Evidence linking APOBEC3B genesis and evolution of innate immune antagonism by gamma-herpesvirus ribonucleotide reductases
Figures
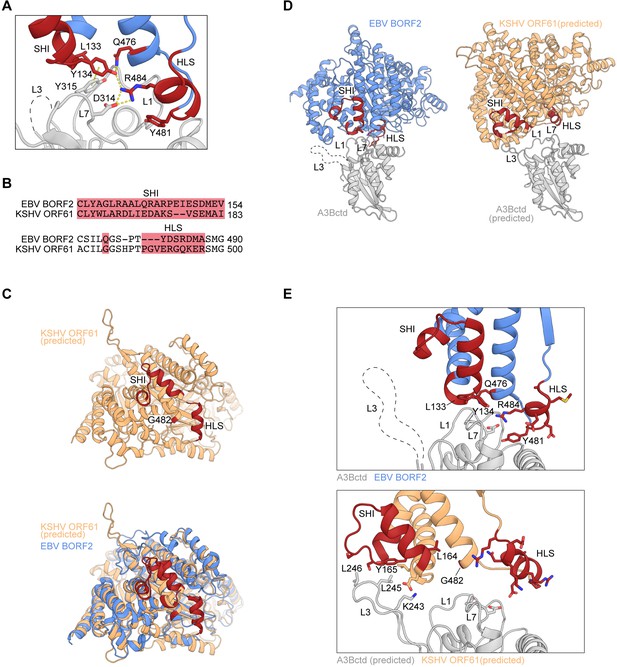
Structural differences and similarities between Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV) ribonucleotide reductase (RNR) large subunits.
(A) Zoom-in of the EBV BORF2-A3Bctd interface (pdb 7rw6, chains A and B) showing a network of interactions between BORF2 (blue) and A3Bctd L7 and L1 regions (gray). BORF2 short helix insertion (SHI), helical loop structure (HLS), and interacting residue Q476 are shown in red. Gray dashed line represents the L3 region not visible by cryo-EM. (B) Amino acid sequence alignment of the SHI and HLS regions of EBV BORF2 and KSHV ORF61 (complete alignment in Figure 1—figure supplement 1A). (C) RoseTTAFold-predicted structure of KSHV ORF61 (orange, above) overlayed with EBV BORF2 (pdb 7rw6, chain A; blue, below). HLS and SHI regions in both proteins and residue G482 in ORF61 are shown in red. (D–E) Cryo-EM structure of EBV BORF2-A3Bctd (pdb 7rw6, chains A and B) and highest-ranked protein-protein docking model of KSHV ORF61-A3Bctd generated using RosettaDock (Figure 1—figure supplement 1B). Panel D shows the full complexes with key regions labeled. Panel E shows a zoom-in of the RNR-A3Bctd interface (BORF2, blue; ORF61, orange; A3Bctd gray; SHI and HLS regions, red). A3Bctd loops are labeled L1, L3, and L7 (gray dashed line represents the L3 region not visible by cryo-EM).
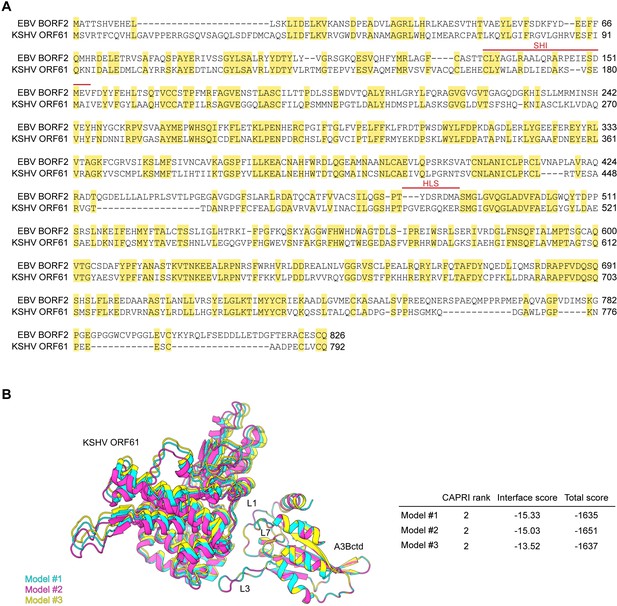
EBV BORF2 and KSHV ORF61 amino acid sequence alignment and summary of protein-protein docking models generated.
(A) Amino acid sequence alignment of Epstein-Barr virus (EBV) BORF2 and Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF61 with identical residues shaded yellow and the short helix insertion (SHI) and helical loop structure (HLS) regions designated by red lines. (B) A superposition of the three highest-ranked protein-protein docking models of the KSHV ORF61-A3Bctd complex generated using RosettaDock (left). The Critical Assessment of Predicted Interactions (CAPRI) model rankings computed by RosettaDock range from 0 (incorrect model) to 3 (highest quality model; detailed in Lensink et al., 2020, with additional metrics shown to right).
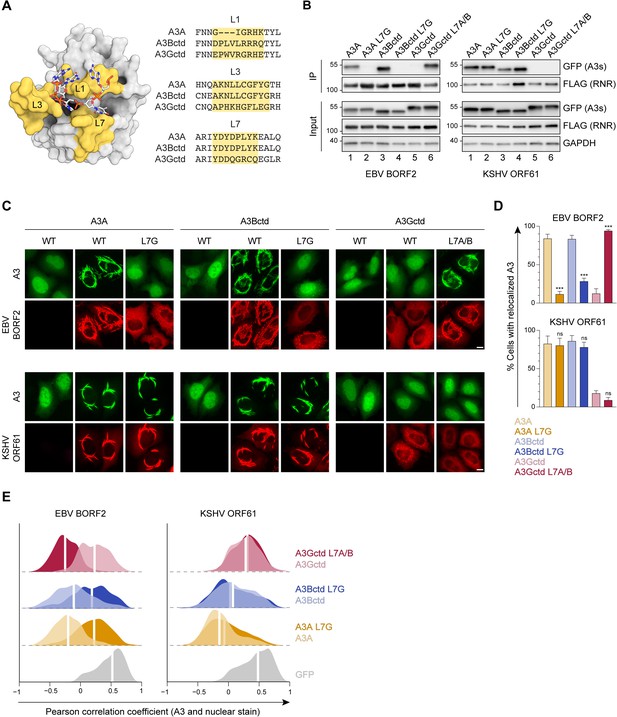
Involvement of the A3B and A3A loop 7 regions in binding to gamma-herpesvirus ribonucleotide reductases (RNRs).
(A) Surface-filled representation of single-stranded DNA (ssDNA) bound by an A3 (pdb 5sww, chains A and E). L1, L3, and L7 regions are colored in yellow with amino acid alignments to the right. (B) Co-immunoprecipitation (Co-IP) of A3 L7 chimeras with Epstein-Barr virus (EBV) BORF2 and Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF61. FLAG-tagged RNR subunits were co-expressed with the indicated A3-EGFP constructs in 293T cells, affinity purified, and analyzed by immunoblotting to detect co-purifying A3 proteins. (C) Representative IF microscopy images of HeLa cells co-transfected with FLAG-tagged EBV BORF2 or KSHV ORF61 (red) and the indicated A3-EGFP constructs (green). Scale = 10 μm. (D–E) Quantification of A3-EGFP relocalization in HeLa cells expressing EBV BORF2 or KSHV ORF61. Panel D shows the percentage of cells exhibiting relocalized A3-EGFP (mean ± standard deviation, n≥100 cells per condition). Statistical analyses were performed using unpaired t-tests to determine significant changes in the relocalization of L7 swapped A3s (darker shade) relative to wild-type (WT) (lighter shade; n=3 independent experimental replicates; ns, not significant p>0.5; *p≤0.5; **p≤0.01; ***p≤0.001). Panel E shows Pearson correlation coefficient (PCC) values for A3-EGFP (or EGFP alone as a control) and Hoechst (nuclear stain). Density curves show the PCC value distribution in each condition, and the white vertical lines indicate the median PCC value (n≥100 cells per condition).
-
Figure 2—source data 1
File contains original immunoblots for Figure 2B.
- https://cdn.elifesciences.org/articles/83893/elife-83893-fig2-data1-v2.zip
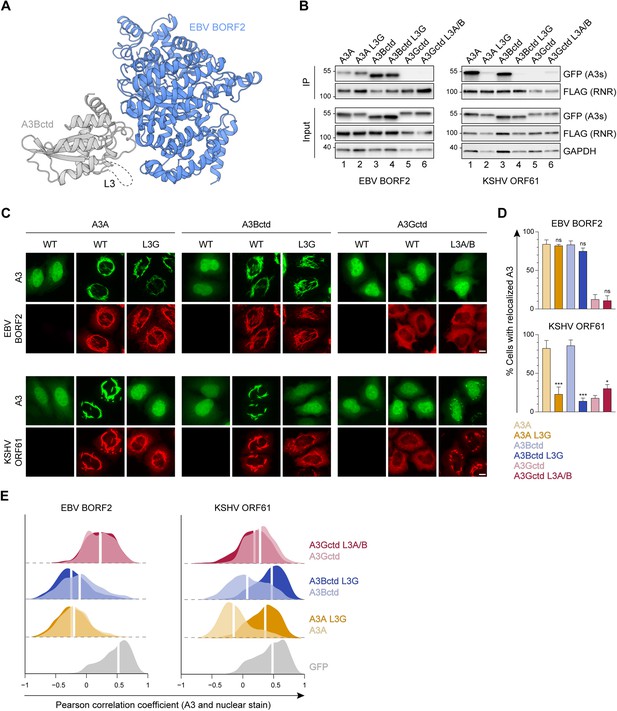
Role of A3B and A3A loop 3 in binding to gamma-herpesvirus ribonucleotide reductases (RNRs).
(A) Ribbon schematic of the Epstein-Barr virus (EBV) BORF2-A3Bctd complex (pdb 7rw6, chains A and B) with BORF2 in blue and A3Bctd in gray. Gray dashed line represents the L3 region not visible by cryo-EM. (B) Co-immunoprecipitation (Co-IP) of A3 L3 chimeras with EBV BORF2 and Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF61. FLAG-tagged RNR subunits were co-expressed with the indicated A3-EGFP constructs in 293T cells, affinity purified, and analyzed by immunoblotting to detect co-purifying A3 proteins. (C) Representative IF microscopy images of HeLa cells co-transfected with FLAG-tagged EBV BORF2 or KSHV ORF61 (red) and the indicated A3-EGFP constructs (green). Scale = 10 μm. (D–E) Quantification of A3-EGFP relocalization in HeLa cells expressing EBV BORF2 or KSHV ORF61. Panel D shows the percentage of cells exhibiting relocalized A3-EGFP (mean ± standard deviation, n≥100 cells per condition). Statistical analyses were performed using unpaired t-tests to determine significant changes in the relocalization of L3 swapped A3s (darker shade) relative to wild-type (WT) (lighter shade; n=3 independent experimental replicates; ns, not significant p>0.5; *p≤0.5; **p≤0.01; ***p≤0.001). Panel E shows Pearson correlation coefficient (PCC) values for A3-EGFP (or EGFP alone as a control) and Hoechst (nuclear stain). Density curves show the PCC value distribution in each condition, and the white vertical lines indicate the median PCC value (n≥100 cells per condition).
-
Figure 3—source data 1
File contains original immunoblots for Figure 3B.
- https://cdn.elifesciences.org/articles/83893/elife-83893-fig3-data1-v2.zip
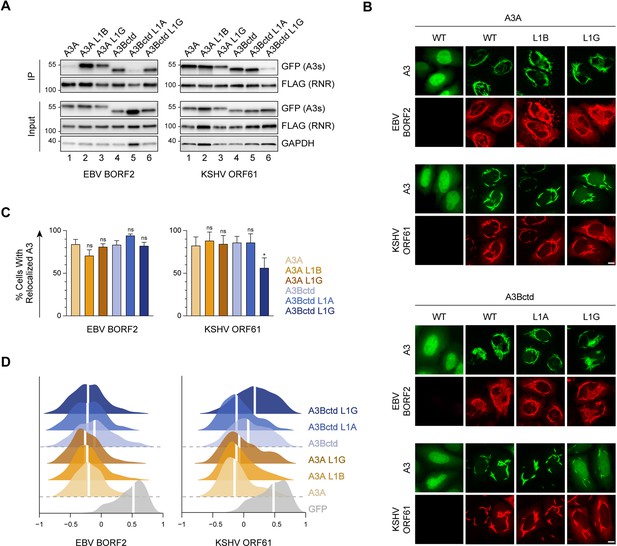
Role of A3B and A3A loop 1 in binding to gamma-herpesvirus ribonucleotide reductases (RNRs).
(A) Co-immunoprecipitation (Co-IP) of A3 L1 chimeras with Epstein-Barr virus (EBV) BORF2 and Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF61. FLAG-tagged RNR subunits were co-expressed with the indicated A3-EGFP constructs in 293T cells, affinity purified, and analyzed by immunoblotting to detect co-purifying A3 proteins. (B) Representative IF microscopy images of HeLa cells co-transfected with FLAG-tagged EBV BORF2 or KSHV ORF61 (red) and the indicated A3-EGFP constructs (green). Scale = 10 μm. (C–D) Quantification of A3-EGFP relocalization in HeLa cells expressing EBV BORF2 or KSHV ORF61. Panel C shows the percentage of cells exhibiting relocalized A3-EGFP (mean ± standard deviation, n≥100 cells per condition). Statistical analyses were performed using unpaired t-tests to determine significant changes in the relocalization of L1 swapped A3s (darker shade) relative to wild-type (WT) (lighter shade; n=3 independent experimental replicates; ns, not significant p>0.5; *p≤0.5; **p≤0.01; ***p≤0.001). Panel D shows Pearson correlation coefficient (PCC) values for A3-EGFP (or EGFP alone as a control) and Hoechst (nuclear stain). Density curves show the PCC value distribution in each condition, and the white vertical lines indicate the median PCC value (n≥100 cells per condition).
-
Figure 4—source data 1
File contains original immunoblots for Figure 4A.
- https://cdn.elifesciences.org/articles/83893/elife-83893-fig4-data1-v2.zip
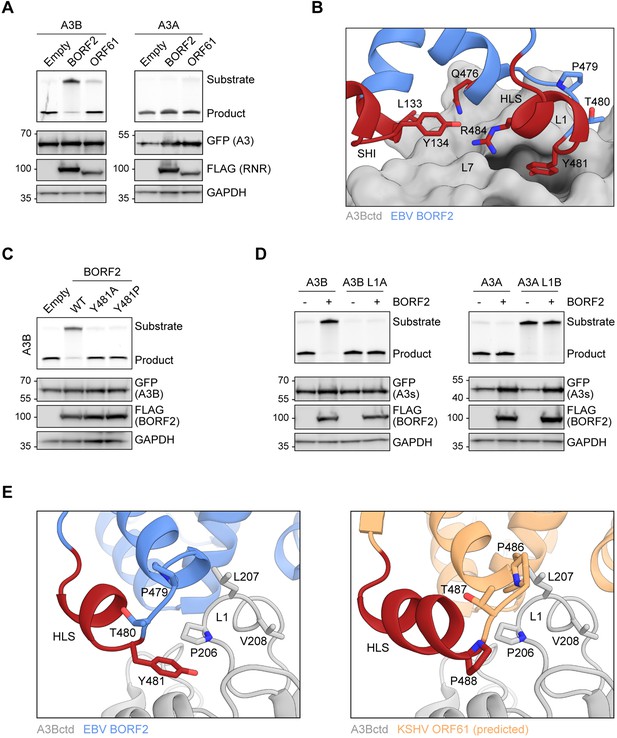
Epstein-Barr virus (EBV) BORF2 interacts with A3Bctd loop 1 to inhibit DNA deaminase activity.
(A) Upper panels show single-stranded DNA (ssDNA) deaminase activity of extracts from HeLa T-REx cells transfected with the indicated FLAG-tagged ribonucleotide reductase (RNR) subunits and incubated with doxycycline to induce the expression of A3B-EGFP or A3A-EGFP. Lower panels show protein expression levels in the same extracts by immunoblotting. (B) Zoom-in of the EBV BORF2-A3Bctd interface (pdb 7rw6, chains A and B) showing a network of interactions between BORF2 (blue, ribbon representation) and A3Bctd L7 and L1 regions (gray, surface-filled representation). BORF2 short helix insertion (SHI), helical loop structure (HLS), and Q476 are shown in red. (C) Upper panels show ssDNA deaminase activity of extracts from HeLa T-REx cells transfected the indicated FLAG-tagged EBV BORF2 constructs and incubated with doxycycline to induce the expression of A3B-EGFP. Lower panels show protein expression levels in the same extracts by immunoblotting. (D) Upper panels show ssDNA deaminase activity of extracts from HeLa cells co-transfected with FLAG-tagged EBV BORF2 and the indicated A3-EGFP constructs. Lower panels show protein expression levels in the same extracts by immunoblotting. (E) Ribbon schematic of the EBV BORF2-A3Bctd interface (pdb 7rw6, chains A and B) highlighting BORF2 tyrosine 481 and A3Bctd L1. BORF2 is shown in blue, the HLS region in red, and A3Bctd in gray. The right schematic shows the same view with the predicted structure of Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF61 (orange) overlayed and BORF2 residues hidden to allow better visualization (full model in Figure 1C).
-
Figure 5—source data 1
File contains original deaminase activity assay gels and immunoblots for Figure 5A.
- https://cdn.elifesciences.org/articles/83893/elife-83893-fig5-data1-v2.zip
-
Figure 5—source data 2
File contains original deaminase activity assay gels and immunoblots for Figure 5C.
- https://cdn.elifesciences.org/articles/83893/elife-83893-fig5-data2-v2.zip
-
Figure 5—source data 3
File contains original deaminase activity assay gels and immunoblots for Figure 5D.
- https://cdn.elifesciences.org/articles/83893/elife-83893-fig5-data3-v2.zip
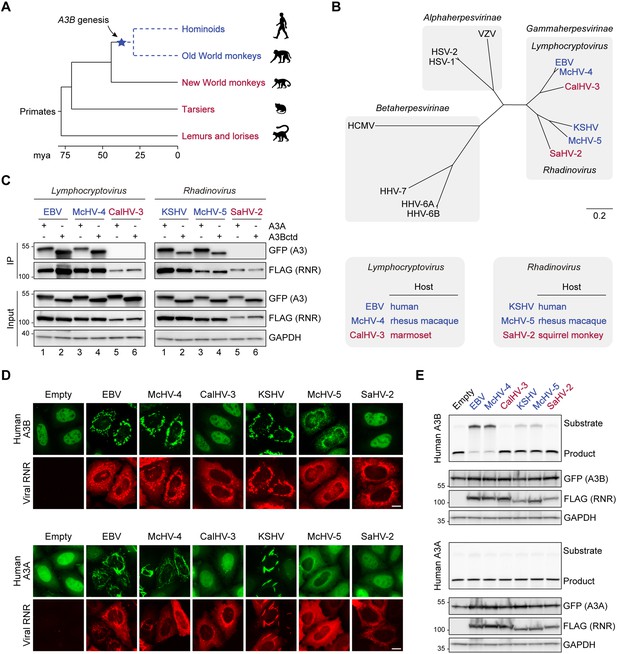
The genesis of A3B may have shaped gamma-herpesvirus ribonucleotide reductases (RNRs) evolution.
(A) Chronogram of primates depicting the generation of A3B in the common ancestor of hominoids and Old World monkeys. Primate groups shown in blue are composed of species that encode A3B and those shown in red lack A3B. Primate phylogeny and divergence intervals were derived from timetree.org (Kumar et al., 2017). Mya = million years ago. (B) Top: Phylogenetic tree of human herpesvirus and indicated non-human primate herpesvirus RNR large subunits. Shaded boxes indicate alpha-, beta-, and gamma-herpesvirus subfamilies. Genus classification is shown for gamma-herpesviruses. Gamma-herpesviruses that infect host species that encode A3B are shown in blue and those that infect host species that lack A3B are shown in red. Bottom: Host species infected by the indicated gamma-herpesviruses. Virus-host pairs are colored according to A3B status, as described for panel A. (C) Co-immunoprecipitation (Co-IP) of human A3A and A3Bctd with the RNR large subunits from the indicated viruses. FLAG-tagged RNR subunits were co-expressed with A3A-EGFP or A3Bctd-EGFP in 293T cells, affinity purified, and analyzed by immunoblotting to detect co-purifying A3 proteins. (D) Representative IF microscopy images of HeLa T-REx cells transfected with the indicated FLAG-tagged RNR subunits (red) and incubated with doxycycline to induce the expression of human A3B-EGFP or A3A-EGFP (green). Scale = 10 μm. (E) Upper panels show single-stranded DNA (ssDNA) deaminase activity of extracts from HeLa T-REx cells transfected with the indicated FLAG-tagged RNR subunits and incubated with doxycycline to induce the expression of A3B-EGFP or A3A-EGFP. Lower panels show protein expression levels in the same extracts by immunoblotting.
-
Figure 6—source data 1
File contains original immunoblots for Figure 6C.
- https://cdn.elifesciences.org/articles/83893/elife-83893-fig6-data1-v2.zip
-
Figure 6—source data 2
File contains original deaminase activity assay gels and immunoblots for Figure 6E.
- https://cdn.elifesciences.org/articles/83893/elife-83893-fig6-data2-v2.zip
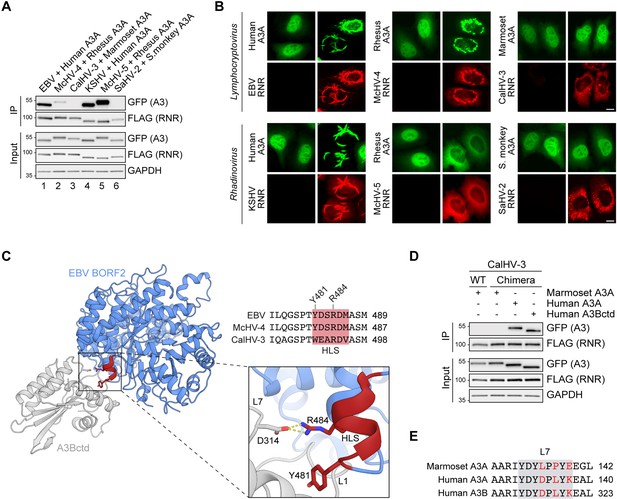
A short helical loop structure from Epstein-Barr virus (EBV) BORF2 enables the marmoset CalHV-3 ribonucleotide reductase (RNR) to bind to human A3B and A3A.
(A) Co-immunoprecipitation (Co-IP) of human, rhesus macaque, marmoset, and squirrel monkey A3As with the indicated gamma-herpesvirus RNR subunits. FLAG-tagged RNR subunits were co-expressed with the indicated A3-EGFP constructs in 293T cells, affinity purified, and analyzed by immunoblotting to detect co-purifying A3 proteins. (B) Representative IF microscopy images of HeLa cells co-transfected with the indicated FLAG-tagged viral RNRs (red) and A3A-EGFP constructs (green). Scale = 10 μm. (C) Ribbon schematic of the EBV BORF2-A3Bctd complex (pdb 7rw6, chains A and B) with BORF2 in blue and A3Bctd in gray. The helical loop structure (HLS) of EBV BORF2 is depicted in red. Bottom-right: zoom-in of the interacting surfaces highlighting BORF2 Y481 and R484 interactions with A3B loops 1 and 7, respectively. Top-right: amino acid alignment of the HLS regions of the EBV, McHV-4, and CalHV-3 RNRs (red boxes) and adjacent residues. (D) Co-IP of marmoset A3A, human A3A, and human A3Bctd with wild-type (WT) or chimeric CalHV-3 RNR containing the HLS region of EBV BORF2. FLAG-tagged RNR subunits were co-expressed with the indicated A3-EGFP constructs in 293T cells, affinity purified, and analyzed by immunoblotting to detect co-purifying A3 proteins. (E) Amino acid alignment of the loop 7 regions of marmoset A3A, human A3A, and human A3B (gray boxes) with non-identical loop 7 residues highlighted in red.
-
Figure 7—source data 1
File contains original immunoblots for Figure 7A.
- https://cdn.elifesciences.org/articles/83893/elife-83893-fig7-data1-v2.zip
-
Figure 7—source data 2
File contains original deaminase activity assay gels and immunoblots for Figure 7D.
- https://cdn.elifesciences.org/articles/83893/elife-83893-fig7-data2-v2.zip
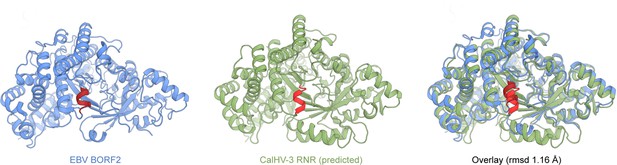
Ribbon schematics of the Epstein-Barr virus (EBV) BORF2 cryo-EM structure (pdb 7rw6, chain A; blue) and the RoseTTAFold-predicted CalHV-3 RNR structure (green).
The helical loop structure (HLS) region of each protein is shaded red and a structural overlay is shown for comparison.
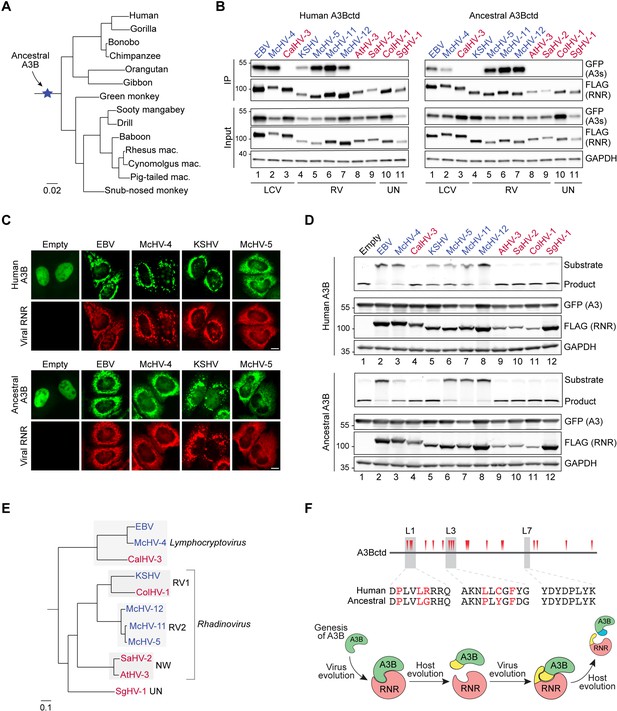
Ancestral A3B is active and antagonized by the ribonucleotide reductases (RNRs) from gamma-herpesviruses that infect A3B-encoding primates.
(A) Phylogenetic tree of primate A3B sequences used in this study to reconstruct ancestral A3B (amino acid sequence alignment in Figure 8—figure supplement 1). (B) Co-immunoprecipitation (Co-IP) of human and ancestral A3Bctd with the indicated gamma-herpesvirus RNR subunits. FLAG-tagged RNR subunits were co-expressed with the indicated A3-EGFP constructs in 293T cells, affinity purified, and analyzed by immunoblotting to detect co-purifying A3 proteins. RNRs from viruses that infect host species that encode A3B are shown in blue, and RNRs from viruses that infect host species that lack A3B are shown in red. LCV = Lymphocryptovirus; RV = Rhadinovirus; UN = unclassified gamma-herpesvirus (no official genus classification from the International Committee on Taxonomy of Viruses; Lefkowitz et al., 2018). (C) Representative IF microscopy images of HeLa cells co-transfected with human or ancestral A3B-EGFP (green) and the indicated FLAG-tagged viral RNRs (red). Scale = 10 μm. See Figure 8—figure supplement 2A for additional RNR data. (D) Upper panels show single-stranded DNA (ssDNA) deaminase activity of extracts from HeLa cells co-transfected with human or ancestral A3B-EGFP and the indicated FLAG-tagged viral RNRs. Lower panels show protein expression levels in the same extracts by immunoblotting. (E) Phylogenetic tree of primate gamma-herpesviruses RNR large subunits. Rhadinovirus RV1, RV2, and NW refer to primate Rhadinovirus lineages (Greensill et al., 2000; Strand et al., 2000; Lacoste et al., 2001; Schultz et al., 2000; Whitby et al., 2003) (UN, unclassified). (F) Top: positively selected sites in A3Bctd identified using mixed effects model of evolution algorithm (MEME) (Murrell et al., 2012) with p<0.1 (red triangles in schematic and red residues in loop region alignments; see Figure 8—figure supplement 4 for details). Bottom: schematic of the proposed genetic conflict between primate gamma-herpesvirus RNRs and cellular A3B.
-
Figure 8—source data 1
File contains original immunoblots for Figure 8B.
- https://cdn.elifesciences.org/articles/83893/elife-83893-fig8-data1-v2.zip
-
Figure 8—source data 2
File contains original deaminase activity assay gels and immunoblots for Figure 8D.
- https://cdn.elifesciences.org/articles/83893/elife-83893-fig8-data2-v2.zip

Amino acid sequence alignment of the indicated primate A3Bs.
A3Bctd regions are shaded yellow and the L1, L3, and L7 regions are highlighted with blue boxes. The predicted ancestral A3B protein sequence is positioned at the bottom of the group.
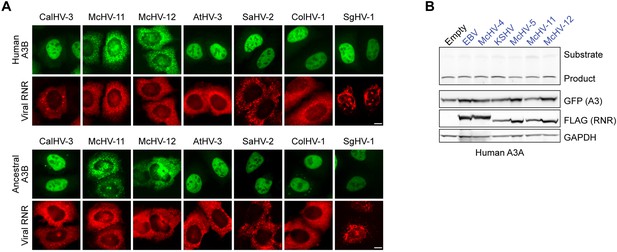
Relocalization of additional viral RNRs by human and ancestral A3B and lack of human A3A inhibition by Catarrhini gamma-herpesvirus RNRs.
(A) Representative IF microscopy images of HeLa cells co-transfected with human or ancestral A3B-EGFP (green) and the indicated FLAG-tagged viral ribonucleotide reductases (RNRs) (red). Scale = 10 μm. (B) Upper panels show single-stranded DNA (ssDNA) deaminase activity of extracts from HeLa cells co-transfected with human A3A-EGFP and the indicated FLAG-tagged viral RNRs. Lower panels show protein expression levels in the same extracts by immunoblotting.
-
Figure 8—figure supplement 2—source data 1
File contains original deaminase activity assay gels and immunoblots for Figure 8—figure supplement 2, panel B.
- https://cdn.elifesciences.org/articles/83893/elife-83893-fig8-figsupp2-data1-v2.zip
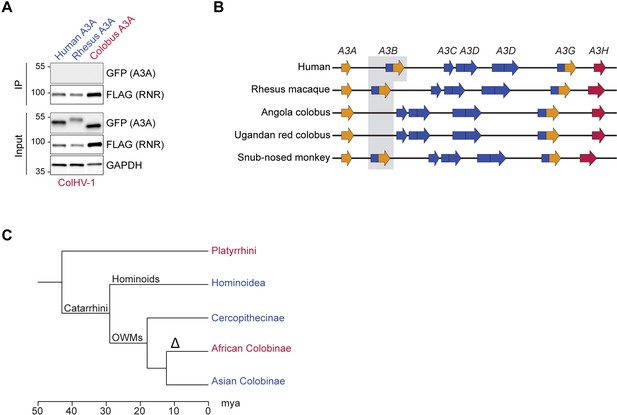
Lack of A3-binding by ColHV-1 RNR may be explained by the apparent loss of A3B in African colobus monkeys.
(A) Co-immunoprecipitation (Co-IP) of human, rhesus macaque, and Angola colobus A3A enzymes with the ColHV-1 ribonucleotide reductase (RNR). FLAG-tagged ColHV-1 RNR was co-expressed with the indicated A3A-EGFP constructs in 293T cells, affinity purified, and analyzed by immunoblotting to detect co-purifying A3 proteins. A3A enzymes from primate species that encode A3B are shown in blue and A3A enzymes from primate species that lack A3B are shown in red. (B) Schematic diagram depicting the APOBEC3 loci from the indicated primate species representative of Hominoidea (human, genome assembly GRCh38.p13), Cercopithecinae (rhesus macaque, genome assembly Mmul_10), African Colobinae (Angola colobus, genome assembly Cang.pa_1.0; Ugandan red colobus, genome assembly ASM277652v2), and Asian Colobinae (golden snub-nosed monkey, genome assembly Rrox_v1). The African Colobinae monkey genomes available via ENSEMBL (Cunningham et al., 2022) indicate that the A3B gene has been lost, likely after the Colobinae subfamily split into the African and Asian tribes (10–14 million years ago) because its closest relatives, the Asian Colobinae (e.g., Golden snub-nosed monkey), still encode A3B. APOBEC3 domains are colored according to Z domain classification (gold = Z1, blue = Z2, red = Z3). (C) Chronogram of primates depicting the loss of A3B following the split of the Colobinae subfamily into the African and Asian tribes (indicated by delta symbol). Primate groups shown in blue are composed of species that encode A3B and primates groups shown in red lack A3B. Primate phylogeny and divergence intervals were obtained from timetree.org (Kumar et al., 2017). Mya = million years ago.
-
Figure 8—figure supplement 3—source data 1
File contains original immunoblots for Figure 8—figure supplement 3, panel A.
- https://cdn.elifesciences.org/articles/83893/elife-83893-fig8-figsupp3-data1-v2.zip
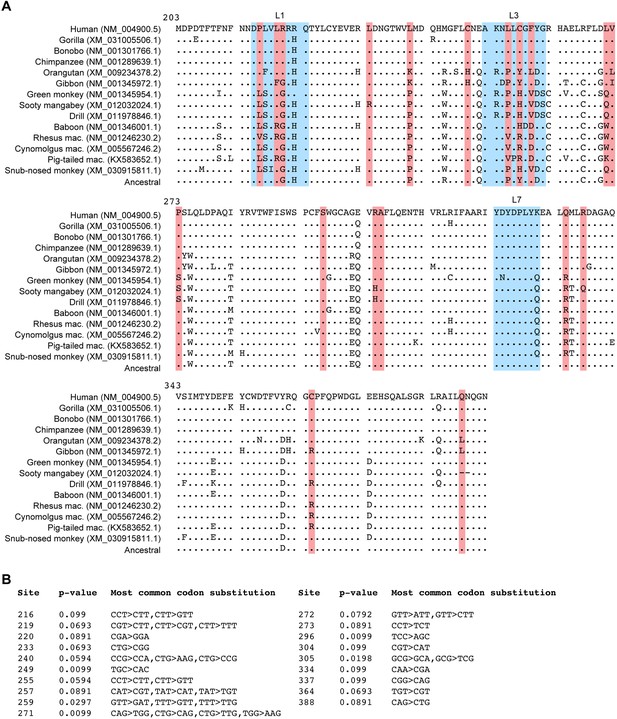
Diversifying positive selection in primate A3B.
(A) Amino acid sequence alignment of the C-terminal catalytic domain of the indicated primate A3B proteins with sites under diversifying positive selection identified using mixed effects model of evolution algorithm MEME (Murrell et al., 2012) with p<0.1 highlighted in red. L1, L3, and L7 regions are colored with blue boxes. (B) Summary of MEME analysis results showing the p-values in dN/dS ratio test and the most common codon substitutions at each codon site.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83893/elife-83893-mdarchecklist1-v2.docx
-
Source data 1
Numerical data for the bar plots and ridgeline plots in Figure 2, Figure 3, and Figure 4.
- https://cdn.elifesciences.org/articles/83893/elife-83893-data1-v2.xlsx





