Allele-specific gene-editing approach for vision loss restoration in RHO-associated retinitis pigmentosa
Figures
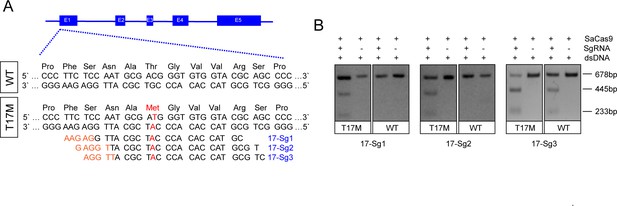
Schematic representation of allele-specific sgRNA design for RHO-T17M mutation.
(A) and confirmation of the specificity achieved using SaCas9 protein complexed with an sgRNA to target the mutant RHO sequence and its corresponding WT sequence in vitro (B), the full-length amplicon was 678 bp, the two truncated amplicons were 445 bp and 233 bp, respectively. The mutation c.50C>T (p.T17M) is indicated in red. PAM sequences were marked in orange. Exons were indicated by closed boxes.
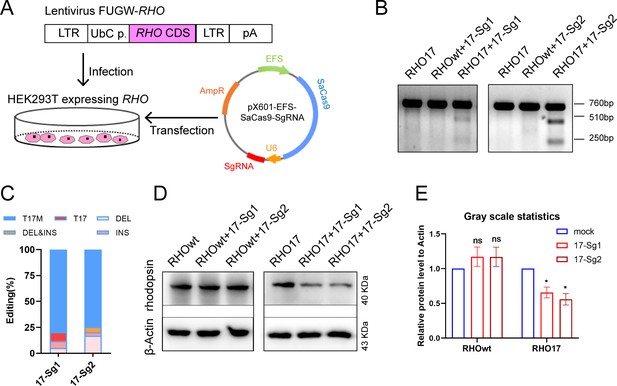
In vitro knockdown of human RHO-T17M expression.
(A) Schematic view of construction of 293T stably expressing human RHO protein and transfection of pX601-EFS-SaCas9-U6-sgRNA (SgRNA) plasmid. (B) T7E1 assay indicated that SaCas9/17-Sg1 and SaCas9/17-Sg2 were appeared to cut the mutant sequence specifically, the full-length amplicon was 760 bp, the two truncated amplicons were 510 bp and 250 bp, respectively. (C) The cutting efficacy of two sgRNAs with SaCas9 determined by TA and Sanger sequencing in 293T cells. (D) Rhodopsin expression reduction was determined by WB in RHO17 cells transfected with 17-Sg1 and -Sg2 plasmid, comparing to the RHOwt cells with 17-Sg1 and -Sg2 plasmid. (E) Densitometric analysis of immunoblots performed on RHOwt and RHO17 cells transfected with 17-Sg1 and -Sg2 plasmid, respectively. The experiment was performed in triplicate and presented as mean ± SEM, the significance was calculated using two-tailed paired t-test, ns = not significant, *p<0.05.
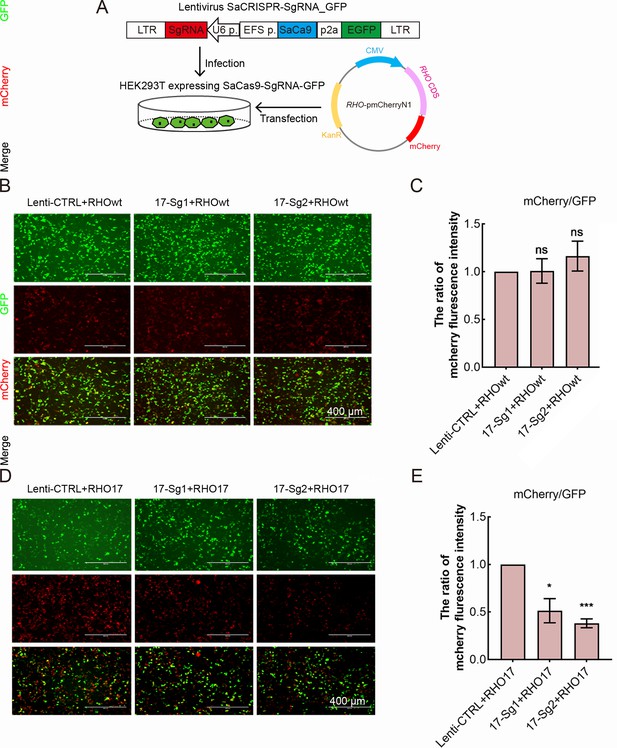
The knockdown of human RHO-T17M gene expression in 293T cells stably expressing SaCas9 and SgRNA.
(A) SaCas9/SgRNA_GFP stable cell line generation of 293T and transfection of human RHO cDNA plasmid. The calculation of mCherry/GFP ratio indicated that SaCas9/SgRNA did not cut the WT RHO allele (B–C). While SaCas9 with 17-Sg1(SaCas9/17-Sg1) and SaCas9/17-Sg2 could significantly reduce mCherry/GFP ratio (D–E). Scale bar = 400 μm. Error bars show SEM, and the significance was calculated using two-tailed paired t-test, ns = not significant, *p<0.05, ***p<0.005.
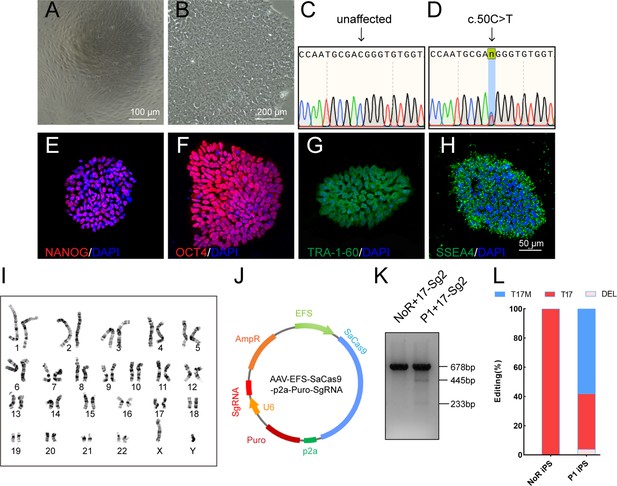
Determination of specificity and cutting efficiency of SaCas9 and sgRNA in patient-specific iPSCs.
(A) Photograph of UCs. Scale bar = 100 μm. (B) Photograph of iPSCs. Scale bar = 200 μm. (C–D) Sanger sequencing results showed that P1 iPS had the heterozygous c.50C>T mutation. IF staining showed the iPSCs expressed human embryonic stem cell-specific surface antigens including NANOG (E), OCT4 (F), TRA-1–60 (G) and SSEA-4 (H) protein. (I) Chromosomal content analysis revealed a normal 46, XY karyotype of patient P1. (J) Schematic view of AAV-EFS-SaCas9-U6-sgRNA-p2a-Puro plasmid. (K) T7E1 assay indicated that SaCas9/17-Sg2 was appeared to cut the mutant sequence specifically. (L) The cutting efficacy of SaCas9/17-Sg2 is determined by TA and Sanger sequencing in iPSCs.
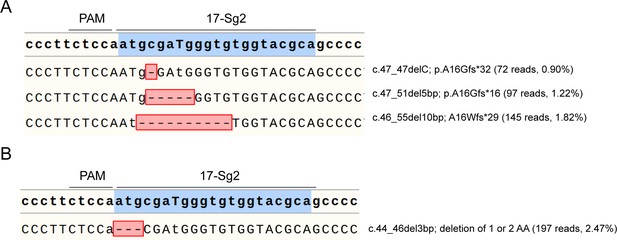
Hi-Tom sequence results of SaCas9/17-Sg2-treated P1 iPS (A) and P1 iPS colony (B).
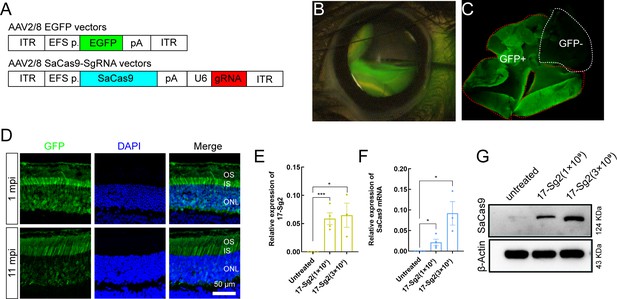
AAV2/8-mediated SaCas9 and sgRNA expression in mouse retinas.
(A) (Top) AAV2/8 vectors expressing EGFP under the control of EFS promoter. (Bottom) AAV2/8 vectors expressing SaCas9 under the control of EFS promoter and sgRNA under the control of U6 promoter were schematized. (B) Fluorescein shows AAV vectors distribution following subretinal injection immediately. (C) Infected areas treated with AAV-EGFP vector at dose of 1×109 dose were labeled by GFP expression (GFP+area). (D) IF staining indicated that GFP expressed in mouse retinas at 1 mpi and 11 mpi treated with AAV-EGFP vector at 3×109 dose. ONL = outer nuclear layer; OS = outer segment; IS = inner segment. Scale bar = 50 μm. (E–F) QPCR analysis indicated the expression of 17-Sg2 and SaCas9 in Rhowt/hum retinas at 3 mpi treated with AAV-SaCas9/17-Sg2 and AAV-EGFP vector (1:1 mixture) at different doses. Error bars show SEM, and the significance was calculated using two-tailed unpaired t-test, *p<0.05; ***p<0.005. (G) Determination of SaCas9 expression by WB analysis at 3 mpi at 1×109 and 3×109 dose.
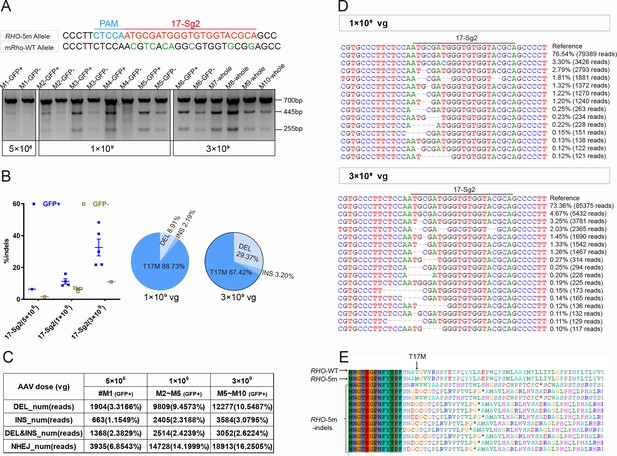
CRISPR/Cas9 targeting of c.50C>T RHO dominant variant encoding p.T17M in Mut-Rhowt/hum mouse retinas.
(A) T7E1 assay was performed on GFP+ and GFP- retinas to detect gene-editing activity of AAV-SaCas9/17-Sg2 in Mut-Rhowt/hum mouse retinas at 5×108, 1×109, and 3×109 doses, the full-length amplicon was 700 bp, the two truncated amplicons were 445 bp and 255 bp, respectively; 17-Sg2 was marked in red; PAM sequence was marked in light blue and mismatches in mouse Rho sequence comparing to 17-Sg2 were indicated in green. Hi-Tom sequencing (B) and PCR-based NGS (C) were used to detect the gene-editing efficiency and types of indels in AAV-SaCas9/17-Sg2-treated Mut-Rhowt/hum mouse retinas at 5×108, 1×109, and 3×109 doses. (D) Graphic representation of indels scored in the target site of Mut-Rhowt/hum mouse retinas treated with AAV-SaCas9/17-Sg2 at 1×109 and 3×109 doses. (E) Analysis of the amino acid sequence of the edited RHO-5m allele, * indicated the PTC.
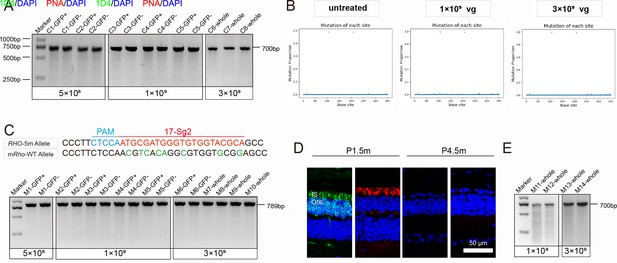
Analysis of gene-editing specificity and efficiency in RHO humanized mouse retinas.
(A) T7E1 assay performed on GFP+ and GFP- retinas indicated undetectable gene-editing activity (gene-editing specificity) of AAV-SaCas9/17-Sg2 in Rhowt/hum mouse at 5×108, 1×109, and 3×109 doses, the full-length amplicon was 700 bp. (B) PCR-based NGS indicated that AAV-SaCas9/17-Sg2 did not cut WT human RHO allele in Rhowt/hum mouse retinas at 1×109 and 3×109 doses and untreated Rhowt/hum retinas. the X axis represented the base site, the Y axis represented the indels proportion in WT human RHO allele of AAV-SaCas9/17-Sg2-treated Rhowt/hum mouse retinas. (C) T7E1 assay performed on GFP+ and GFP- retinas indicated that AAV-SaCas9/17-Sg2 did not cut WT mouse Rho allele in Mut-Rhowt/hum mouse retinas at 5×108, 1×109, and 3×109 doses, the full-length amplicon was 789 bp, 17-Sg2 was marked in red; PAM sequence was marked in light blue and mismatches in mouse Rho sequence comparing to 17-Sg2 were indicated in green. (D) IF staining indicated rapid loss of photoreceptor cells in Mut-Rhohum/hum mice, rhodopsin staining (1D4) was shown in green, PNA staining was shown in red, nuclei were stained blue by DAPI. IS = inner segment, ONL = outer nuclear layer. P1.5/4.5m = postnatal 1.5/4.5 month. Scale bar = 50 μm. (E) T7E1 assay indicated undetectable cutting activity in AAV-SaCas9/17-Sg2-treated Mut-Rhohum/hum mouse retinas at 1×109 dose and 3×109 dose.
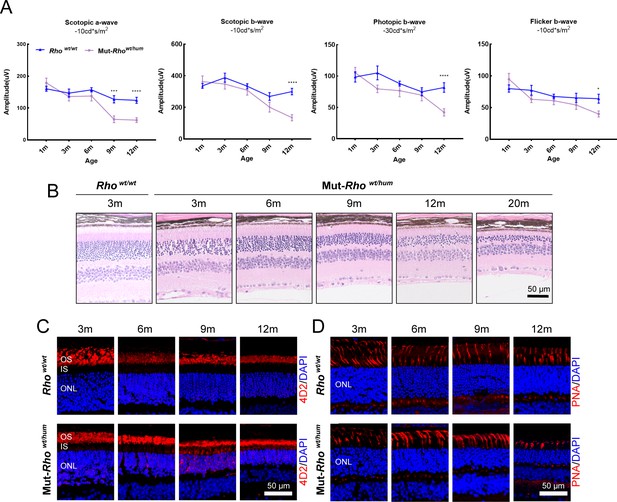
Phenotypes of the humanized mouse models.
(A) Scotopic and photopic ERG results of Rhowt/wt and Mut-Rhowt/hum mice with increasing age. Error bars show SEM, and the significance was calculated using two-tailed paired t-test, *p<0.05; ***p<0.005; ****p<0.001. (B) Histologic sections of Mut-Rhowt/hum mice and WT controls. A gradual loss in the number of photoreceptor cells in the ONL was identified with age in Mut-Rhowt/hum mice. Scale bar = 50 μm. IF staining of rhodopsin (C), PNA (D) in Mut-Rhowt/hum mice and WT controls. Rhodopsin (4D2 antibody) and PNA staining were shown in red. Nuclei were stained blue by DAPI. OS = outer segment, ONL = outer nuclear layer. Scale bar = 50 μm.
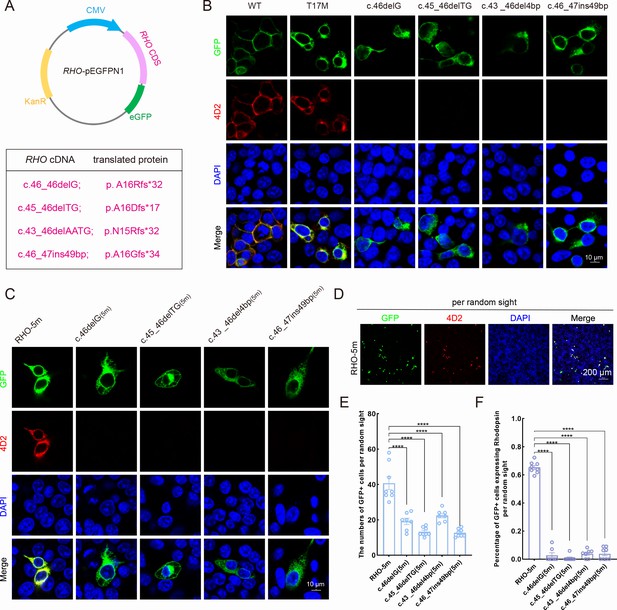
Expression of the mutant human RHO allele after gene editing with SaCas9/17-Sg2 in vitro.
(A) Schematic view of the different human RHO gene variants created by gene editing. (Top) Map of the pEGFPN1 vector used to overexpress these variants. (Bottom) The description of variants at DNA and protein level. (B) Colocalization of GFP and rhodopsin (4D2, red) in 293T cells transfected with pEGFPN1 vector carrying RHO-WT, RHO-T17M, and four edited RHO-T17M variants, 1 week after transfection. Scale bar = 10 μm. (C) Colocalization of GFP and rhodopsin (4D2, red) in 293T cells transfected with pEGFPN1 vector carrying RHO-5m and four edited RHO-5m variants, 1 week after transfection. Scale bar = 10 μm. (D–F) The number of GFP+ cells and percentage of GFP+ cells expressing rhodopsin per random sight. Nuclei were stained blue by DAPI. Scale bar = 200 μm.
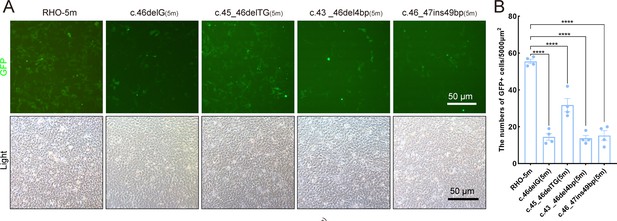
Expression of the mutant human RHO allele after gene editing with SaCas9/17Sg-2 in vitro.
(A–B) GFP expression in 293T cells transfected with pEGFPN1 vector carrying RHO-5m (five variants in RHO CDS) and four edited variants, 24 hr after transfection. Scale bar = 50 μm.
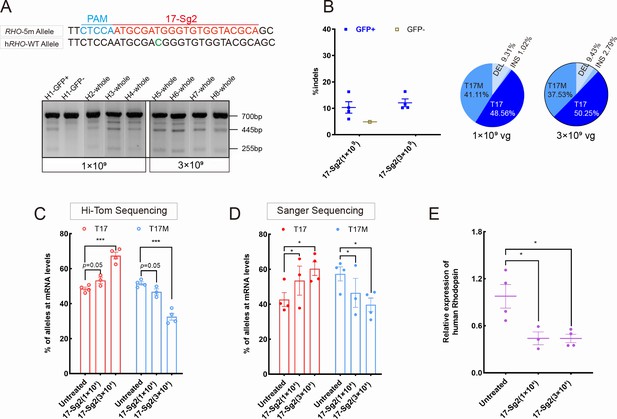
Expression of the RHO-5m allele after gene editing with AAV-based SaCas9/17-Sg2 in Rhohum/m-hum retinas.
(A) T7E1 assay was performed on GFP+ and GFP- retinas to detect cutting activity of AAV-SaCas9/17-Sg2 in Rhohum/m-hum retinas at 3 mpi, the full-length amplicon was 700 bp, the two truncated amplicons were 445 bp and 255 bp, respectively; 17-Sg2 was marked in red; PAM sequence was marked in light blue and mismatches in WT human RHO sequence comparing to 17-Sg2 were indicated in green. (B) Hi-Tom sequencing was used to detect the gene-editing efficiency and types of indels in AAV-SaCas9/17-Sg2-treated Rhohum/m-hum retinas at 1×109 and 3×109 doses. Relative RNA level of RHO-WT and RHO-5m alleles in SaCas9/17-Sg2-treated retinas (n=3 at 1×109 dose, n=4 at 3×109 dose) versus age-matched untreated retinas (n=4) determined by Hi-Tom sequencing (C) and TA and Sanger sequencing (D). (E) Relative mRNA levels of human RHO in SaCas9/17-Sg2-treated retinas (n=3 at 1×109 dose, n=4 at 3×109 dose) versus age-matched untreated retinas (n=4) determined by qPCR analysis. Error bars show SEM, and the significance was calculated using two-tailed unpaired t-test, *p<0.05; ***p<0.005.
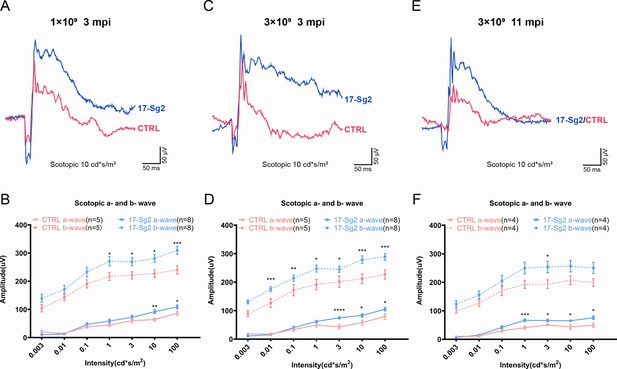
Significant improvement of retinal functions with AAV-based SaCas9/17-Sg2 in Mut-Rhowt/hum mice determined by full-field ERG.
Representative scotopic ERG (at the flash intensity of 10 cd*s/m2) waveforms of Mut-Rhowt/hum mice treated with AAV-SaCas9/17-Sg2 (blue) and AAV-SaCas9/CTRL (red) at 1×109 dose (3 mpi, A), 3×109 dose (3 mpi, C), and at 3×109 dose (11 mpi, E). Light dependence profile of scotopic a- and b-wave amplitudes of Mut-Rhowt/hum mice treated with AAV-SaCas9/17-Sg2 (blue) and AAV-SaCas9/CTRL (red) at 1×109 dose (B, 3 mpi), 3×109 dose (D, 3 mpi) and at 3×109 dose (F, 11 mpi). Error bars show SEM, and the significance was calculated using two-tailed unpaired t-test, *p<0.05; **p<0.01; ***p<0.005.
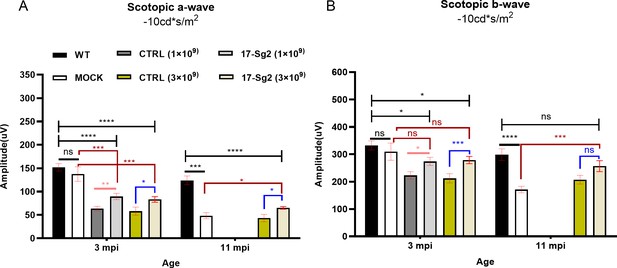
The ERG results of AAV-SaCas9/17-Sg12-treated Mut-Rhowt/hum mice (n=8), AAV-SaCas9/CTRL-treated mice (n=5), untreated Mut-Rhowt/hum mice (n=5), and untreated WT mice (n=7).
The mean amplitudes of scotopic a-wave (B) and b-wave (B) at 10 cd*s/m2. Error bars show SEM, and the significance was calculated using two-tailed unpaired t-test, *p<0.05; **p<0.01; ***p<0.005; ****p<0.001; ns = not significant.
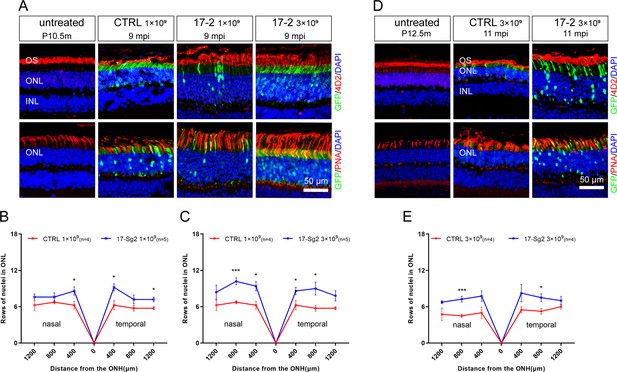
Photoreceptor cell preservation after gene editing with AAV-based SaCas9/17-Sg2 in Mut-Rhowt/hum mice.
(A) At 9 mpi, IF staining of rhodopsin (4D2, red) and PNA (red) from age-matched untreated mouse retina, the AAV-SaCas9/CTRL-treated and AAV-SaCas9/17-Sg2-treated retinal regions (GFP+) in Mut-Rhowt/hum mice at 1×109 and 3×109. The graph revealed the average number of rows of ONL nuclei in AAV-SaCas9/17-Sg2-treated (blue) and AAV-SaCas9/CTRL-treated (red) Mut-Rhowt/hum mice at 1×109 (B) and 3×109 (C) dose at 9 mpi. (D) At 11 mpi, IF staining of rhodopsin (4D2, red) and PNA (red) from age-matched untreated mouse retina, the AAV-SaCas9/CTRL-treated and AAV-SaCas9/17-Sg2-treated retinal regions (GFP+) in Mut-Rhowt/hum mice at 3×109. (E) The graph revealed the average number of rows of ONL nuclei in AAV-SaCas9/17-Sg2-treated (blue) and AAV-SaCas9/CTRL-treated (red) Mut-Rhowt/hum mice at 3×109 dose at 11 mpi. Error bars show SEM, and the significance was calculated using two-tailed unpaired t-test, *p<0.05; ***p<0.005. Rhodopsin and PNA were indicated in red, nuclei were stained blue by DAPI. ONL = outer nuclear layer; OS = outer segment; IS = inner segment. P10.5/12.5m = postnatal 10.5/12.5 month. Scale bar = 50 μm.
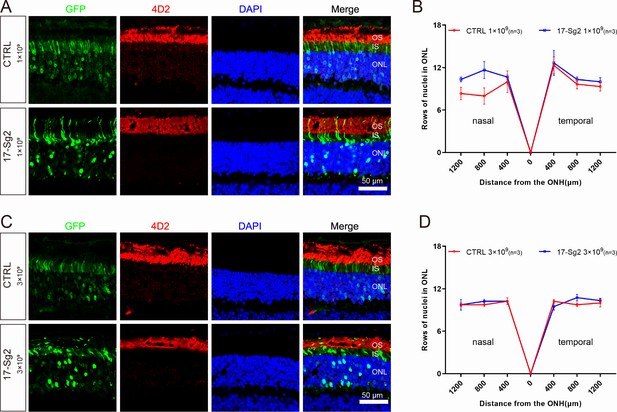
Retinal images after gene editing with AAV-based SaCas9/17-Sg2 in Mut-Rhowt/hum mice at 4 mpi.
Retinal images from the treated (GFP+) regions in Mut-Rhowt/hum mice at 1×109 dose (A) and 3×109 dose (C). (Upper) Representative image of mouse retina treated with AAV-SaCas9/CTRL. (Lower) Representative image of mouse retina treated with AAV-SaCas9/17-Sg2. The graph reveals the average number of rows of ONL nuclei in AAV-SaCas9/17-Sg2-treated (blue) mice and AAV-SaCas9/CTRL-treated (red) mice at 1×109 dose (B) and 3×109 dose (D) at 4 mpi. Error bars show SEM, and the significance was calculated using two-tailed unpaired t-test. Rhodopsin (4D2) was indicated in red, nuclei were stained blue by DAPI. ONL = outer nuclear layer; OS = outer segment; IS = inner segment. Scale bar = 50 μm.
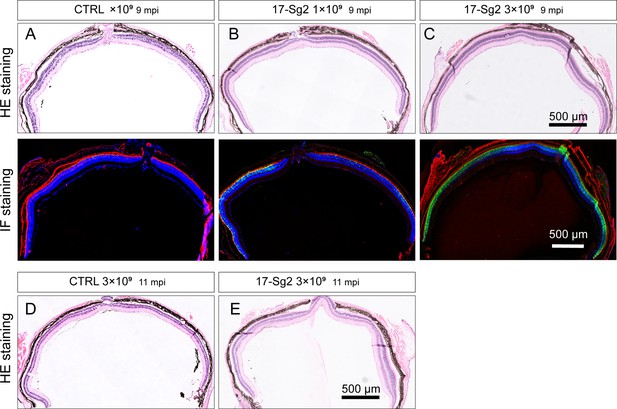
Flat-mount images of Mut-Rhowt/hum retinas after treatment.
At 9 mpi, HE and IF staining of AAV-SaCas9/CTRL-treated (A) and AAV-SaCas9/17-Sg2-treated retinas in Rhowt/hum mice at 1×109 (B) and 3×109 dose (C).At 11 mpi, HE staining of AAV-SaCas9/CTRL-treated (D) and AAV-SaCas9/17-Sg2-treated (E) retinas in Rhowt/hum mice at 3×109 dose.
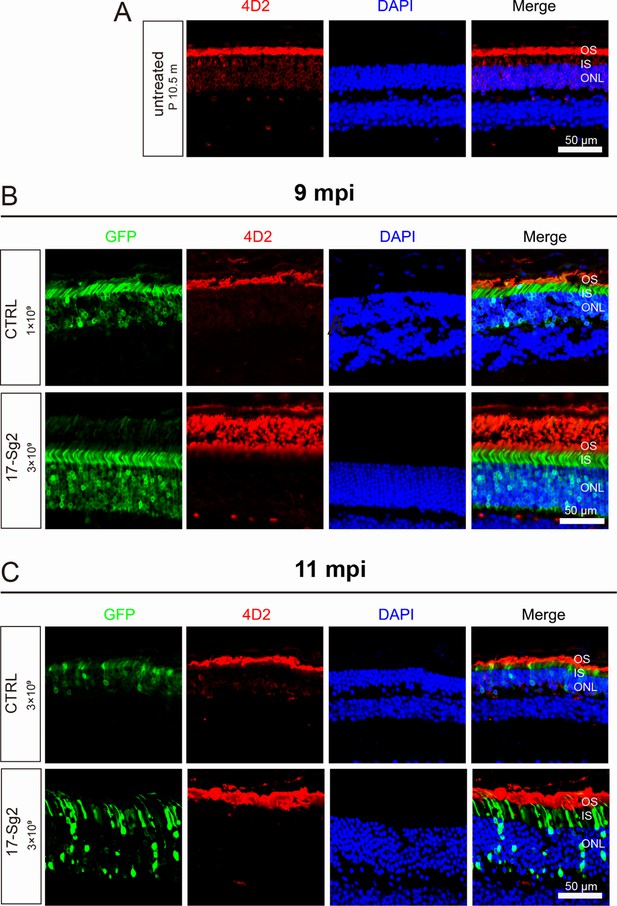
Retinal images after gene editing with AAV-based SaCas9/17-Sg2 in Mut-Rhowt/hum mice.
(A) Retinal images of untreated Mut-Rhowt/hum mice (P10.5m), mutant rhodopsin was retained in RIS and cell bodies (4D2, red). Retinal images from the treated (GFP+) regions in Mut-Rhowt/hum mice at 9 mpi (B) and 11 mpi (C). (Upper) Representative image of mouse retina treated with AAV-SaCas9/CTRL. (Lower) Representative image of mouse retina treated with AAV-SaCas9/17-Sg2. Rhodopsin (4D2) was indicated in red, nuclei were stained blue by DAPI. ONL = outer nuclear layer; OS = outer segment; IS = inner segment. P10.5m = postnatal 10.5/12.5 month. Scale bar = 50 μm.
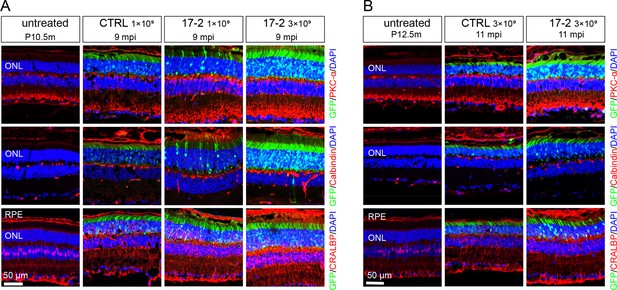
Inner retinal cells preservation after gene editing with AAV-based SaCas9/17-Sg2 in Mut-Rhowt/hum mice.
At 9mpi (A) and 11 mpi (B), IF staining of PKC-α (red), calbindin (red), CRALBP (red) from age-matched untreated mouse retina, the AAV-SaCas9/CTRL-treated and AAV-SaCas9/17-Sg2-treated retinal regions (GFP+) in Mut-Rhowt/hum mice at 1×109 and 3×109 dose. Blue cones and rod bipolar cells are labeled with PKC-α; calbindin immunofluorescence is prominent in the cell bodies of horizontal cells; CRALBP labeling is robust in RPE and Müller cells. Nuclei were stained blue by DAPI. ONL = outer nuclear layer; RPE = retinal pigment epithelium. P10.5/12.5m = postnatal 10.5/12.5 month. Scale bar = 50 μm.
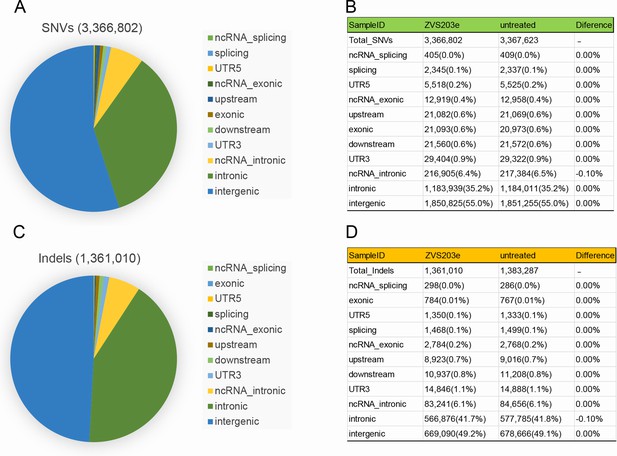
Examination of SaCas9/17-Sg2 off-target effects in human gDNA using WGS.
Identification of SNVs (A) and indels (C) in 293T cells transfected with 17-Sg2 plasmid at the WGS level. The type of SNVs (B) and indels (D) in 293T cells transfected with 17-Sg2 plasmid and untreated cells at the WGS level.
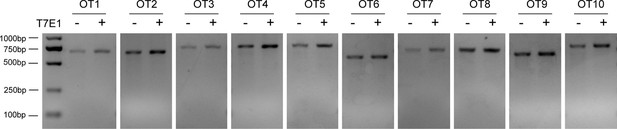
Analysis of SaCas9/17-Sg2 off-target activity.
T7E1 assay showed undetectable off-target activity for all candidate sites for 17-Sg2 (OT1-OT10).
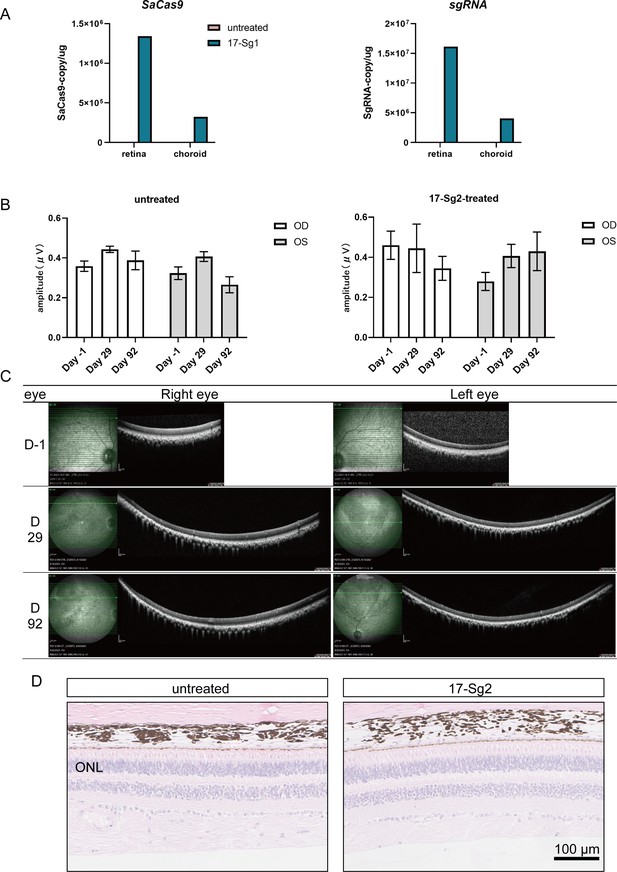
Safety analysis of AAV-SaCas9/17-Sg2 in NHP.
(A) The qPCR result indicated that AAV-SaCas9/17-Sg2 could express in NHP retina. (B) Multifocal ERG results of untreated NHP and an AAV-SaCas9/17-Sg2-treated NHP. The multifocal ERG amplitudes of supratemporal, suprasporal, suprasporal, and supratemporal retinas in each NHP were calculated. OD = right eye, OS = left eye. Error bars show SEM, and the significance was calculated using two-tailed unpaired t-test. (C) The OCT results of NHP retinas before and after treatment. (D) Representative HE image of untreated NHP retinas (left) and NHP retinas (right) treated with AAV-SaCas9/17-Sg2. Scale bar = 100 μm.
Additional files
-
Supplementary file 1
Supplementary tables.
(a) Results of TA cloning and Sanger sequencing in 293T cells. (b) A comprehensive summary of mouse experiments. (c) Results of TA cloning and Sanger sequencing in AAV-based SaCas9/17-Sg2-treated Mut-Rhowt/hum retinas. (d) Off-target sites of 17-Sg2 obtained from Benchling (https://www.benchling.com/). (e) Off-target sites of 17-Sg2 obtained from Cas-OFFinder (http://www.rgenome.net/cas-offinder) and NGS results. (f) Off-target sites of 17-Sg2 obtained from Cas-OFFinder (http://www.rgenome.net/cas-offinder) and NGS results. (g) The number of off-target sites of 17-Sg2 obtained from Cas-OFFinder (http://www.rgenome.net/cas-offinder). (h) Whole-genome sequencing results of off-target activity for SaCas9/17Sg-2. (i) List of primers.
- https://cdn.elifesciences.org/articles/84065/elife-84065-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84065/elife-84065-mdarchecklist1-v2.pdf
-
Source data 1
Source data files of the raw unedited gels or blots.
- https://cdn.elifesciences.org/articles/84065/elife-84065-data1-v2.zip
-
Source data 2
Source data files of the gels or blots with the relevant bands clearly labelled.
- https://cdn.elifesciences.org/articles/84065/elife-84065-data2-v2.zip
-
Source data 3
Source data files of NGS results and ERG statistical data.
- https://cdn.elifesciences.org/articles/84065/elife-84065-data3-v2.zip





