Pederick et al., 2021. (B) Deletion of Lphn2 from CA1 leads to distal CA1 axons mistargeting to distal subiculum, suggesting that Lphn2 acts cell-autonomously as a repulsive receptor. (C) Deletion of Lphn2 from proximal subiculum results in proximal CA1 axon mistargeting to proximal subiculum, suggesting Lphn2 acts cell-non-autonomously as a repulsive ligand. Figures (A–C) are based on Pederick et al., 2021. (D) Experimental design of Lphn2 misexpression assay in proximal CA1. At postnatal day (P) 0, lentivirus expressing Cre or Cre and Lphn2 was injected into CA1. This was followed by injection at P42 of Cre-dependent membrane bound mCherry (mCh) into proximal CA1 as an axon tracer. (E and F) Representative images of AAV-DIO-mCh (magenta; mCh expression in a Cre-dependent manner) injections in proximal CA1 (top) and corresponding projections in the subiculum (bottom). (G) A representative image of the subiculum with proximal subiculum (pSub), mid subiculum (mSub), and distal subiculum (dSub) regions highlighted. (H) The fraction of total axon intensity within proximal, mid, and distal subiculum. Cre: N=5 and Cre-Lphn2: N=5. Means ± SEM; two-way ANOVA with Sidak’s multiple comparisons test. Injection sites of all subjects are shown in Figure 1—figure supplement 3. Scale bars represent 200 μm.
-
Figure 1—source data 1
Misexpression of latrophilin-2 (Lphn2) in proximal CA1 axons causes axon mistargeting to the proximal subiculum.
- https://cdn.elifesciences.org/articles/83529/elife-83529-fig1-data1-v2.zip
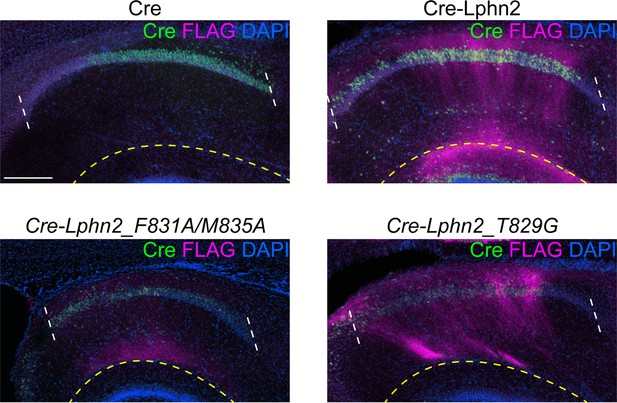
In vivo expression of lentivirus used in Figures 1, 3 and 4.
Representative images of Cre and FLAG immunostaining in CA1 of P9 mice injected with lentiviruses used in Figures 1, 3 and 4. The region between the white dashed line is CA1. The region below the yellow dashed line is the molecular layer of dentate gyrus. FLAG staining below the dashed yellow line is coming from dentate gyrus and not CA1 infected cells. Scale bar represents 200 μm. FLAG staining in Cre-Lphn2_F831A/M835A appears lower than Cre-Lphn- and Cre-Lphn2_T829G due to a lower efficiency injection site. Please see Figure 1—figure supplement 2 for further quantification of expression levels of latrophilin-2 (Lphn2, Lphn2_F831A/M835A, and Lphn2_T829G).
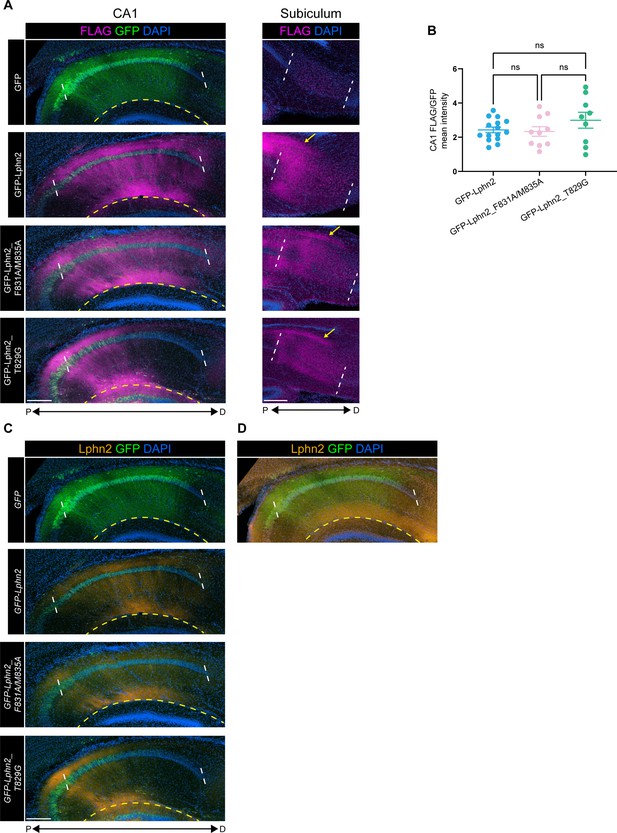
Quantification of latrophilin-2 (Lphn2), Lphn2_F831A/M835A, and Lphn2_T829G expression in CA1.
(A) Representative images of GFP and FLAG staining in CA1 of P9 mice injected with LV-GFP, LV-GFP-P2A-Lphn2-FLAG, LV-GFP-P2A-Lphn2_F831A/M835A-FLAG, or LV-GFP-P2A-Lphn2_T829G-FLAG (left). Representative images of FLAG staining in the subiculum of mice injected with LV-GFP, LV-GFP-P2A-Lphn2-FLAG, LV-GFP-P2A-Lphn2_F831A/M835A-FLAG, or LV-GFP-P2A-Lphn2_T829G-FLAG in CA1 (Right). FLAG staining present in the subiculum originates from CA1 axons (arrows). (B) Mean expression of FLAG immunostaining relative to mean expression of GFP within the same section. FLAG signal was measured from CA1 molecular layer and GFP signal was measured from CA1 cell body layer. FLAG/GFP mean intensity was used to normalize the variation between injections. The section with the highest mean expression of GFP was used from each animal. GFP-Lphn2: N=15, GFP-Lphn2_F831A/M835A: N=10 and GFP-Lphn2_T829G: N=9. Means ± SEM. One-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test. (C) Representative images of GFP and latrophilin-2 (Lphn2) immunostaining in the same sections as (A). (D) GFP and Lphn2 immunostaining in the same section as top (C) with 19 x higher laser power. At increased laser power endogenous Lphn2 can be visualized in the molecular layer of CA1. We found that the laser power required to visualize endogenous Lphn2 in the molecular layer of CA1 was ~19 x of that required to observed overexpressed wild type Lphn2 and mutant Lphn2. This suggests that even with variations of injection efficiency, the levels of overexpressed Lphn2 are very likely above that of endogenous Lphn2. The region between the white dashed line is CA1. The region below the yellow dashed line is the molecular layer of dentate gyrus. Immunostaining signal below the dashed yellow line is coming from dentate gyrus and not CA1 infected cells. Scale bars represent 200 μm.
-
Figure 1—figure supplement 2—source data 1
Quantification of latrophilin-2 (Lphn2), Lphn2_F831A/M835A, and Lphn2_T829G expression in CA1.
- https://cdn.elifesciences.org/articles/83529/elife-83529-fig1-figsupp2-data1-v2.zip
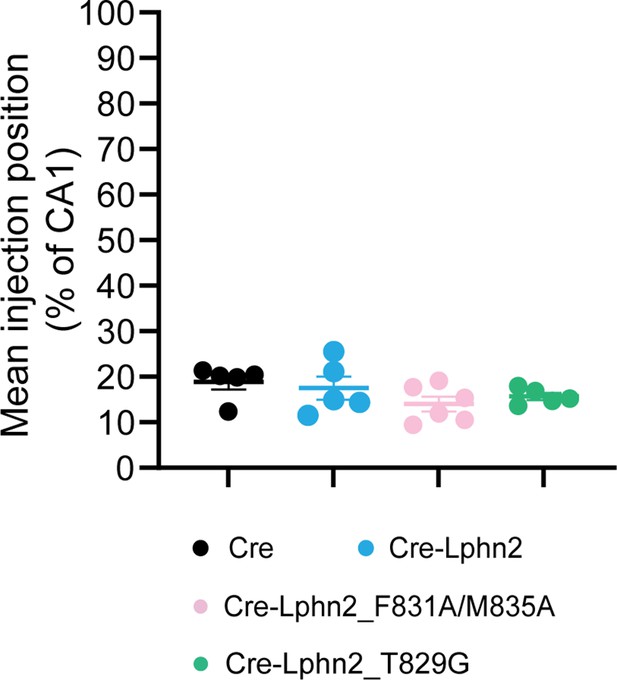
Mean injection site positions for proximal CA1 axon tracing in Figures 1, 3 and 4.
All injection sites are in the most proximal 30% of CA1. The most proximal 30% of CA1 is the Lphn2-low region of CA1 (Pederick et al., 2021) and was, therefore, targeted for latrophilin-2 (Lphn2) misexpression.
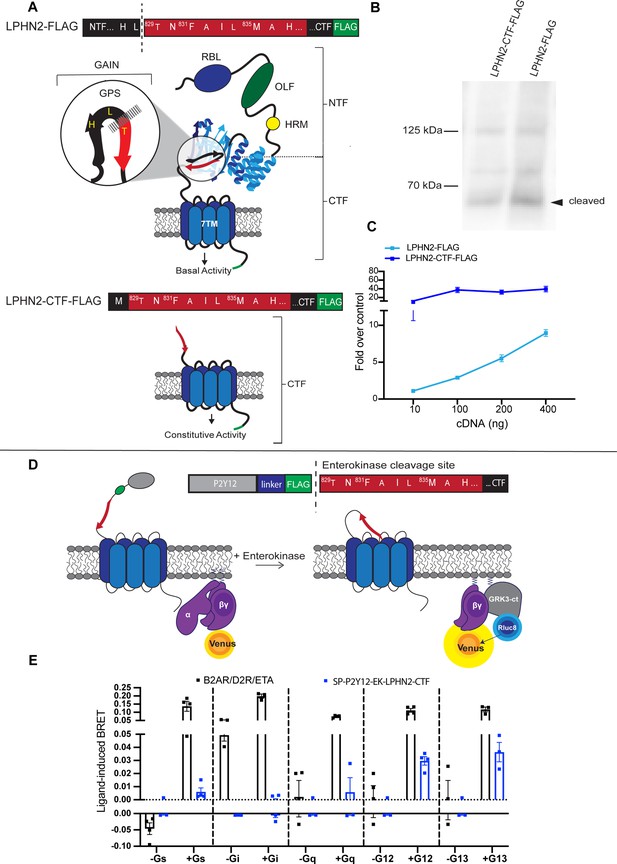
Exposure of the latrophilin-2 (Lphn2) tethered agonist (TA) promotes intracellular signaling through Gα12/13.
(A) Cartoon representations of full-length and tethered agonist-exposed (CTF) Lphn2 with detailed amino acid sequences for the TA. The extracellular domain of Lphn2 comprises an N-terminal rhamnose-binding lectin domain (RBL), an olfactomedin-like domain (OLF), a serine/threonine-rich region, and a HormR domain (HRM). It also contains the GPCR autoproteolysis-inducing (GAIN) domain necessary for autoproteolytic cleavage. This cleavage divides the aGPCR into two polypeptide chains: an N-terminal fragment (NTF) and a C-terminal fragment (CTF). The peptide stretch directly following the proteolytic cleavage site is known as the ‘Stachel’ or tethered agonist. Exposure of the tethered agonist results in aGPCR activation and downstream signaling. (B) Representative immunoblot analysis (N=3) of wild-type Lphn2 and Lphn2-CTF expression in HEK293T cells using a primary antibody against FLAG (1:500, ThermoFisher, PA1-984B). Expected bands for full-length Lphn2-FLAG and Lphn2-CTF-FLAG are 164 kDa and 72 kDa, respectively. (C) Serum response element (SRE) luciferase reporter assay for Lphn2 and Lphn2-CTF shows that removing the entire NTF up to the GPS cleavage site constitutively enhances SRE signaling (N=3 biological replicates, 9 technical replicates). (D) Schematic outlining the Gβγ-release bioluminescence resonance energy transfer (BRET) assay. The Lphn2 tethered agonist is capped with an enterokinase cleavage site (EK) preceded by a hemagglutinin signal peptide (SP), the P2Y12 N-terminal extracellular sequence, and a flexible linker (Lizano et al., 2021). Addition of 10 nM enterokinase generates a tethered agonist neoepitope identical to activated endogenous Lphn2. Lphn2 activation results in G protein dissociation, allowing Gβγ-Venus to associate with the C-terminus of GPCR kinase 3 (GRK3-ct) (Hollins et al., 2009). (E) Gβγ-release BRET assay testing SP-P2Y12-EK-Lphn2-CTF activation of Gαs, Gαi, Gαq, Gα12, and Gα13 in HEKΔ7 cells (N=3–4 biological replicates, 9–12 technical replicates). β2-adrenergic receptor (β2AR) with 1 µM isoproteronal, dopamine receptor D2 (D2R) with 10 µM quinpirole, and endothelin receptor (ETA) with 100 nM ligand ET-1 were used as positive controls, for Gαs, Gαi and Gαq/12/13 ,respectively. Means ± SEM; Multiple unpaired t tests between no G protein and G protein conditions; **p<0.01; ****p<0.0001.
-
Figure 2—source data 1
Full raw unedited blot of latrophilin-2 (Lphn2) expression in HEK293T cells.
- https://cdn.elifesciences.org/articles/83529/elife-83529-fig2-data1-v2.zip
-
Figure 2—source data 2
Uncropped immunoblot analysis of latrophilin-2 (Lphn2) expression in HEK293T cells.
Primary antibody against Flag (1:500, ThermoFisher, PA1-984B), secondary antibody anti-rabbit HRP (1:10,000, ThermoFisher, Cat #31458). Blue arrows indicate bands of interest for Lphn2-CTF-Flag and full-length Lphn2-Flag.
- https://cdn.elifesciences.org/articles/83529/elife-83529-fig2-data2-v2.zip
-
Figure 2—source data 3
Replicates of the immunoblot assay.
- https://cdn.elifesciences.org/articles/83529/elife-83529-fig2-data3-v2.zip
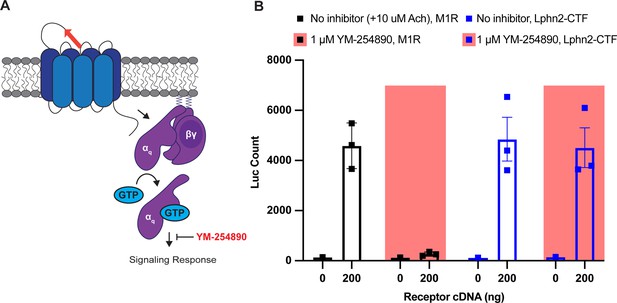
Gαq-inhibitor YM-254890 does not impair serum response element (SRE) luciferase response of Lphn2-CTF.
(A) Schematic of YM-254890 activity in the gene reporter assay. YM-254890 inhibits signaling responses downstream of Gαq activation. (B) 1 µM YM-254890 inhibited muscarinic acetylcholine receptor M1 (M1R) signaling in the SRE luciferase assay (N=3 biological replicates, 9 technical replicates). For the M1R, 10 µM acetylcholine (Ach) was added for 6 hr prior to reading the luminescence signal. 1 µM YM-254890 did not inhibit Lphn2-CTF signaling in the SRE luciferase assay.
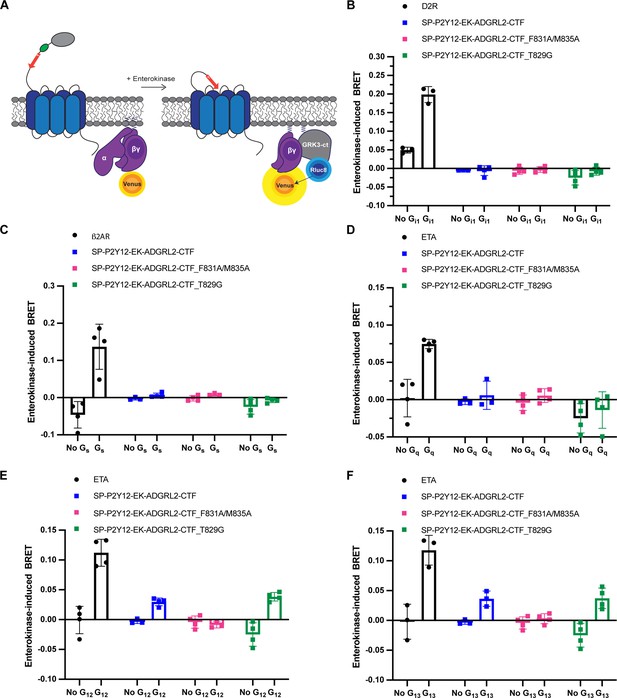
Gβγ-release bioluminescence resonance energy transfer (BRET) assay shows latrophilin-2 (Lphn2) couples to Gα12 and Gα13.
(A) Schematic outlining the Gβγ-release BRET assay. The Lphn2 tethered agonist is capped with an enterokinase cleavage site (EK) preceded by a hemagglutinin signal peptide (SP), P2Y12 N-terminal extracellular sequence, and flexible linker (Lizano et al., 2021). Addition of 10 nM enterokinase generates a tethered agonist neoepitope identical to activated endogenous Lphn2. Lphn2 activation results in G protein dissociation, allowing Gβγ-Venus to associate with the C-terminus of GPCR kinase 3 (GRK3-ct) (Hollins et al., 2009). Testing activation of SP-P2Y12-EK-Lphn2-CTF constructs with (B) Gαi,(C) Gαs, (D) Gαq, (E) Gα12, and (F) Gα13 in HEKΔ7 cells (N=3–4 biological replicates, 9–12 technical replicates). Dopamine receptor D2 (D2R) with 10 µM quinpirole, β2-adrenergic receptor (β2AR) with 1 µM isoproterenol, and endothelin receptor (ETA) with 100 nM ligand ET-1 were used as positive controls.
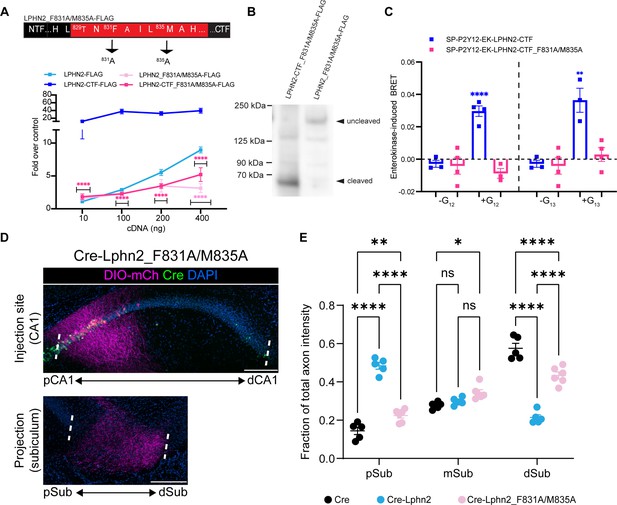
Lphn2_F831A/M835A has impaired G protein coupling activity and autoproteolytic cleavage and fails to misdirect proximal CA1 (pCA1) axons to the proximal subiculum (pSub) when misexpressed.
(A) Schematic of the mutated tethered agonist for Lphn2_F381A/M835A. The serum response element (SRE) luciferase reporter assay shows that both the full-length Lphn2_F831A/M835A and the Lphn2-CTF_F831A/M835A have impaired signaling (N=3 biological replicates, 9 technical replicates). Means ± SEM; Multiple unpaired t-tests between full-length latrophilin-2 (Lphn2) and Lphn2_F831A/M835A and Lphn2-CTF and Lphn2-CTF_F831A/M835A constructs; ****p<0.0001. (B) Representative immunoblot analysis (N=3) of TA-dead Lphn2 and TA-dead Lphn2-CTF expression in HEK293T cells using a primary antibody against FLAG (1:500, ThermoFisher, PA1-984B). Expected bands for full-length Lphn2_F831A/M835A-FLAG and Lphn2_F831A/M835A-CTF-FLAG are 164 kDa and 72 kDa, respectively. (C) Gβγ-release BRET assay testing SP-P2Y12-EK-Lphn2-CTF_F831A/M835A activation of Gα12 and Gα13 in HEKΔ7 cells (N=3–4 biological replicates, 9–12 technical replicates). SP-P2Y12-EK-Lphn2-CTF signaling is shown for comparison. Means ± SEM; Multiple unpaired t tests between no G protein and G protein conditions; *p<0.05, **p<0.01; ****p<0.0001. (D) Representative images of AAV-DIO-mCh (magenta; mCh expression in a Cre-dependent manner) injections in proximal CA1 (top) and corresponding projections in the subiculum (bottom). (E) Fraction of total axon intensity within proximal, mid, and distal subiculum. Cre: N=5, Cre-Lphn2: N=5 and Cre-Lphn2_F831A/M835A: N=6. Means ± SEM; two-way ANOVA with Sidak’s multiple comparisons test. Injection sites of all subjects are shown in Figure 1—figure supplement 3. Scale bars represent 200 μm.
-
Figure 3—source data 1
Uncropped immunoblot analysis of latrophilin-2 (Lphn2) expression in HEK293T cells.
Primary antibody against Flag (1:500, ThermoFisher, PA1-984B), secondary antibody anti-rabbit HRP (1:10,000, ThermoFisher, Cat #31458). Magenta arrows indicate bands of interest for Lphn2-F831A/M835A-CTF-Flag and full-length Lphn2-F831A/M835A-Flag.
- https://cdn.elifesciences.org/articles/83529/elife-83529-fig3-data1-v2.zip
-
Figure 3—source data 2
Lphn2_F831A/M835A has impaired G protein coupling activity and autoproteolytic cleavage and fails to misdirect proximal CA1 (pCA1) axons to the proximal subiculum (pSub) when misexpressed.
- https://cdn.elifesciences.org/articles/83529/elife-83529-fig3-data2-v2.zip
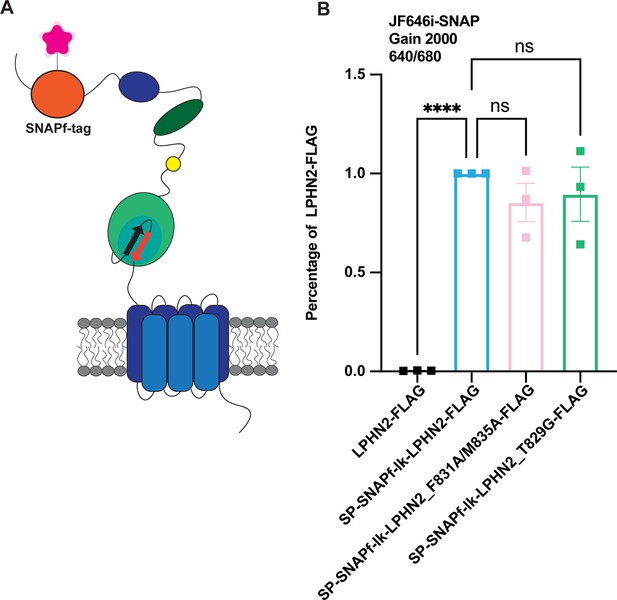
Latrophilin-2 (Lphn2), Lphn2_F831A/M835A, and Lphn2_T829G are expressed at the cell surface at comparable levels.
(A) Schematic of Lphn2 constructs with an additional SNAPfast-tag on the N terminus. (B) Lphn2 constructs tagged with a SNAPfast-tag were expressed in HEK293T cells for 24 hr. Transfected cells were incubated for 30 min with 1 µM membrane-impermeant Janelia Fluor 646, a SNAP-tag substrate. Emission was read using the filter 640/680 at a gain of 2000 using a PHERAstar FS microplate reader. Expression was calculated as a percentage of wild-type Lphn2. Statistical analysis was conducted using a one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparisons test (N=3 biological replicates, nine technical replicates; ****p<0.0001).
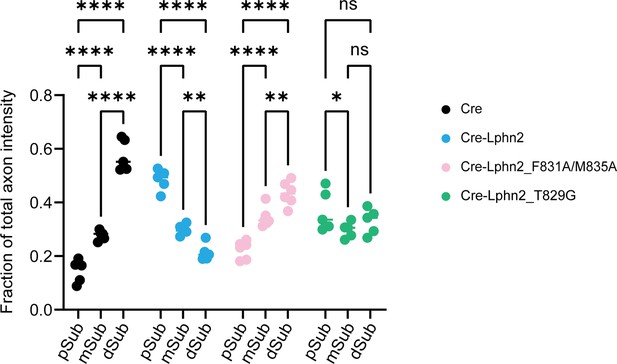
Comparison of fraction of total axon intensity across the subiculum within the same experimental condition.
Cre animals have low axon intensity in proximal subiculum (pSub) and high axon intensity in distal subiculum (dSub). Cre-Lphn2 animals have high axon intensity in pSub and low axon intensity in dSub. Cre-Lphn2_F831A/M835A animals show a similar pattern to LV-Cre with increasing axon intensity from pSub to dSub. Cre-Lphn2_T829G shows moderate axon enrichment in pSub but overall the total axon intensity is not biased towards any one region of subiculum. Means ± SEM; two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons test.
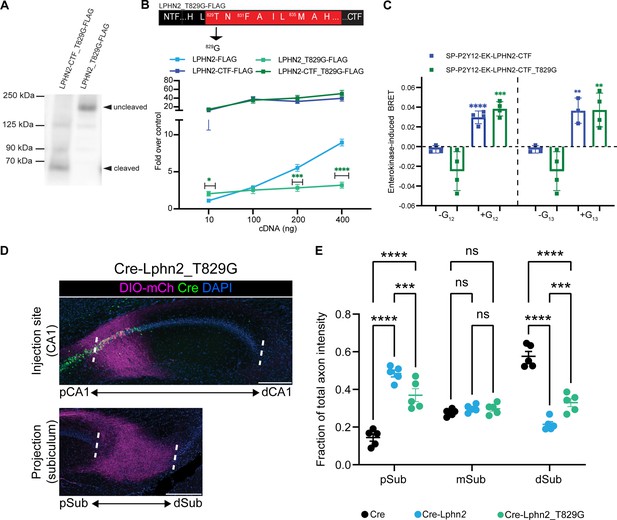
Lphn2_T829G impairs autoproteolytic cleavage, retains G protein activity in the truncated receptor, and misdirects axons to the proximal subiculum (pSub) when misexpressed.
(A) Representative immunoblot analysis (N=3) of Lphn2_T829G and Lphn2-CTF_T829G expression in HEK293T cells using a primary antibody against FLAG (1:500, ThermoFisher, PA1-984B). Expected bands for full-length Lphn2_T829G-FLAG and Lphn2-CTF_T829G-FLAG are 164 kDa and 72 kDa, respectively. (B) Schematic of the mutated tethered agonist for Lphn2_T829G. The serum response element (SRE) luciferase reporter assay shows that the full-length Lphn2_T829G has impaired SRE levels while the Lphn2_T829G truncated up to the GPS cleavage site has SRE levels comparable to Lphn2_CTF (N=3 biological replicates, nine technical replicates). Means ± SEM; Multiple unpaired t tests between full-length Lphn2 and Lphn2_T829G and Lphn2-CTF and Lphn2-CTF_T829G constructs; *p<0.05; ***p<0.001; ****p<0.0001. (C) Gβγ-release BRET assay testing SP-P2Y12-EK-Lphn2-CTF_T829G activation of Gα12 and Gα13 in HEKΔ7 cells (N=3–4 biological replicates, 9–12 technical replicates). SP-P2Y12-EK-Lphn2-CTF signaling is shown for comparison. Means ± SEM; Multiple unpaired t-tests between no G protein and G protein conditions; ***p<0.001; ****p<0.0001. (D) Representative images of AAV-DIO-mCh (magenta; mCh expression in a Cre-dependent manner) injections in proximal CA1 (top) and corresponding projections in the subiculum (bottom). (E) Fraction of total axon intensity within proximal, mid, and distal subiculum. Cre: n=5, Cre-Lphn2: n=5 and Cre-Lphn2_T829G: n=5. Means ± SEM; two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons test. Injection sites of all subjects are shown in Figure 1—figure supplement 3. Scale bars represent 200 μm.
-
Figure 4—source data 1
Uncropped immunoblot analysis of latrophilin-2 (Lphn2) expression in HEK293T cells.
Primary antibody against Flag (1:500, ThermoFisher, PA1-984B), secondary antibody anti-rabbit HRP (1:10,000, ThermoFisher, Cat #31458). Green arrows indicate bands of interest for Lphn2-T829G-CTF-Flag and full-length Lphn2-T829G-Flag.
- https://cdn.elifesciences.org/articles/83529/elife-83529-fig4-data1-v2.zip
-
Figure 4—source data 2
Lphn2_T829G impairs autoproteolytic cleavage, retains G protein activity in the truncated receptor, and misdirects axons to the proximal subiculum (pSub) when misexpressed.
- https://cdn.elifesciences.org/articles/83529/elife-83529-fig4-data2-v2.zip
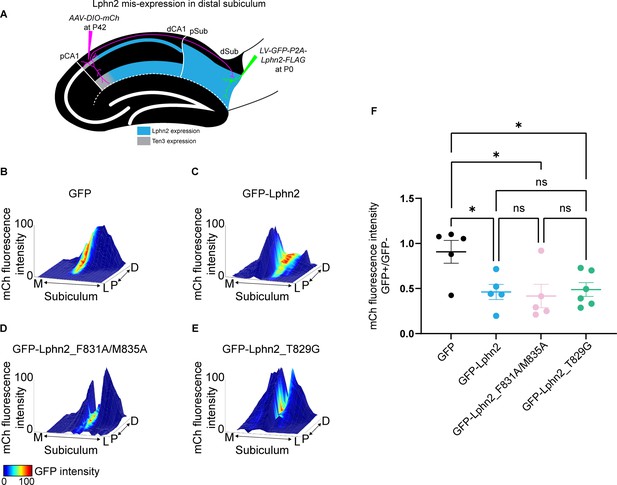
Misexpression of latrophilin-2 (Lphn2) mutants in target distal subiculum neurons does not cause mistargeting of proximal CA1 axons.
(A) Experimental design of Lphn2 misexpression assay in distal subiculum. (B to E). Representative mountain plots showing normalized mCh fluorescence as height (proximal CA1 axon projections in subiculum) and normalized GFP fluorescence as color (lentivirus expression). P, proximal; D, distal; M, medial; L, lateral. (F) Ratio of mCh fluorescence intensity (from proximal CA1 axons) in GFP+ versus GFP– regions of the subiculum. GFP: N=5, GFP-Lphn2: N=5, GFP-Lphn2_F831A/M835A: N=5 and GFP-Lphn2_T829G: N=6. Means ± SEM. One-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test.
© 2021, AAAS. Data from panels B, C and F (left two columns) are respectively reproduced from Figure S8B (top left panel), Figure S8E (bottom right panel) and Figure 2G (left two columns) of Pederick et al., 2021, reprinted with permission from AAAS. The panels from B, C and F are therefore not covered by the CC-BY 4.0 license and further reproduction would need permission from the copyright holder
-
Figure 5—source data 1
Misexpression of Lphn2 mutants in target distal subiculum neurons does not cause mistargeting of proximal CA1 axons.
- https://cdn.elifesciences.org/articles/83529/elife-83529-fig5-data1-v2.zip
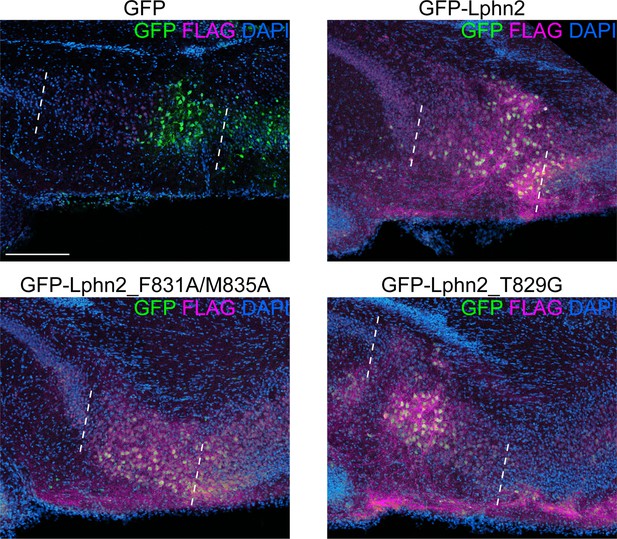
In vivo expression of lentivirus used in Figure 5.
Representative images of GFP and FLAG immunostaining in P8 subiculum of mice injected with lentiviruses used in Figure 5. The region between the white dashed line is subiculum. Scale bar represents 200 μm.
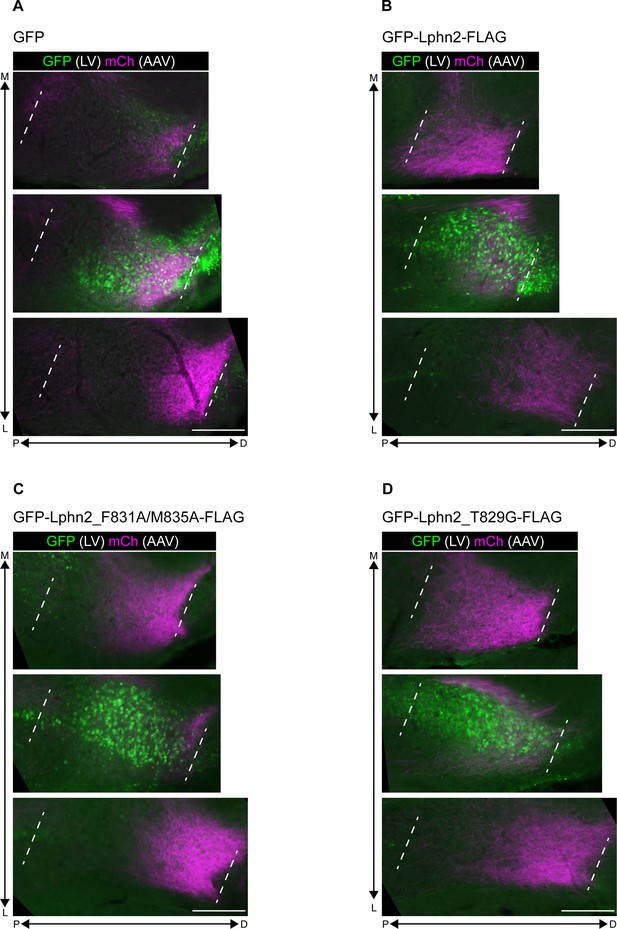
Representative images corresponding to Figure 5B–E.
Representative images from mice injected with LV-GFP (A) and LV-GFP-P2A-Lphn2-FLAG (B), LV-GFP-P2A-Lphn2_F831A/M835A-FLAG (C), or LV-GFP-P2A-Lphn2_T829G-FLAG (D) at P0 subiculum, followed by AAV-mCh injection in adult proximal CA1, showing axons from proximal CA1 neurons (magenta) and LV injection site (green) in subiculum.
Three 60 μm sections that contain proximal CA1 axons at different positions along the medial–lateral (M–L) axis are shown, with the center section overlapping with the LV injection site. These images correspond to mice analyzed in Figure 5B–E. Scale bar represents 200 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HEK293T (epithelial, kidney) | ATCC | RRID:CVCL_0063 | |
| Cell line (Homo sapiens) | HEKΔ7 | Alvarez-Curto et al., 2016, PMCID:PMC5207144 | JBC | HEK293 cells with targeted deletion via CRISPR-Cas9 of GNAS, GNAL, GNAQ, GNA11, GNA12, GNA13, and GNAZ |
| Antibody | anti-FLAG (rabbit polyclonal) | ThermoFisher, PA1-984B | RRID:AB_347227 | IB: 1:500 |
| Antibody | anti-rabbit HRP (donkey polyclonal) | ThermoFisher, Cat #31458 | RRID:AB_228213 | IB: 1:10,000 |
| Recombinant DNA reagent | SRE-luciferase/glo (plasmid) | Nazarko et al., 2018, PMCID:PMC6137404 | iScience | |
| Recombinant DNA reagent | Lphn2-Flag (plasmid) | This paper | pCDNA3.1 with Kozak (GCC) and C-terminal Flag tag | |
| Recombinant DNA reagent | Lphn2-CTF-Flag (plasmid) | This paper | pCDNA3.1 with Kozak (GCC), Lphn2 C-terminal fragment and C-terminal Flag | |
| Recombinant DNA reagent | SP-P2Y12-EK-Lphn2-CTF (plasmid) | This paper | Enterokinase cleavage site based on Lizano et al., 2021, Lphn2-CTF | |
| Recombinant DNA reagent | Lphn2-F831A/M835A-Flag (plasmid) | This paper | pCDNA3.1 with kozak (GCC), Lphn2, F831A and M835A, and C-terminal Flag | |
| Recombinant DNA reagent | Lphn2-F831A/M835A-CTF-Flag (plasmid) | This paper | pCDNA3.1 with kozak (GCC), Lphn2, F831A and M835A C-terminal fragment, and C-terminal Flag | |
| Recombinant DNA reagent | SP-P2Y12-EK-Lphn2-F831A/M835A-CTF (plasmid) | This paper | Enterokinase cleavage site based on Lizano et al., 2021, Lphn2-F831A/M835A-CTF | |
| Recombinant DNA reagent | Lphn2-T829G-Flag (plasmid) | This paper | pCDNA3.1 with kozak (GCC), Lphn2, T829G, and C-terminal Flag | |
| Recombinant DNA reagent | Lphn2-T829G-CTF-Flag (plasmid) | This paper | pCDNA3.1 with kozak (GCC), Lphn2, T829G C-terminal fragment, and C-terminal Flag | |
| Recombinant DNA reagent | SP-P2Y12-EK-Lphn2-T829G-CTF (plasmid) | This paper | Enterokinase cleavage site based on Lizano et al., 2021, Lphn2-T829G-CTF | |
| Recombinant DNA reagent | Gαs/Gαi/ Gαq/ Gα12/Gα13 (plasmid) | Hollins et al., 2009, PMCID:PMC2668204 | Cell Signal | |
| Recombinant DNA reagent | Gβ1 | Hollins et al., 2009, PMCID:PMC2668204 | Cell Signal | |
| Recombinant DNA reagent | Gγ2-Venus | Hollins et al., 2009, PMCID:PMC2668204 | Cell Signal | |
| Recombinant DNA reagent | GRK3ct-Rluc8 | Hollins et al., 2009, PMCID:PMC2668204 | Cell Signal | |
| Chemical compound, drug | Firefly D-luciferin | NanoLight Technology | Cat #306 | |
| Chemical compound, drug | Coelenterazine-h | NanoLight Technology | Cat #301 | |
| Chemical compound, drug | YM-254890 | AdipoGene Life Sciences | CAS 568580-02-9 | |
| Chemical compound, drug | Janelia Fluor 646 | Lavis Lab, Howard Hughes Medical Institute Janelia Research Campus | ||
| Genetic reagent (Mus musculus) | CD-1 | Charles River Laboratory | CD-1 mice were used for all animal experiments | |
| Recombinant DNA reagent | Lentiviral UbC-Cre | This paper | Lentivirus co-expressing Cre for in vivo experiments | |
| Recombinant DNA reagent | Lentiviral UbC-Cre-P2A-Lphn2-FLAG | This paper | Lentivirus co-expressing Cre-P2A-Lphn2-FLAG for in vivo experiments | |
| Recombinant DNA reagent | Lentiviral UbC-Cre-P2A Lphn2_F831A/M835A-FLAG | This paper | Lentivirus co-expressing Cre-P2A-Lphn2_F831A/M835A-FLAG for in vivo experiments | |
| Recombinant DNA reagent | Lentiviral UbC-Cre-P2A-Lphn2_T828G-FLAG | This paper | Lentivirus co-expressing Cre-P2A-Lphn2_T829G-FLAG for in vivo experiments | |
| Recombinant DNA reagent | Lentiviral UbC-GFP | Pederick et al., 2021, PMCID:PMC8830376 | Science | Lentivirus co-expressing GFP for in vivo experiments |
| Recombinant DNA reagent | Lentiviral UbC-GFP-P2A-Lphn2-FLAG | Pederick et al., 2021, PMCID:PMC8830376 | Science | Lentivirus co-expressing GFP-P2A-Lphn2-FLAG for in vivo experiments |
| Recombinant DNA reagent | Lentiviral UbC-GFP-P2A-Lphn2_F831A/M835A-FLAG | This paper | Lentivirus co-expressing GFP-P2A-Lphn2_F831A/M835A-FLAG for in vivo experiments | |
| Recombinant DNA reagent | Lentiviral UbC-GFP-P2A-Lphn2_T828G-FLAG | This paper | Lentivirus co-expressing GFP-P2A-Lphn2_F831A/M835A-FLAG for in vivo experiments | |
| Recombinant DNA reagent | AAV8-EF1a-DIO-ChR2-mCh | Addgene plasmid 20297 | AAV used to label axons of Cre expressing neurons with mCherry | |
| Recombinant DNA reagent | AAV8-CaMKIIa-ChR2-mCh | Addgene plasmid26975 | AAV used to label axons of neurons with mCherry | |
| Chemical compound, drug | DAPI | ThermoFisher | D1306 | 1:10,000 |
| Antibody | anti-mCherry (rat monoclonal) | ThermoFisher, M11217 | RRID:AB_2536611 | Immunohistochemistry 1:1,000 |
| Antibody | anti-Cre (Rabbit polyclonal) | Synaptic Systems, 257 003, | RRID:AB_2619968 | Immunohistochemistry 1:500 |
| Antibody | anti-GFP (chicken polyclonal) | Aves Labs, GFP-1020 | RRID:AB_10000240 | Immunohistochemistry 1:2,500 |
| Antibody | anti-FLAG (goat polyclonal) | Abcam, ab95045 | RRID:AB_10676074 | Immunohistochemistry 1:3000 |
| Antibody | anti-Lphn2 (Rabbit polyclonal) | Novus Biologicals, nbp2-58704 | Immunohistochemistry 1:500 | |
| Software, algorithm | Zen | Zeiss | Previously existing | |
| Software, algorithm | ImageJ | National Institutes of Health | Previously existing | |
| Software, algorithm | Adobe Photoshop | Adobe | Previously existing | |
| Software, algorithm | Adobe Illustrator | Adobe | Previously existing | |
| Software, algorithm | Matlab | MathWorks | Previously existing | |
| Software, algorithm | GraphPad Prism 9 | GraphPad Software | Previously existing |





