Single-cell analysis reveals dynamics of human B cell differentiation and identifies novel B and antibody-secreting cell intermediates
Figures
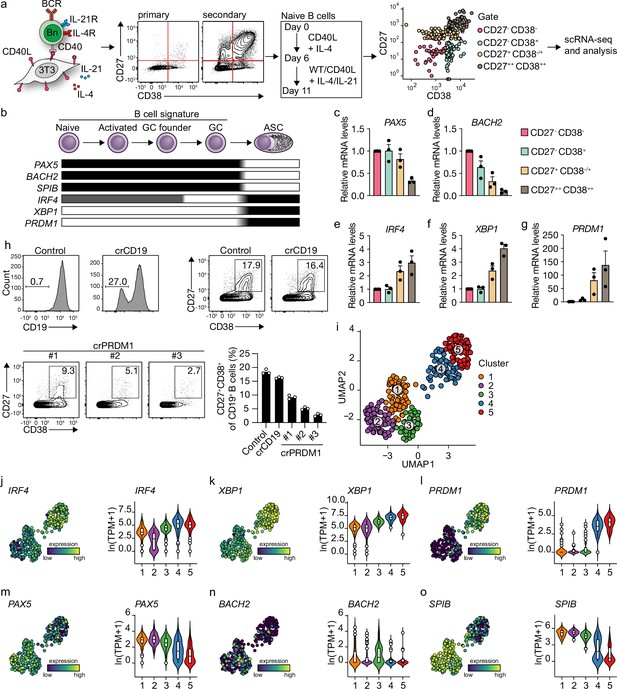
Unbiased analysis of in vitro-induced human naive B cell differentiation by scRNA-seq reveals five transcriptionally distinct B cell clusters.
(a) Overview of the scRNA-seq experiment in short human naive B cells was isolated and cultured with a human CD40L-expressing mouse fibroblast (3T3) cell line and recombinant IL-4. Secondary cultures were initiated on day 6 using 9:1 wild type (WT) 3T3 to CD40L-expressing 3T3 cells and IL-4/IL-21 as this was necessary to induce substantial differentiation into the CD27++CD38++ ASC subset. On day 11, 384 cells were single-cell sorted based on the expression of CD27 and CD38 and sequenced using the Smart-seq2 method. (b) Overview of the cellular stages and important transcription factors involved in B cells differentiation from naive to antibody-secreting cell (GC, germinal center; ASC, antibody-secreting cell). (c–g) Expression of PAX5 (c), BACH2 (d), IRF4 (e), XBP1 (f), and PRMD1 (g) mRNA in sorted populations was analyzed by qPCR and related to levels present in CD27-CD38- cells. Each data point represents the mean of an individual experiment (n = 3) with triplicate measurements. Mean values are represented as bars. (h) FACS analysis of surface CD19 levels (top left) and ASC differentiation (top right) in cultured primary human B cells expressing the indicated control or CD19-targeting RNP. FACS analysis of ASC differentiation (bottom left) and quantification (bottom right) in cultured primary human B cells expressing the indicated control or three different PRDM1-targeting RNPs. Representative of three independent experiments with triplicate measurements. (i) Uniform Manifold Approximation and Projection (UMAP) projection of single-cell transcriptomes of in vitro differentiated human naive B cells (276 high-quality cells). Each point represents one cell, and colors indicate graph-based cluster assignments. (j–o) UMAP projection as in (i) colored by the transcriptional regulators IRF4 (j), XBP1 (k) and PRMD1 (l), PAX5 (m), BACH2 (n), SPIB (o), which are important in B cell differentiation (left graph of each panel), along with corresponding distribution of average expression levels (ln(TPM+1)) across the B cell clusters (1, 2, and 3) and the ASC clusters (4 and 5) (right graph of each panel).
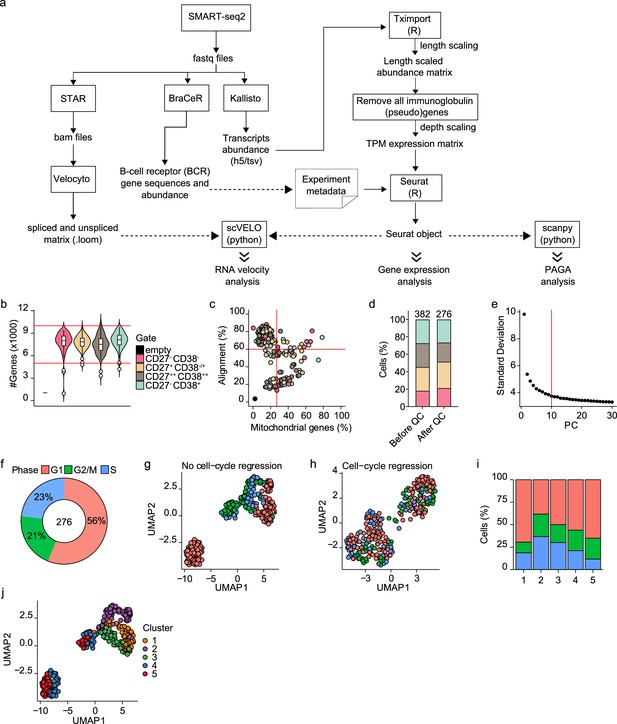
scRNA-seq analysis of differentiating human B cells.
(a) Schematic overview of the analysis pipeline. (b) Violin plot depicting the number of genes in each of the four sorted populations based on CD27 and CD38 expression. (c) Scatter plot showing the percentage alignment and the proportion of mitochondrial genes per single cell. (d) Stacked bars denote the frequency of the four sorted populations based on CD27 and CD38 expression before and after quality control. (e) The number of principal components (PC; x-axis) with the variance in gene expression explained by each component. The top 10 PCs were selected for further analysis. (f) Distribution of the different cell-cycle phases (Gap 1(G1), Gap 2/mitosis (G2/M), and synthesis (S)) among the 276 high-quality single cells. (g–h) Uniform Manifold Approximation and Projection (UMAP) projection of single-cell transcriptomes of in vitro differentiated human naive B cells without (g) and with (h) cell-cycle regression. Each point represents one cell, and colors indicate the cell-cycle phase. (i) Mean cell-cycle phase frequencies within each of the B cell subsets that were identified after cell-cycle regression. (j) UMAP projection of single-cell transcriptomes of in vitro differentiated human naive B cells without cell-cycle regression. Each point represents one cell, and colors indicate graph-based cluster assignments identified with cell-cycle regression.
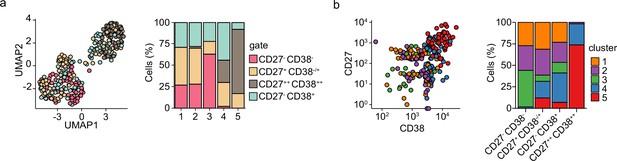
Subset distribution of differentiating primary human B cells.
(a) Uniform Manifold Approximation and Projection (UMAP) projection (left) as in Figure 1i colored by the four sorted populations based on CD27 and CD38 expression. Barplot show the relative abundance of CD27-CD38- CD27+CD38-/+, CD27++CD38++, CD27-CD38+ cells among the five transcriptionally distinct clusters (right). (b) Dot plot showing combined CD27 and CD38 protein expression values of cells from each cluster (left). Barplot show the relative abundance of the five transcriptionally distinct clusters within the CD27-CD38- CD27+CD38-/+, CD27++CD38++, and CD27-CD38+ populations (right).
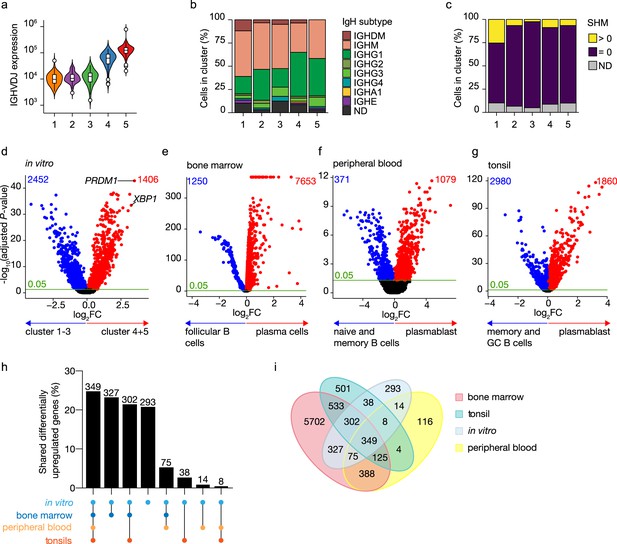
Identification of antibody-secreting cells with substantial gene expression overlap as compared to ex vivo-derived plasmablast/plasma cells among the in vitro-differentiated human naive B cells.
(a) Violin plot of the reconstructed B cell receptor immunoglobulins gene expression as determined by BraCeR within each B cell cluster. (b) Stacked bars denotes the frequency of isotype analysis of the reconstructed immunoglobulin heavy chain within each cluster. (c) Stacked bars showing the percentage of cells with a given B cell receptor mutation count within each B cell cluster. (d–g) Volcano plot depicting significantly (adjusted p-value <0.05) down- and upregulated genes. (d) In vitro-generated antibody-secreting cells (clusters 4 and 5), ex vivo bone marrow-derived plasma cells (e), ex vivo peripheral blood-derived plasmablast (f), and ex vivo tonsil-derived plasmablast/plasma cells (g) were compared to each specific B cell counterparts. (h, i) UpSetR plot (h) and Venn diagram (i) depict the intersection among all upregulated genes identified in in vitro and ex vivo antibody-secreting cells.
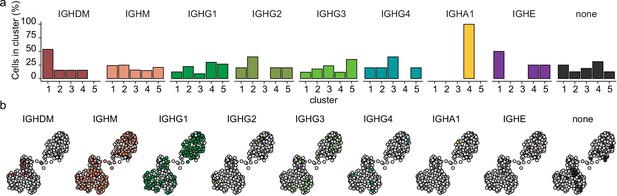
The distribution of heavy immunoglobulin gene in differentiating primary human B cells.
(a) Mean antibody subclass frequencies within each B cell subset. (b) Uniform Manifold Approximation and Projection (UMAP) projection as in Figure 1i colored by the reconstructed immunoglobulin heavy chain isotype.
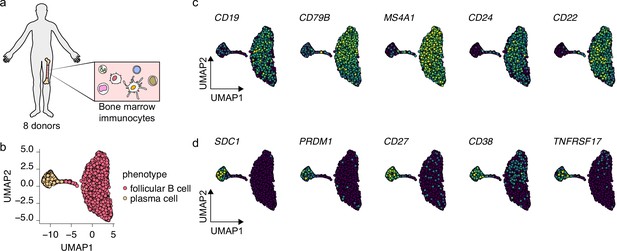
Selection of bone marrow-derived mature B cells and antibody-secreting cells.
(a) Schematics of scRNA-seq on bone marrow-derived immunocytes derived from eight human donors. (b) Uniform Manifold Approximation and Projection (UMAP) plot of single-cell transcriptomes of bone marrow-derived B cells and antibody-secreting plasma cells. Each point is one cell, and colors indicate graph-based cluster assignments. (c, d) UMAP plot of single-cell transcriptomes of bone marrow-derived B cells and antibody-secreting plasma cells, colored by expression of indicated B cell (c) and plasma cell (d) marker genes.
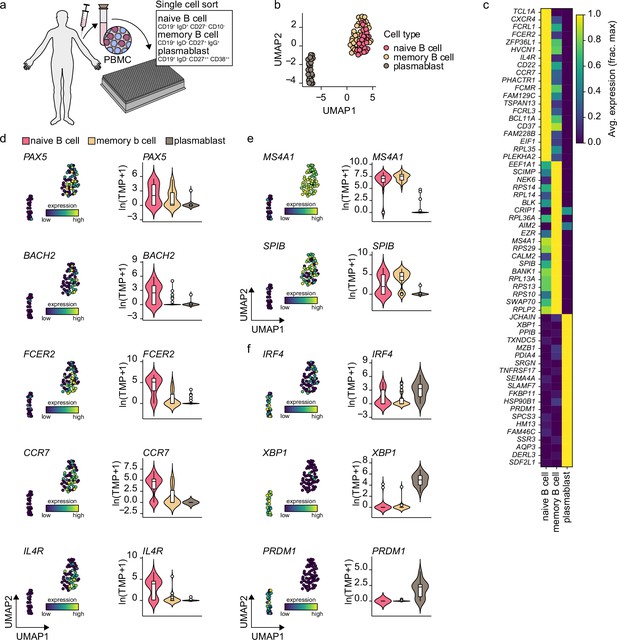
Gene expression characteristics of peripheral blood naive B cells, memory B cells, and plasmablast.
(a) Schematics of scRNA-seq on sort purified naive B cells, memory b cells, and plasmablast derived from healthy human peripheral blood. (b) Uniform Manifold Approximation and Projection (UMAP) plot of single-cell transcriptomes of peripheral human naive B cells and plasmablast. Each point is one cell, and colors indicate graph-based cluster assignments. (c) Heatmap of the 15 most differentially expressed genes significantly enriched in each cell cluster. (d-f) Violin plots of previously characterized naive B cell (d), memory b cell (e), and plasmablast (f) specific marker genes.
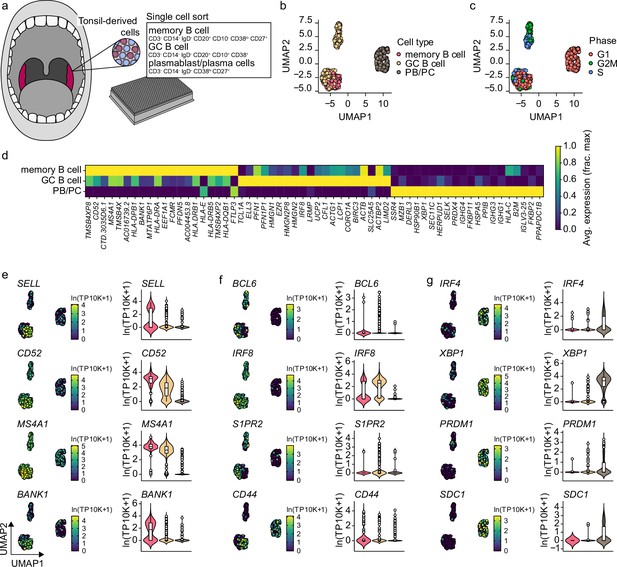
Gene expression characteristics of tonsillar memory B cells, germinal center (GC) B cells, and plasmablast/plasma cells.
(a) Schematics of scRNA-seq on sort purified memory b cells, GC B cells, and plasmablast/plasma cells derived from human tonsils. (b) Uniform Manifold Approximation and Projection (UMAP) plot of single-cell transcriptomes of tonsillar memory b cells, GC B cells, and plasmablast/plasma cells. Each point is one cell, and colors indicate graph-based cluster assignments. (c) UMAP plot as in b annotated by the cell-cycle phase. (d) Heatmap of the 20 most differentially expressed genes significantly enriched in each cell cluster. (e–g) Violin plots of previously characterized memory B cell (e), GC b cell (f), and plasmablast/plasma cell (g) specific marker genes. UMAP projection (left), along with corresponding distribution of average expression levels (ln(TP10k+1)), across the different B cell subsets (right).
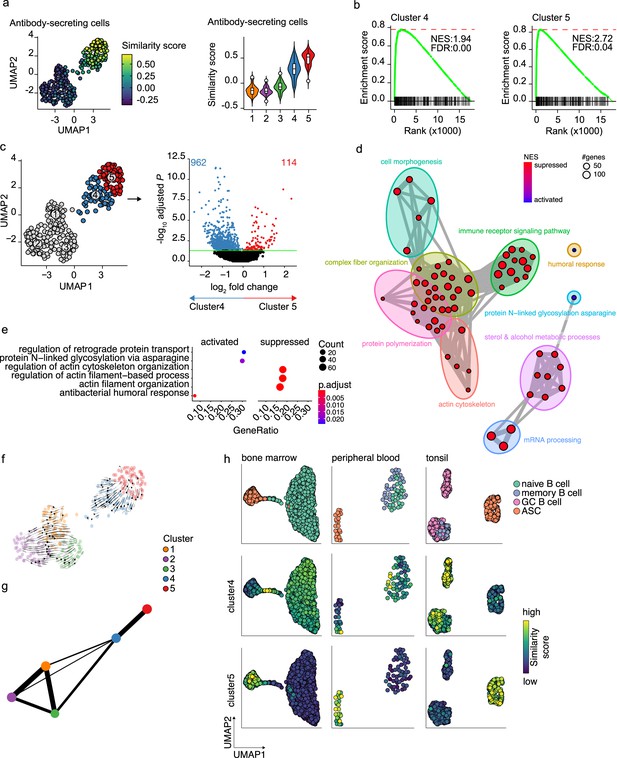
Pre- and terminally differentiated antibody-secreting cells (ASCs) separated in different clusters.
(a) Uniform Manifold Approximation and Projection (UMAP) projection colored by ASC similarity score (left), along with corresponding distribution of the similar score within each cluster (right). (b) Gene Set Enrichment Analysis (GSEA) enrichment plots for ASC gene signature in cells from clusters 4 (left) and 5 (right) compared to the other clusters (NES, normalized enrichment score; FDR, false discovery rate). (c) UMAP projection as in Figure 1i colored by clusters 4 and 5 membership (left) and volcano plot depicting significantly (adjusted p-value<0.05) down- and upregulated genes in cluster 5 compared to cluster 4. (d) Functional grouping network diagram of GSEA comparing clusters 4 and 5. The dot size and the dot color represent the number of genes in the pathway and the NES, respectively. (e) The top 3 most significantly differentially activated pathways as determined by GO enrichment analysis comparing clusters 4 and 5. The dot size and the dot color represent the number of genes in the pathway and the adjusted p-value, respectively. (f) Velocyto force field showing the average differentiation trajectories (velocity) for cells located in different parts of the UMAP plot. Arrow size conveys the strength of predicted directionality. (g) PAGA graph showing the connectivity between the clusters. Each node corresponds to each of the clusters identified using Seurat. The most probable path connecting the clusters is plotted with thicker edges. (h) UMAP projection of ex vivo B cells and ASC derived from bone marrow, peripheral blood, and tonsil colored by cluster 4 and 5 similarity score.
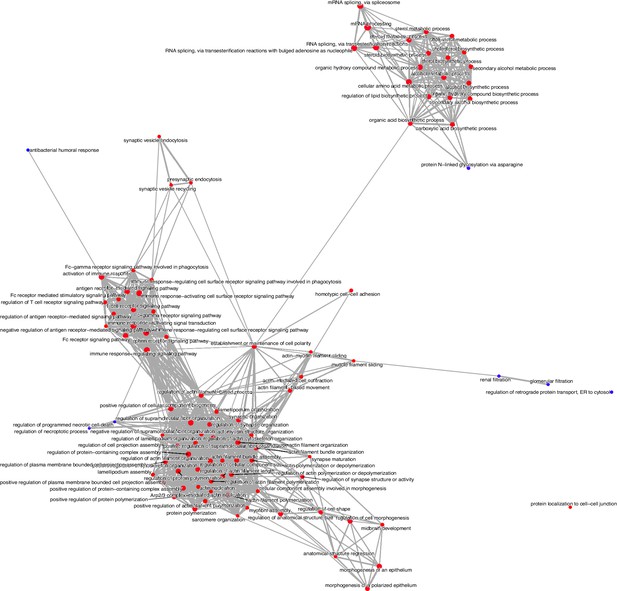
Enrichment map of Gene Set Enrichment Analysis comparing clusters 4 and 5.
The dot size and the dot color represent the number of genes in the pathway and the normalized enrichment score (NES), respectively.
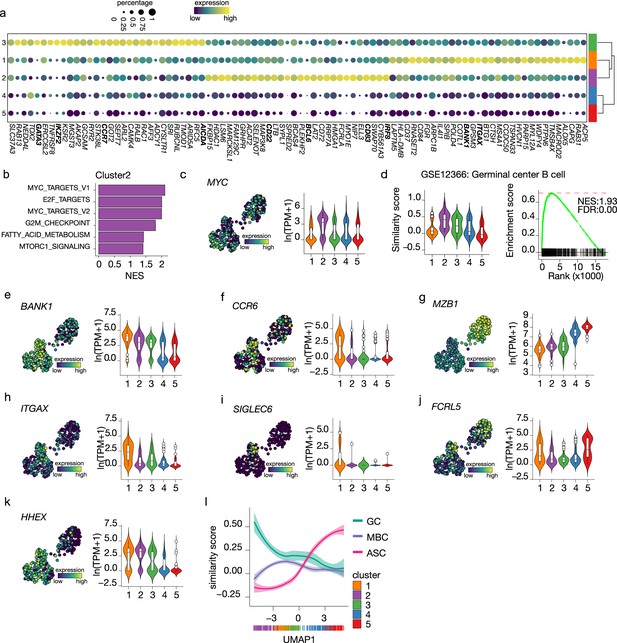
Terminal differentiation into antibody-secreting cells (ASCs) starts from germinal center (GC)-like cells.
(a) Top 30 differentially expressed genes identified in clusters 1–3. The size of the dot encodes the percentage of cells within a cluster, while the color encodes the average gene expression across all cells within a cluster. (b) Significantly enriched hallmark gene sets overlapping with the differentially expressed genes identified in cluster 2. (c) Uniform Manifold Approximation and Projection (UMAP) projection colored by expression of the transcription factors MYC (left), along with corresponding distribution of expression levels (ln(TPM+1)) within each cluster (right). (d) Corresponding distribution of the GC similarly score within each cluster (left) and Gene Set Enrichment Analysis (GSEA) enrichment plot (right) for GC B cell gene signature in cells from cluster 2 (NES, normalized enrichment score; FDR, false discovery rate). (e–k) UMAP projection colored by expression of the BANK1 (e), CCR6 (f), MZB1 (g), ITGAX (h), SIGLEC6 (i), FCRL5 (j), HHEX (k; left), along with corresponding distribution of expression levels (ln(TPM+1)) within each cluster (right). (l) The similarity score of ex vivo germinal center B cell (GC), memory B cell (MBC), and ASC gene sets ordered by UMAP1.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83578/elife-83578-mdarchecklist1-v2.pdf
-
Supplementary file 1
Primer sequences.
- https://cdn.elifesciences.org/articles/83578/elife-83578-supp1-v2.docx





