A comprehensive survey of C. elegans argonaute proteins reveals organism-wide gene regulatory networks and functions
Figures
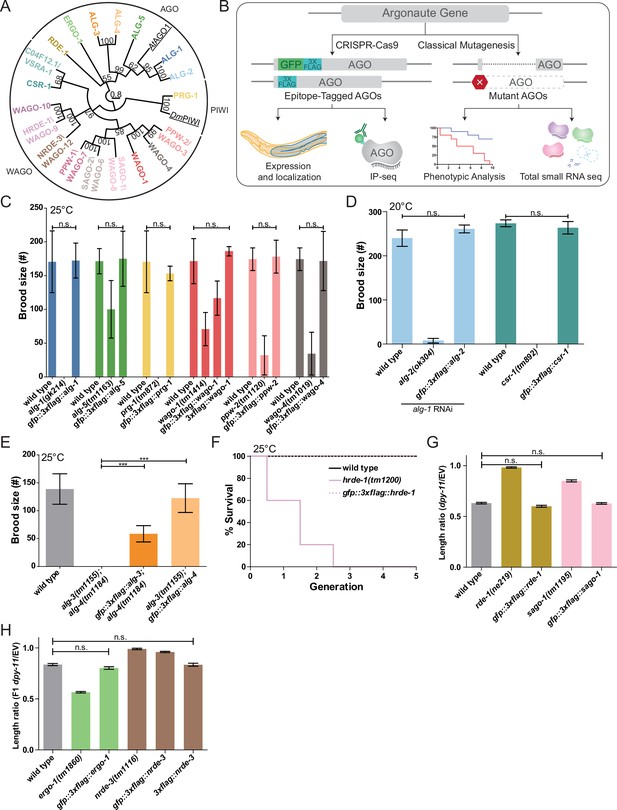
Functional validations of GFP::3xFLAG and 3xFLAG tagged Argonautes.
(A) Maximum likelihood evolutionary tree of A. thaliana AGO1 (AtAGO1), D. melanogaster PIWI (DmPIWI), and C. elegans Argonautes. (B) Workflow for characterizing C. elegans Argonautes. (C) Functional validations of tagged ALG-1, ALG-5, PRG-1, WAGO-1, PPW-2, and WAGO-4 strains. Brood size was determined at 25°C for each indicated genotype. N ≧ 5 worms per condition. (D) Functional validation of tagged ALG-2 and CSR-1 strains. Brood size was determined at 20°C for each indicated genotype. For the ALG-2 tag validations, the brood size was determined when worms were fed dsRNA of alg-1. N = 5 worms per condition. (E) Functional validations of tagged ALG-3 and ALG-4 strains. Brood size was determined at 25°C for each indicated genotype. N = 10 worms per condition. (F) Functional validation of the tagged HRDE-1 strain via a Mortal germline assay at 25°C. N = 5 worms per condition. (G) Functional validations of tagged RDE-1 and SAGO-1 strains. Worms were fed bacteria expressing dsRNA of dpy-11 or an empty vector (EV) RNAi control. The length ratio of dpy-11 dsRNA fed P0 worms compared to the average length on EV was determined. N = 30 worms per condition. (H) Functional validations of tagged ERGO-1 and NRDE-3 strains. Worms were fed bacteria expressing dsRNA of dpy-11 or an EV control. The length ratio of the F1s of the dpy-11 dsRNA fed worms compared to the average length on EV was determined. N = 30 worms per condition. (C–H) *p-value<0.05, **p-value<0.01, ***p-value, n.s. = not significant. One-way ANOVA with Tukey’s post hoc multiple comparison test. All error bars represent standard deviation.
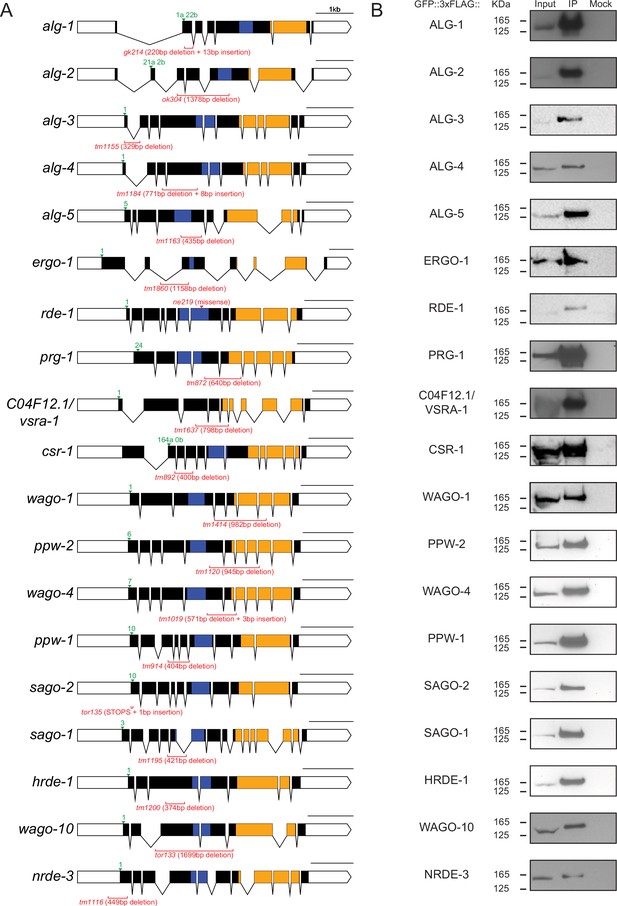
Epitope tag locations and Western blot validations.
(A) Gene diagrams of each Argonaute. The green arrow points to the GFP::3xFLAG tag insertion site. The number corresponds to the distance in amino acids from the first methionine. The letter corresponds to the isoform if there is more than one. Genotypes of mutants that were used in this study are indicated in red. (B) Western blots of GFP::3xFLAG::AGO IPs. Inputs are 100 µg of total protein lysate. Mock conditions are IPs that were conducted using RFP antibodies.
-
Figure 1—figure supplement 1—source data 1
This file contains original western blots of AGO IPs used in creating Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/83853/elife-83853-fig1-figsupp1-data1-v2.zip
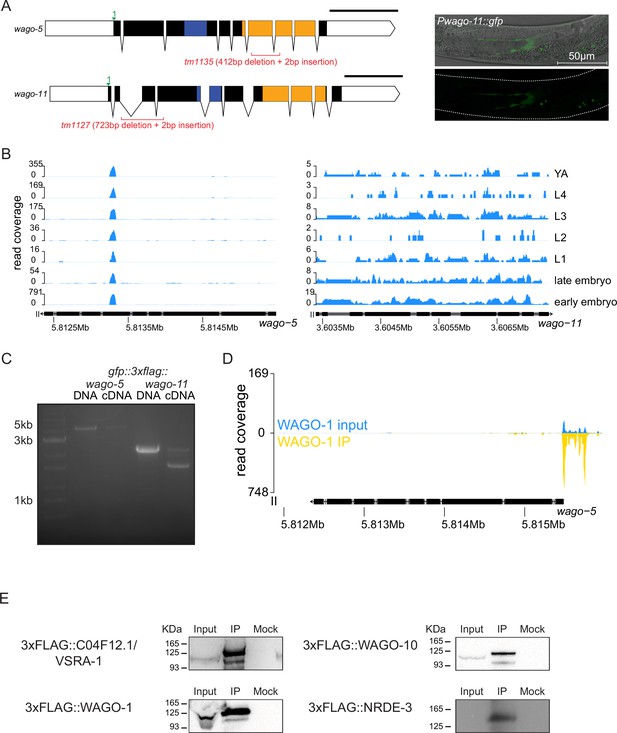
Additional analyses of tagged Argonautes.
(A) Gene diagrams of WAGO-5 and WAGO-11 as in Figure 1—figure supplement 1 (left). GFP driven under the promoter of WAGO-11 (right). GFP is observed in the distal gonadal sheath of L4 hermaphrodites. (B) Genome browser tracks of mRNA tiling array sequencing at the wago-5 (left) and wago-11 (right) loci from modENCODE. (C) RT-PCR and gel electrophoresis analysis of gfp::3xflag::wago-5 and gfp::3xflag::wago-11. Only the gfp::3xflag::wago-11 cDNA is detected by PCR. (D) Genome browser tracks of small RNAs targeting the wago-5 locus in WAGO-1 IP. (E) Western blots of 3xFLAG tagged AGO IPs. Inputs are 100 ug of total protein lysate. Mock conditions are IPs conducted using non-antibody conjugated beads. 3xFLAG tag insertion sites are at the same position as GFP::3xFLAG tags shown in Figure 1—figure supplement 1.
-
Figure 1—figure supplement 2—source data 1
This file contains original western blots of AGO IPs used in creating Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/83853/elife-83853-fig1-figsupp2-data1-v2.zip
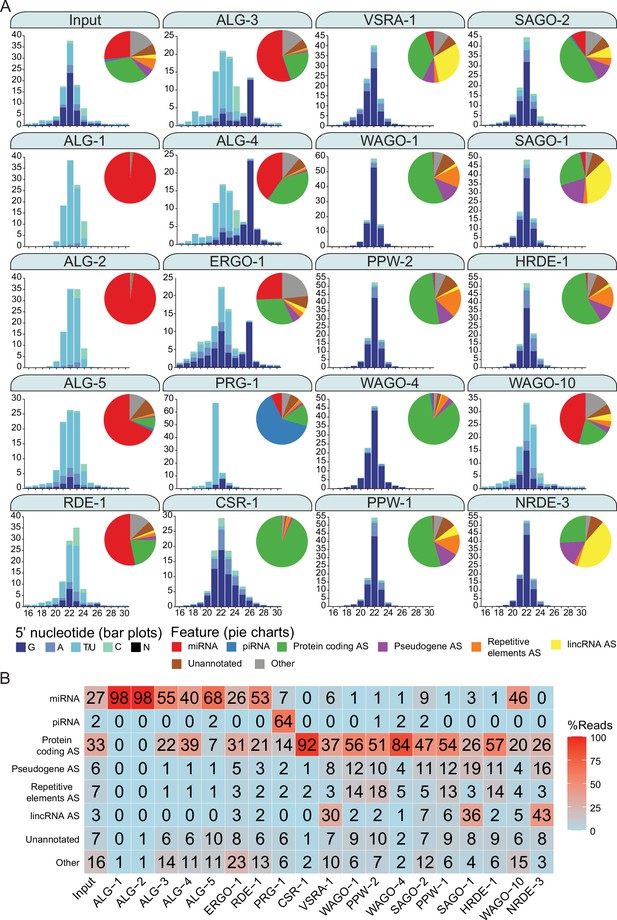
Argonautes associate with different types of sRNAs and target different categories of genetic features.
(A) 5′ nucleotide and length of sRNAs present in each Argonaute IP shown in bar graph form. The pie charts depict which type of genetic element (biotype) the sRNAs correspond to, as listed. AS = antisense, S = sense. The ‘Other’ category encompasses: miRNA AS, piRNA AS, protein-coding S, pseudogene S, repetitive elements S, lincRNA S, rRNA S/AS, snoRNA S/AS, snRNA S/AS, tRNA S/AS, ncRNA S/AS, and antisense lncRNAs (ancRNAs/anr loci) (Nam and Bartel, 2012). The average of two biological replicates is shown. The GFP::3xFLAG tagged Argonautes were used for IPs except for C04F12.1/VSRA-1, WAGO-10, and NRDE-3, where a 3xFLAG tag was used. All IPs were performed on Young Adult samples except for ALG-3, ALG-4, and WAGO-10, which were conducted on L4 staged animals. The CSR-1 strain tags both isoforms. (B) A table summarizing the percentage of reads in each set of AGO IPs corresponding to genetic element types in (A).
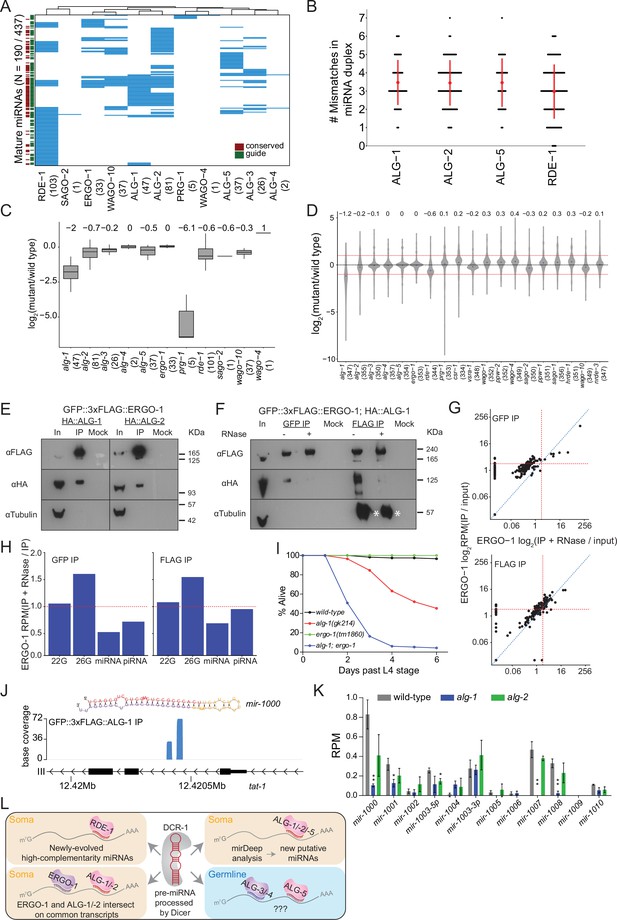
Analysis of miRNAs in AGO IPs and ago mutants reveals novel miRNAs.
(A) Clustering diagram of miRNAs enriched in AGOs. Each blue line represents an individual mature miRNA sequence. miRNAs are categorized by conservation, with those conserved to C. briggsae designated in burgundy on the left, and by whether they are the canonical guide strand, as designated in green on the left. (B) The number of mismatches in the precursor miRNA sequences for which a mature miRNA was enriched in the indicated AGOs. Each black dot represents a single precursor miRNA duplex. The red dot and lines indicate the average and standard deviation. (C) Fold change of miRNAs enriched in AGO IPs in the corresponding ago mutant. (D) Fold change of all detected miRNAs in ago mutants compared to wild-type. (E) Western blots of co-IP experiments of ERGO-1 and ALG-1 and ALG-2. GFP::3xFLAG::ERGO-1 was crossed to HA::ALG-1 or HA::ALG-2 strains and IPed using anti-GFP antibodies. (F) As in (E) but GFP::3xFLAG::ERGO-1 IPs were conducted either with anti-GFP or anti-FLAG antibodies with or without RNase treatment. Asterisks indicate IgG. (G) Scatter plots showing enrichment of miRNAs in IP and IP +RNase-treated samples of GFP::3xFLAG::ERGO-1. The top graph shows the results of an anti-GFP IP and the bottom graph shows the results of an anti-FLAG IP (one replicate per condition). (H) Bar plots showing quantification of sRNA types in IP and IP +RNase-treated samples of GFP::3xFLAG::ERGO-1 (anti-GFP IP on the left; anti-FLAP IP on the right). (I) Survival of worms of the indicated genotype beyond the L4 stage (bottom). N ≧ 100. (J) An example of a novel miRNA within an intron of the gene tat-1 as determined by mirDeep2 analysis of ALG-1 IPs. (K) Analysis of the levels of predicted novel miRNAs in wild-type, alg-1 and alg-2 mutants. Predicted novel miRNAs are provisionally named. Error bars represent standard deviation. (L) A summary of miRNA pathway observations.
-
Figure 3—source data 1
This file contains original western blots of AGO IPs used in creating Figure 3.
- https://cdn.elifesciences.org/articles/83853/elife-83853-fig3-data1-v2.zip
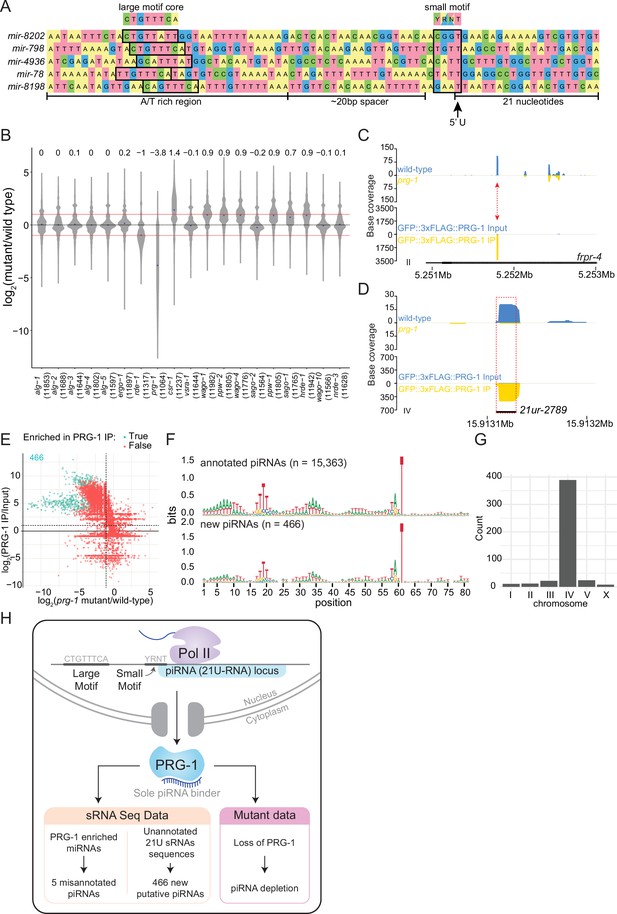
Analysis of piRNAs in AGO IPs and ago mutants reveals new piRNAs.
(A) Genomic loci of annotated miRNAs that are enriched in PRG-1 IPs and suspected to be piRNAs. Ruby motifs highlighted above. (B) Violin plots depicting the fold change of all detected piRNAs in ago mutants compared to wild-type. (C) An example of a novel piRNA sequence, where a 21U sRNA sequence that was enriched in PRG-1 IPs and depleted in prg-1 mutants originated from the intron of the gene frpr-4 in the antisense orientation (red arrow). Note the difference in scales. (D) An example of a novel piRNA sequence originating from a shift of 3 nt from the annotated 21ur-2789 piRNA (dotted red box, black line). Note the difference in scales. (E) Scatter plot showing individual expression levels of 21U sRNAs that are not annotated as piRNAs. The y-axis shows enrichment in PRG-1 IPs and the x-axis shows depletion in prg-1 mutants. The cyan dots represent individual 21U sRNAs which are twofold enriched in PRG-1 IPs. (F) Sequence logo analysis of annotated piRNA loci (top) and the new 466 piRNA loci (bottom). (G) Chromosome distribution of the 466 putative piRNA sequences. (H) A summary of piRNA pathway observations.
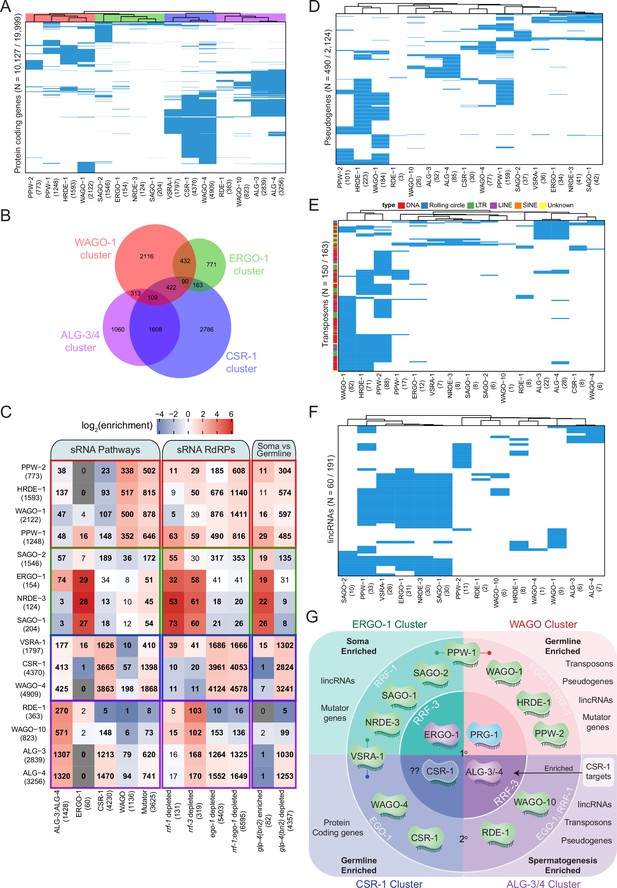
Analysis of endo-siRNA binding AGOs reveals functional categorization of AGOs to regulate distinct genetic elements.
(A) Clustering diagram of AGO protein-coding gene targets. Each blue line represents a transcript for which endo-sRNAs were enriched twofold or more relative to input in both IPs. 22G-RNAs (defined as 20–24 nt with no 5′ nucleotide bias) were considered for all AGOs except ALG-3, ALG-4, and ERGO-1, for which only 26G-RNAs (defined as 25–27 nt with no 5′ nucleotide bias) were considered. (B) A Venn diagram showing the overlaps of protein–coding gene targets of the AGO clusters as highlighted by the color scheme in (A). (C) Enrichment analysis of the AGO protein–coding gene targets in previously described datasets. (D) Clustering diagram of AGO pseudogene targets as in (A). (E) Clustering diagram of AGO transposon targets as in (A). (F) Clustering diagram of AGO lincRNA targets as in (A). (G) A schematic summary highlighting the major AGO/sRNAs networks uncovered by endo-siRNA analysis.
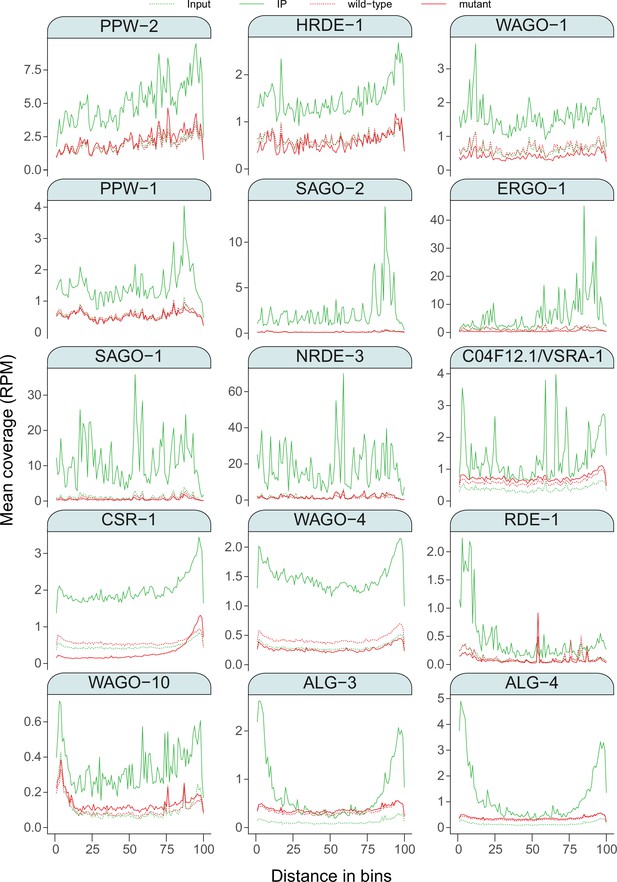
Metagene analysis of sRNAs.
Metagene profiles for the protein-coding gene targeting sRNAs enriched in association with each AGO. The number of targets is indicated in parenthesis. To account for differences in the size of targets, each gene was partitioned into 100 bins and the mean coverage in RPM was determined for each bin. Green line = AGO IP, green dashed line = input, red line = ago mutant, red dashed line = wild type.
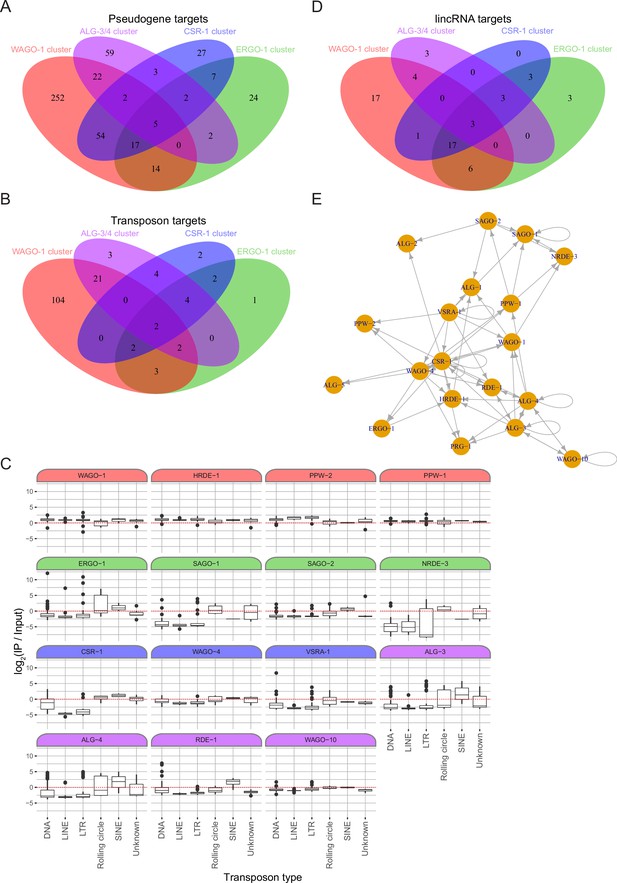
Additional analysis of the endo-siRNA binding AGO IPs.
(A) A Venn diagram showing the overlaps of pseudogene targets of the AGO clusters as highlighted by the color scheme in Figure 5A. (B) A Venn diagram showing the overlaps of transposon targets of the AGO clusters as highlighted by the color scheme in Figure 5A. (C) Box plots showing the enrichment levels (IP/input) of sRNAs targeting each transposon broken down by transposon family in each AGO IP. (D) A Venn diagram showing the overlaps of lincRNA targets of the AGO clusters as highlighted by the color scheme in Figure 5A. (E) Network analysis showing which AGO is enriched for sRNAs targeting another AGO. Arrow direction indicates the direction of regulation; that is the AGO at the blunt end regulates the AGO at the arrow tip.
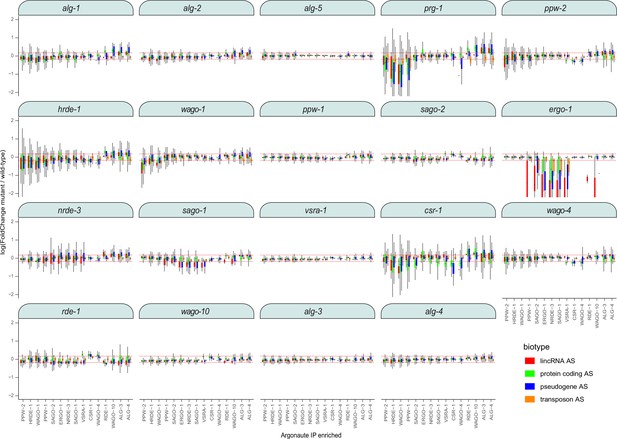
Loss of ago genes results in differential effects of sRNA levels associated with each AGO.
Box plots showing the expression levels (y-axis) of the sRNAs that target the protein-coding targets for which sRNAs were enriched in each AGO IP (x-axis) in the indicated argonaute mutant (panels). Red dashed lines indicate twofold change cutoff.
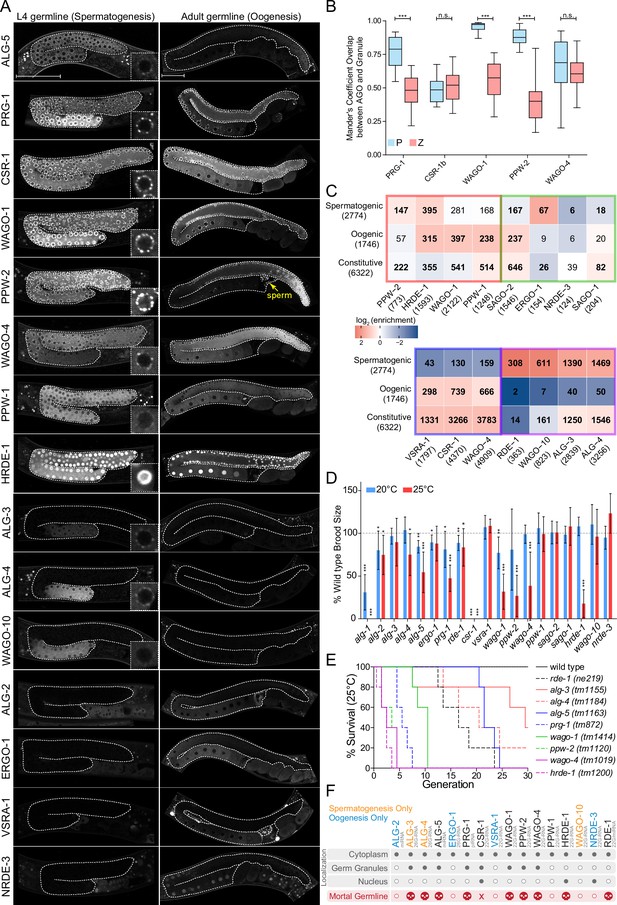
AGOs are differentially expressed in the germline and differentially regulate germline gene expression to promote fertility.
(A) Expression patterns of germline AGOs in L4 (left) and adult (right) germlines. Inset image zoomed in at an individual germ cell nucleus. Yellow arrow points to sperm within the spermatheca. Scale bar represents 50μm. Due to low levels of expression, RDE-1 is not shown. See Figure 7—figure supplement 8 for expression of the rde-1p::gfp transcriptional reporter. (B) Quantification of the number of GFP::AGO pixels that overlap with PGL-1::mRFP (blue) or HA::TagRFP::ZNFX-1 (coral) pixels using Mander’s correlation. For each data set, five Z stacks of proximal germline regions from six different animals per strain were counted (N = 30 slices, approximately 80–100 nuclei per worm). ***p-value<0.001, n.s. = not significant. One-way ANOVA with Bonferroni’s post hoc multiple comparison test. (C) Analysis of the enriched sRNA targets in each of the AGOs in comparison to germline constitutive, oogenic, and spermatogenic expressed genes (Ortiz et al., 2014). Bold numbers indicate significant enrichment or depletion (p<0.05), Fisher’s exact test. The colored borders represent the AGO clusters as defined in Figure 5A. (D) Brood size analysis of all ago mutants at 20°C and 25°C. Data was aggregated from different experiments and normalized to the mean of wild-type control samples. *p<0.05, **p<0.01, ***p<0.001, two-sided t-test. N ≧ 10 P0 worms. Error bars represent standard deviation. (E) Mortal germline assay of ago mutants showing a Mrt phenotype in (D). N = 5 P0 worms. (F) A summary of the spatial and temporal localization of AGOs in the germline and Mrt phenotypes. CSR-1 has an 'X' to indicate it is essential. AGOs in black are expressed in both spermatogenesis and oogenesis.
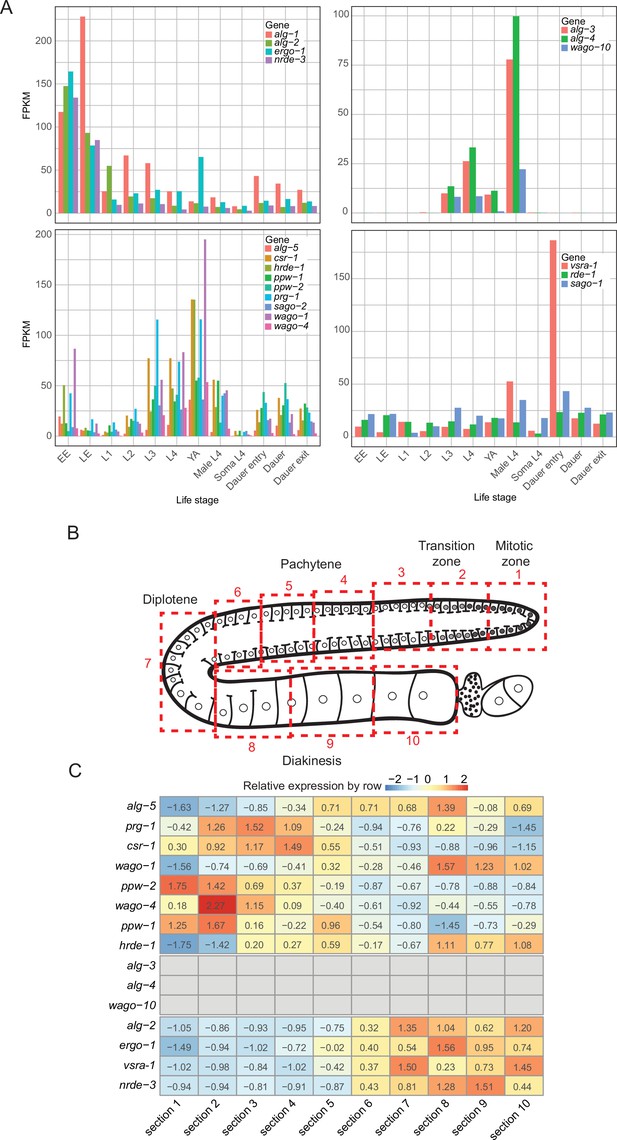
Additional analysis of AGO mRNA expression.
(A) mRNA levels of AGOs in different developmental stages. Data is an aggregate of public datasets available on WormBase version WS273. (B) Illustration of hermaphrodite germline with sections that were sequenced in Tzur et al., 2018. (C) Table showing in heatmap form the relative expression of each argonaute mRNA in each germline section. Note that alg-3, alg-4, and wago-10 were not detected, and alg-2, ergo-1, C04F12.1/vsra-1, and nrde-3 show increasing expression in oocyte sections, consistent with expression patterns.
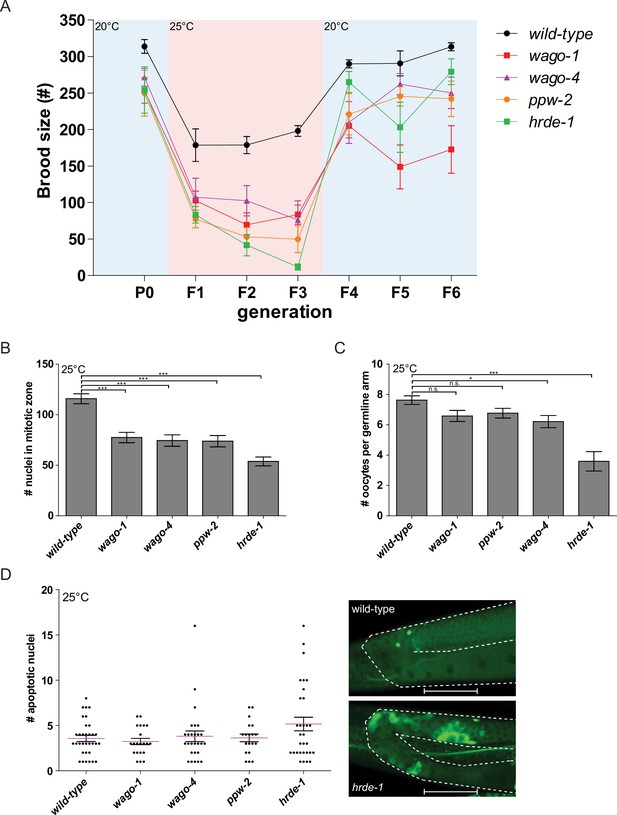
The mortal germline of wago mutants is reversible and associated with reduced germline proliferation.
(A) A temperature shift experiment where wago mutants were placed at 20°C for one generation, then transferred to 25°C for three generations and then back to 20°C. Brood size was measured every generation (N = 10 per genotype). (B) Count of nuclei at the mitotic zone (N = 5). ***p-value<0.001, Two-way ANOVA with Dunnet’s post hoc multiple comparison test. Error bars represent standard deviation. (C) Count of oocytes in germline arms (N ≥ 22). *p-value<0.05, ***p-value<0.001, Two-way ANOVA with Dunnet’s post hoc multiple comparison test. Error bars represent standard deviation. (D) Count of apoptotic nuclei in germlines of acridine orange stained wago mutants (N ≥ 19; left), and an example of acridine orange staining in wild-type vs. hrde-1 mutant worms (right). Scale bars represent 50 μm.
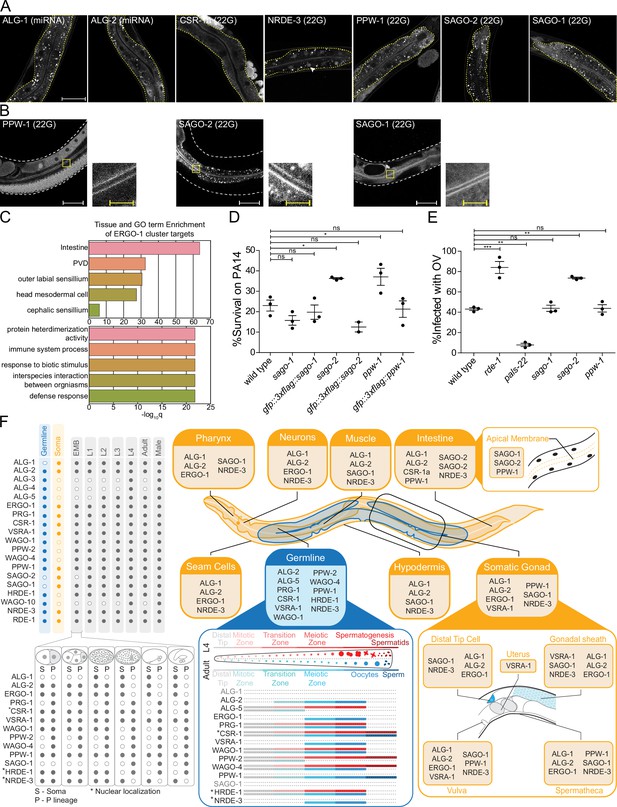
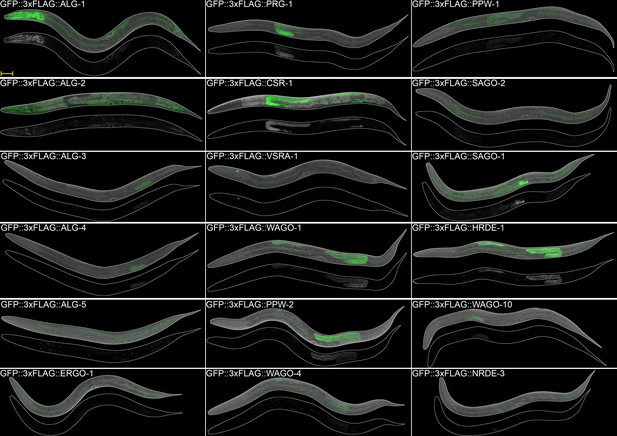
AGOs are expressed in multiple somatic tissues and several are required for normal immunity towards pathogens.
(A) AGOs expressed in the intestine. Adult worms shown. Brackets indicate the type of sRNA AGOs associate with. Intestines are outlined in yellow. Arrowhead indicates intestine cell nuclei. Scale bar represents 50 μm for all images. Bright foci throughout intestinal tissue are autofluorescent gut granules. (B) Apical intestinal membrane localization of PPW-1, SAGO-2, and SAGO-1. Worm body is outlined in white. Zoomed-in panels are indicated with a yellow box. White scale bar represents 50 μm. Yellow scale bar represents 10 μm. Note that PPW-1 is also expressed in the germline. (C) Tissue enrichment analysis (top) and Gene Ontology analysis (bottom) of the ERGO-1 cluster sRNA targets. (D) Percent of worms alive after 72 hr of exposure to a P. aeruginosa PA14 lawn is shown for each strain. This is a representative experimental run out of three conducted. Each dot represents 50 worms. *p-value<0.05, n.s. = not significant. One-way ANOVA with Dunnett’s post hoc multiple comparison test. (E) Percent of worms infected with Orsay virus (OV) 16 hr post infection is shown for each strain. Each dot represents ≥ 100 worms. N = 3. **p-value<0.01, ***p-value<0.001, n.s. = not significant. One-way ANOVA with Tukey post hoc multiple comparison test. (F) A summary of the expression patterns of all C. elegans AGOs throughout development.

AGO expression in embryos.
Images of embryos from left to right: 2 cell stage, 4 cell stage, ~40 cell stage, ~200 cell stage (end of gastrulation), comma/bean elongation stage, threefold stage/pretzel stage. Scale bar = 50 µm.
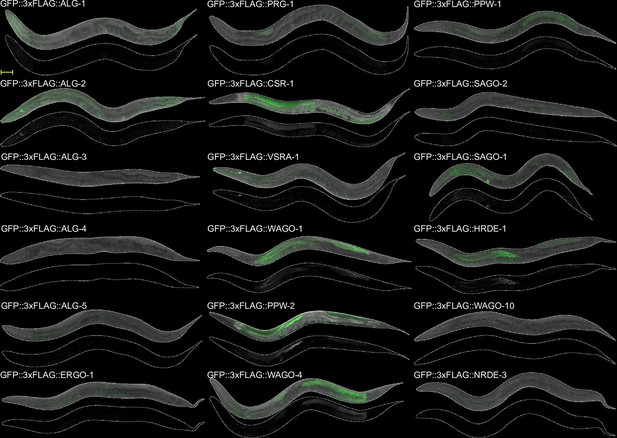
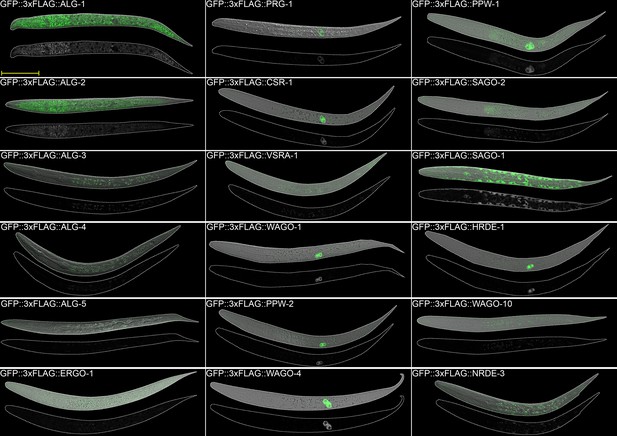
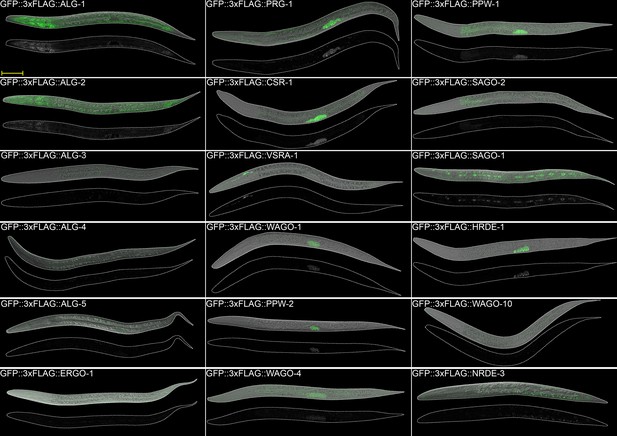
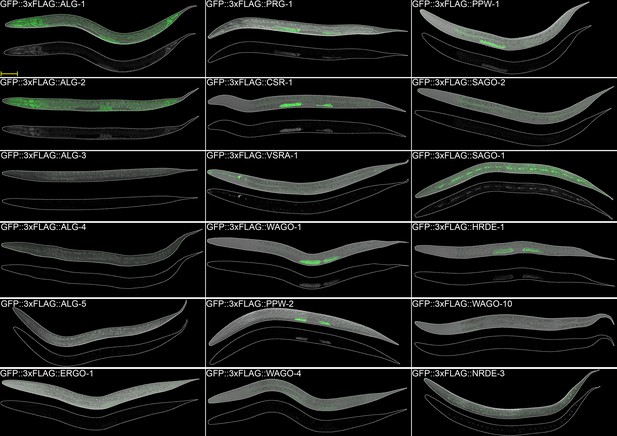
AGO expression in adult worms.
Images of adult worms. Scale bar = 50 µm.
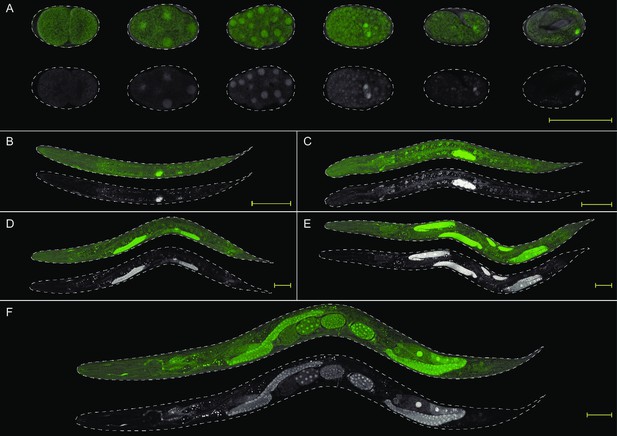
Representative images of developmental stages of the RDE-1 transcriptional reporter.
Top row of each panel is DIC + GFP images; bottom row is GFP images shown in grayscale. (A) embryos, (B) L1, (C) L2, (D) L3, (E) L4, and (F) adult. Scale bar = 50 µm. Note that nuclear localization was observed only for this transcriptional fusion and not for the GFP::3xFLAG::RDE-1 translational fusion.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| gene (C. elegans) | rde-1 | WormBase | WBGene00004323 | |
| gene (C. elegans) | alg-1 | WormBase | WBGene00000105 | |
| gene (C. elegans) | alg-2 | WormBase | WBGene00000106 | |
| gene (C. elegans) | alg-3 | WormBase | WBGene00011910 | |
| gene (C. elegans) | alg-4 | WormBase | WBGene00006449 | |
| gene (C. elegans) | alg-5 | WormBase | WBGene00011945 | |
| gene (C. elegans) | wago-1 | WormBase | WBGene00011061 | |
| gene (C. elegans) | wago-3/ppw-1 | WormBase | WBGene00004094 | |
| gene (C. elegans) | wago-4 | WormBase | WBGene00010263 | |
| gene (C. elegans) | wago-5 | WormBase | WBGene00022877 | |
| gene (C. elegans) | wago-6/sago-2 | WormBase | WBGene00018921 | |
| gene (C. elegans) | wago-7/ppw-1 | WormBase | WBGene00004093 | |
| gene (C. elegans) | wago-8/sago-1 | WormBase | WBGene00019666 | |
| gene (C. elegans) | wago-9/hrde-1 | WormBase | WBGene00007624 | |
| gene (C. elegans) | wago-10 | WormBase | WBGene00020707 | |
| gene (C. elegans) | wago-11 | WormBase | WBGene00021711 | |
| gene (C. elegans) | wago-12/nrde-3 | WormBase | WBGene00019862 | |
| gene (C. elegans) | csr-1 | WormBase | WBGene00017641 | |
| gene (C. elegans) | C04F12.1/vsra-1 | WormBase | WBGene00007297 | |
| strain, strain background (C. elegans; hermaprhodites and males) | Bristol N2 | WormBase | ||
| strain, strain background (E. coli) | OP50 | Caenorhabditis Genetics Center | ||
| strain, strain background (E. coli) | HT115 | Caenorhabditis Genetics Centers | ||
| genetic reagent (C. elegans) | List of strains | This study, Supplementary file 8; Caenorhabditis Genetics Center | ||
| antibody | Anti-mouse IgG, HRP-linked Antibody (horse polyclonal) | Cell Signaling Technology | 7076 S | 1:1000 for western blots |
| antibody | Monoclonal ANTI-FLAG M2 antibody (mouse monoclonal) | Sigma | F1804 | 1:2000 for western blots 5 μg per 50 μl of Dynabeads in 200 μl for IPs |
| antibody | GFP-Trap_MA (alpaca recombinant nanobody) | ChromoTek | gtma | 20 μl of beads per 5 mg total protein in 500 μl |
| antibody | RFP-Trap_MA (alpaca recombinant nanobody) | ChromoTek | rtma | 20 μl of beads per 5 mg total protein in 500 μl |
| sequence-based reagent | List of oligonucleotides | This study, Supplementary file 8 | ||
| sequence-based reagent | High Throughput Sequencing Data (Conine et al., 2010) | https://www.ncbi.nlm.nih.gov/geo/ | GSE18731 | |
| sequence-based reagent | High Throughput Sequencing Data (Vasale et al., 2010) | https://www.ncbi.nlm.nih.gov/geo/ | GSE18714 | |
| sequence-based reagent | High Throughput Sequencing Data (Claycomb et al., 2009) | https://www.ncbi.nlm.nih.gov/geo/ | GSE18165 | |
| sequence-based reagent | High Throughput Sequencing Data (Gu et al., 2009) | https://www.ncbi.nlm.nih.gov/geo/ | GSE18215 | |
| sequence-based reagent | High Throughput Sequencing Data (Sapetschnig et al., 2015) | https://www.ncbi.nlm.nih.gov/geo/ | GSE66344 | |
| sequence-based reagent | High Throughput Sequencing Data | Phillips et al., 2012 | Table S3 | |
| sequence-based reagent | High Throughput Sequencing Data | Ortiz et al., 2014 | Table S1 | |
| sequence-based reagent | High Throughput Sequencing Data | Tzur et al., 2018 | Table S3 | |
| sequence-based reagent | High Throughput Sequencing Data | This study; https://www.ncbi.nlm.nih.gov/geo/ | GSE208702 | |
| commercial assay or kit | NEBuilder HiFi DNA Assembly Cloning Kit | New England Biolabs | E5520 | |
| commercial assay or kit | NEBNext Multiplex Small RNA Library Prep Kit for Illumina | New England Biolabs | E7560 | |
| chemical compound, drug | Q5 High-Fidelity DNA Polymerase | New England Biolabs | M0491 | |
| chemical compound, drug | T4 DNA Ligase | New England Biolabs | M0202 | |
| chemical compound, drug | Tri Reagent | Molecular Research Centre | TR118 | |
| chemical compound, drug | Phenol Chloroform Isoamyl Alcohol | Sigma-Aldrich | P2069 | |
| chemical compound, drug | cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail | Roche | 11836170001 | |
| chemical compound, drug | RNA 5' Polyphosphatase | Epicentre | RP8092H | |
| chemical compound, drug | Phosphatase Inhibitor Cocktail 2 | Sigma-Aldrich | P5726 | |
| chemical compound, drug | Phosphatase Inhibitor Cocktail 3 | Sigma-Aldrich | P0044 | |
| chemical compound, drug | DTT | BioShop Canada | DTT001 | |
| chemical compound, drug | NP-40 | BioBasic | NDB0385-500ML | |
| chemical compound, drug | Levamisole hydrochloride | Fisher Scientific | AC187870100 | |
| chemical compound, drug | Bovine Serum Albumin | BioBasic | 9048-46-8 | |
| chemical compound, drug | SUPERase• In RNase Inhibitor | ThermoFisher Scientific/Invitrogen | AM2696 | |
| chemical compound, drug | T4 Polynucleotide Kinase | New England Biolabs | M0201 | |
| chemical compound, drug | Glycogen | Ambion | AM9510 | |
| chemical compound, drug | RNAse and DNAse Away | BioBasic | DB0339 | |
| chemical compound, drug | Dynabeads Protein G | ThermoFisher Scientific/Invitrogen | 10003D | |
| chemical compound, drug | GB-Magic Protein A/G Immunprecipitation Magnetic Beads | GeneBiosystems | 22202B | |
| chemical compound, drug | Protein Assay Reagent A | Bio-Rad | 5000113 | |
| chemical compound, drug | Protein Assay Reagent B | Bio-Rad | 5000114 | |
| chemical compound, drug | Protein Assay Reagent S | Bio-Rad | 5000115 | |
| chemical compound, drug | Nitrocellulose blotting membrane | GE Healthcare | 10600016 | |
| chemical compound, drug | Luminata Classico Western HRP substrate | Millipore | WBLUC0500 | |
| chemical compound, drug | RNase I | ThermoFisher Scientific/Ambion | AM2295 | |
| software, algorithm | MEGA X | https://www.megasoftware.net/ | ||
| software, algorithm | CRISPOR | http://crispor.tefor.net/ | ||
| software, algorithm | SnapGene | https://www.snapgene.com/ | ||
| software, algorithm | GraphPad Prism | https://www.graphpad.com/scientific-software/prism/ | ||
| software, algorithm | STAR | Dobin et al., 2013 | ||
| software, algorithm | Rstudio | https://www.rstudio.com/ | ||
| software, algorithm | Custom Computational Pipeline | https://github.com/ | https://github.com/ClaycombLab/Seroussi_2022 | |
| other | Bolt Precast Bis-Tris Plus Gradient Gels (4–12%) | ThermoFisher Scientific | NW04120BOX | This study: Materials and Methods |
| other | Hybond C Membrane | GE/Amersham Biosciences | CA95038-380L | This study: Materials and Methods |
| other | Wheaton Steel Dounce Homogenizer | VWR | 62400–675 | This study: Materials and Methods |
Additional files
-
Supplementary file 1
A table summarizing previously published data on C. elegans AGOs.
- https://cdn.elifesciences.org/articles/83853/elife-83853-supp1-v2.xlsx
-
Supplementary file 2
A table summarizing small RNA library information.
- https://cdn.elifesciences.org/articles/83853/elife-83853-supp2-v2.xlsx
-
Supplementary file 3
Tables summarizing the enrichment of reads, biotypes, and AGO targets in all libraries.
- https://cdn.elifesciences.org/articles/83853/elife-83853-supp3-v2.xlsb
-
Supplementary file 4
A table summarizing sRNAs depleted in ago mutants.
- https://cdn.elifesciences.org/articles/83853/elife-83853-supp4-v2.xlsx
-
Supplementary file 5
A table summarizing the mirDeep2 analysis results in predicting novel high confidence miRNAs.
- https://cdn.elifesciences.org/articles/83853/elife-83853-supp5-v2.xlsx
-
Supplementary file 6
A table summarizing the 466 unannotated 21U sRNA sequences enriched in PRG-1 IPs and depleted in prg-1 mutants.
- https://cdn.elifesciences.org/articles/83853/elife-83853-supp6-v2.xlsx
-
Supplementary file 7
Gene Ontology analysis results of AGO targets.
- https://cdn.elifesciences.org/articles/83853/elife-83853-supp7-v2.xlsx
-
Supplementary file 8
Strains and primers used in this study.
- https://cdn.elifesciences.org/articles/83853/elife-83853-supp8-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83853/elife-83853-mdarchecklist1-v2.pdf