Identification of a weight loss-associated causal eQTL in MTIF3 and the effects of MTIF3 deficiency on human adipocyte function
Figures
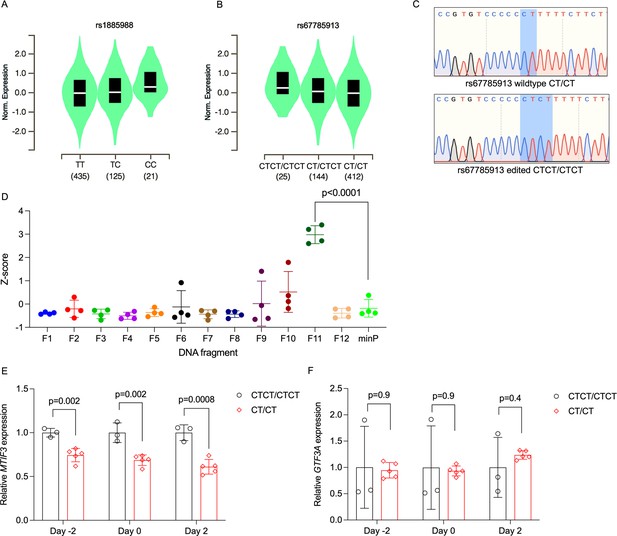
Identification of rs67785913 as a causal cis-eQTL for MTIF3.
(A) Violin plot of MTIF3 expression in subcutaneous adipose tissue for rs1885988 from Genotype-Tissue Expression (GTEx) Project eQTL. (B) Same as in (A), but for rs67785913. (C) Representative Sanger sequencing traces of rs67785913 CTCT/CTCT and CT/CT clones obtained after CRISPR/Cas9-mediated allele editing and single-cell cloning. (D) Normalized Z-score plot of luciferase reporter assays using vectors carrying different DNA fragments of the MTIF3 gene cloned into pGL4.23 luciferase reporter vector. Hypothesis testing was performed by comparing the transcriptional enhancer activity of each of the 12 vectors (F1–12) to the empty vector (minP). All data were plotted as mean ± standard deviation (SD), n = 4 independent experiments, p values are presented in each graph; ordinary one-way analysis of variance (ANOVA) was used for statistical analysis. (E) Relative MTIF3 expression (mRNA) in rs67785913 allele-edited cells 2 days before, at, or 2 days post-differentiation induction (day −2, 0, and 2, respectively). n = 3 clonal populations for CTCT/CTCT genotype, n = 5 clonal populations for CT/CT genotype, error bars show SD. (F) as in (E), but for GTF3A (mRNA) expression. Two-tailed Student’s t-test was used; p values are presented in each graph.
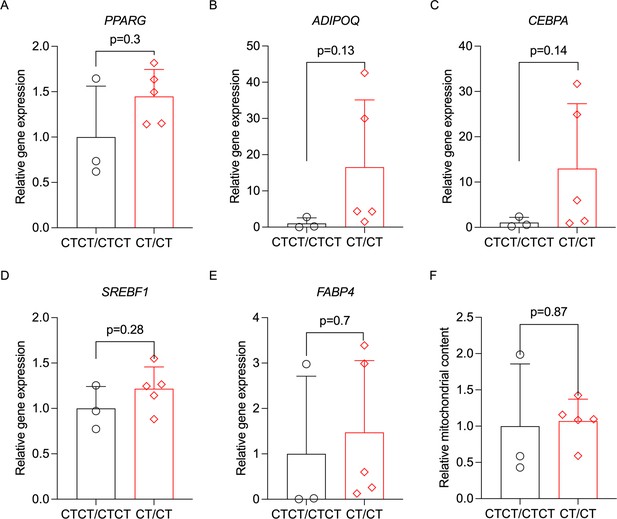
To test if rs67785913 affects adipogenic differentiation in hWAs cells, we differentiated the rs67785913 allele-edited cells (CTCT/CTCT vs. CT/CT) for 12 days.
We then used qPCR to quantify adipogenesis marker gene expression and mitochondrial content (mtDNA). Neither PPARG, ADIPOQ, CEBPA, SREBF1, and FABP4 gene expression (panels A–E) or mitochondrial content (panel F) were significantly different between the two rs67785913 genotypes. n = 3 clonal cell lines with CTCT/CTCT genotype, n = 5 clonal cell lines for CT/CT genotype. Error bars show standard deviation. Statistical analyses were performed using paired Student’s t-test, and p values are presented in the graphs.
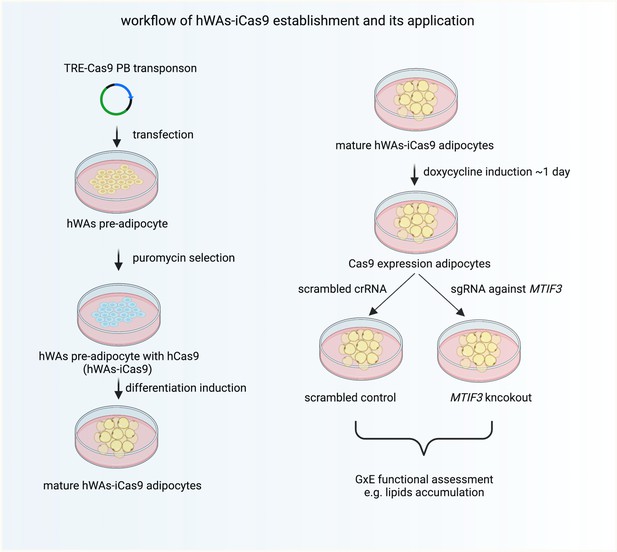
The workflow of establishing hWAs-iCas9 cell line and its application in studying MTIF3 and environment interactions in vitro.
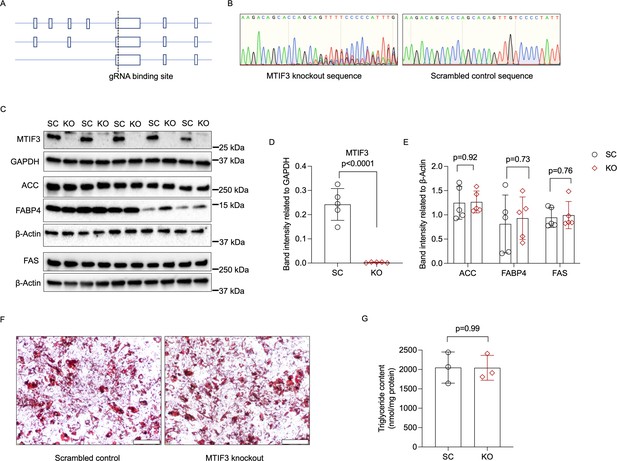
MTIF3 perturbation in mature adipocytes does not affect adipocyte-specific protein expression or total triglyceride content.
(A) An illustration of Cas9-specific single guide RNA (sgRNA)-binding site in the exon expressed in all three MTIF3 protein-encoding transcripts. (B) Representative Sanger sequencing of control and knockout hWAs mature adipocytes. (C) Immunoblots of adipocyte markers in scrambled control and MTIF3 knockout adipocytes, n = 5 independent experiments. (D) Quantitative analysis of MTIF3 band densities in (C). (E) Quantitative analysis of ACC, FABP4, and FAS band densities in (C). (F) Representative Oil-red O staining images of control and MTIF3 knockout in hWAs mature adipocytes. Scale bar is 200 µm. (G) Total triglyceride content in scrambled control (SC) and MTIF3 knockout (KO) cells. n = 3 independent experiments. Error bars show standard deviation in all plots. Statistical analysis was performed using two-tailed Student’s t-test, p values are presented in each graph. Uncropped blot images for (C) and raw.scn data files can be found in Figure 3—source data 1.
-
Figure 3—source data 1
Raw data files for western blots shown in Figure 3C.
- https://cdn.elifesciences.org/articles/84168/elife-84168-fig3-data1-v2.zip
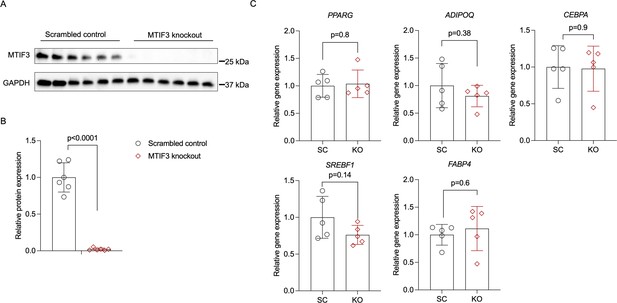
To test the effects of MTIF3 knockout on adipogenic differentiation in hWAs-iCas9 cell line, we first induced MTIF3 knockout in hWAs-iCas9 pre-adipocytes, then differentiated them using standard adipogenic differentiation cocktail.
After 12 days of differentiation, we assessed MTIF3 knockout efficiency using western blots, and examined adipogenic marker gene expression by RT-qPCR. MTIF3 expression was efficiently decreased in the knockout cells in n = 6 independent experiments (panels A, B). There were no significant differences between scrambled control and MTIF3 knockout cells on adipogenesis markers PPARG, ADIPOQ, CEBPA, SREBF1, and FABP4 expression (panel C); n = 5 independent experiments. Error bars show standard deviation. Statistical analyses were performed using paired Student’s t-test, and p values are presented in the graphs. Uncropped blot images for panel A and raw.scn data files can be found in Figure 3—figure supplement 1—source data 1.
-
Figure 3—figure supplement 1—source data 1
Raw data files for western blots shown in Figure 3—figure supplement 1A.
- https://cdn.elifesciences.org/articles/84168/elife-84168-fig3-figsupp1-data1-v2.zip
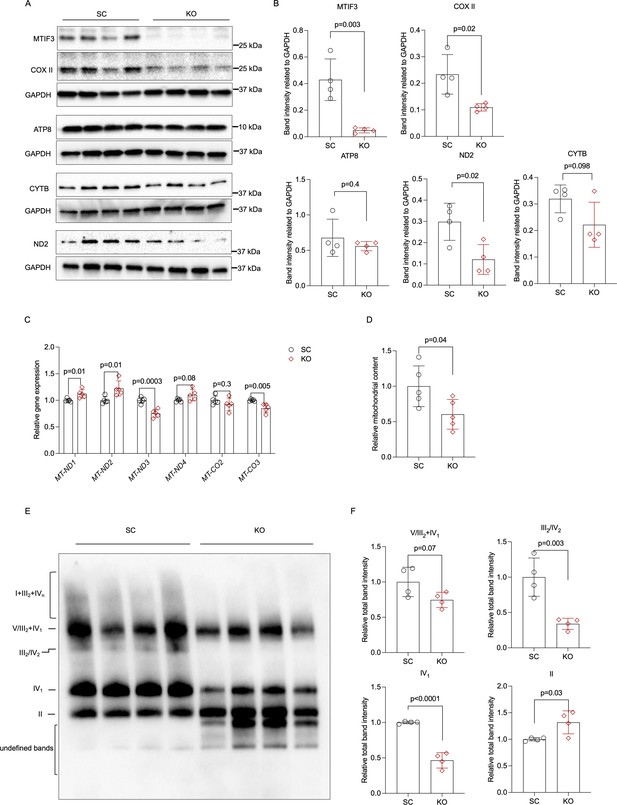
MTIF3 perturbation in mature adipocytes disrupts mitochondrial gene expression and OXPHOS complex assembly.
(A) Immunoblots of mitochondrial genome-encoded proteins in scrambled control and MTIF3 knockout adipocytes. (B) Quantitative analysis of band densities in (A). (C) qPCR for mitochondrial gene expression in scrambled control and MTIF3 knockout adipocytes, n = 5 independent experiments. (D) Relative mitochondrial DNA content in scrambled control and MTIF3 knockout adipocytes, n = 5 independent experiments. (E) Immunoblots of mitochondrial OXPHOS complex assembly after Blue Native-PAGE electrophoresis, n = 4 independent experiments. (F) Quantitative analysis of band densities in (E). Error bars show standard deviation in all plots. Statistical analysis was performed using two-tailed Student’s t-test, p values are presented in each graph. Uncropped blot images for (A) and raw.scn data files can be found in Figure 4—source data 1. Uncropped blot images for (E) and raw.scn data files can be found in Figure 4—source data 2.
-
Figure 4—source data 1
Raw data files for western blots shown in Figure 4A.
- https://cdn.elifesciences.org/articles/84168/elife-84168-fig4-data1-v2.zip
-
Figure 4—source data 2
Raw data files for western blots shown in Figure 4E.
- https://cdn.elifesciences.org/articles/84168/elife-84168-fig4-data2-v2.zip
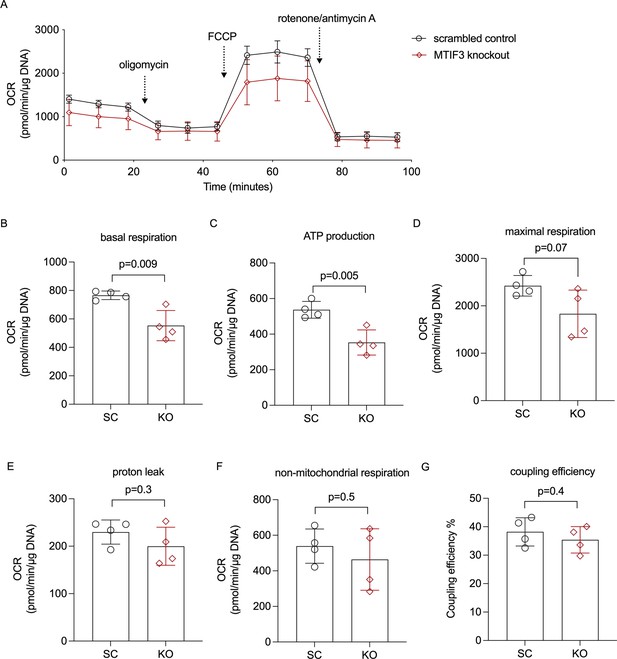
Cellular mitochondrial respiration in hWAs adipocytes.
(A) The average oxygen consumption rate (OCR) traces during basal respiration, and after addition of oligomycin, FCCP, and rotenone/antimycin A. (B) Basal respiration OCR, n = 4 different cell passages. (C) ATP production OCR, n = 4 different cell passages. (D) Maximal respiration OCR, n = 4 different cell passages. (E) Proton leak OCR, n = 4 different cell passages. (F) Non-mitochondrial respiration OCR, n = 4 different cell passages. (G) Coupling efficiency, n = 4 different cell passages. Error bars show standard deviation. Statistical analyses were performed using paired Student’s t-test in each condition, p values are presented in each graph.
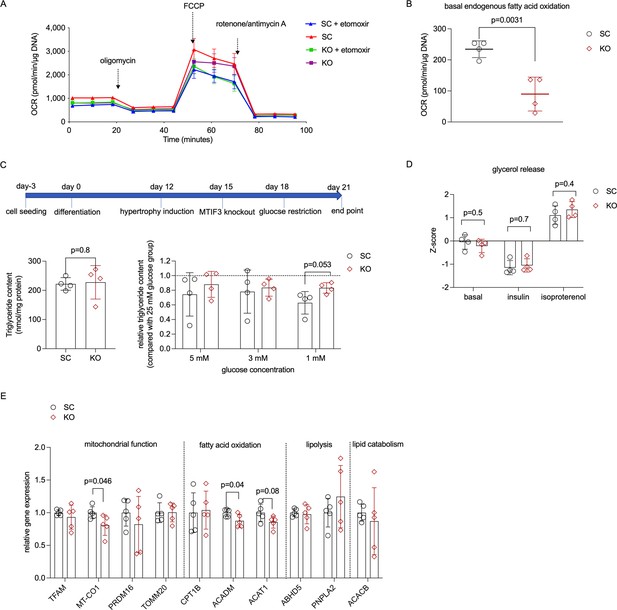
MTIF3 perturbation affects adipocyte fatty acid oxidation.
(A) A representative Seahorse oxygen consumption rate (OCR) trace for endogenous fatty acid oxidation assay. MTIF3 knockout and scrambled control adipocytes were treated with or without etomoxir for 15 min before the assay. Following the basal OCR measurement, oligomycin, FCCP (carbonyl cyanide-p-trifluoromethoxyphenylhydrazone), and rotenone + antimycin A were added sequentially to measure the detection of ATP production OCR, maximal respiration OCR and non-mitochondrial respiration OCR. (B) Basal endogenous fatty acid oxidation OCR in scrambled control (SC) and MTIF3 knockout (KO) adipocytes, n = 4 independent experiments. (C) Upper panel: workflow of glucose restriction in differentiated adipocytes; Lower left panel: total triglyceride content in scrambled control (SC) and MTIF3 knockout (KO) adipocytes in 25 mM glucose conditions; Lower right panel: triglyceride content in adipocytes cultured in glucose-restricted conditions (5, 3, and 1 mM) relative to adipocytes cultured in 25 mM glucose, n = 4 independent experiments. (D) Z-score-normalized data for glycerol release in scrambled control and MTIF3 knockout adipocytes under basal, insulin-stimulated, and isoproterenol-stimulated conditions, n = 4 independent experiments. (E) qPCR for mitochondrial and adipocyte-related gene expression in scrambled control and MTIF3 knockout adipocytes. Error bars show standard deviation in all plots. Statistical analysis was performed using two-tailed Student’s t-test, p values are presented in each graph.
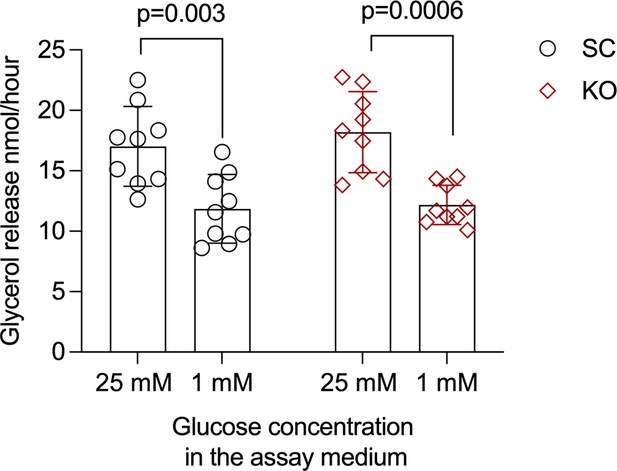
MTIF3 knockout does not affect mature adipocyte glycerol release at either 25 or 1 mM glucose condition.
To assay this, we incubated the differentiated adipocytes in glycerol release assay medium supplemented with either 25 or 1 mM glucose. We then determined glycerol content in each sample after 2-hr incubation. We found no differences in glycerol release between scrambled control and MTIF3 knockout cells cultured in the same glucose concentration. Notably, both scrambled control and MTIF3 knockout cells had significantly decreased glycerol release in 1 mM glucose restriction condition. n = 3 independent experiments, with 3 replicates per group and experiment. Error bars show standard deviation. Statistical analyses were performed using paired Student’s t-test, and p values are presented in the graph.
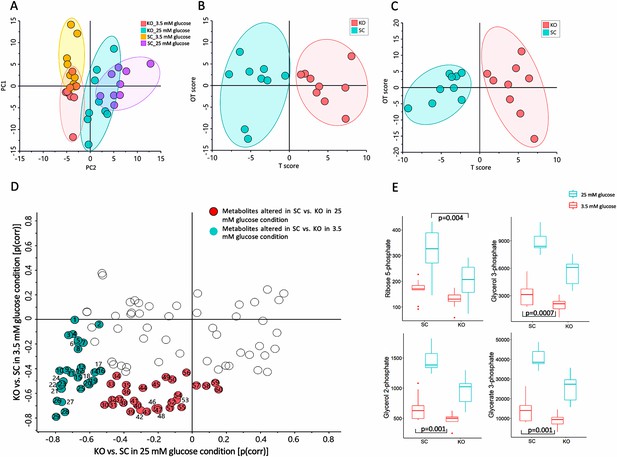
Mass spectrometry-based metabolomics data for control (SC) and MTIF3 knockout (KO) cells in 25 mM glucose (NF, normal feeding) and 5 mM glucose (GR, glucose-restricted) conditions.
(A) Principal component analysis (PCA) score plot displaying the discrimination between MTIF3 knockout and control cells in normal and glucose-restricted conditions (PC1: 28%, PC2: 19%). (B) Orthogonal projections to latent structures discriminant analysis (OPLS-DA) score plot showing classification of MTIF3 knockout and control cells in 25 mM glucose condition. (C) OPLS-DA score plots showing classification of MTIF3 knockout and control cells in glucose-restricted condition. (D) Shared and unique structures (SUS) plot, based on OPLS-DA models in (B, C), showing glucose concentration-dependent differences between MTIF3 knockout and control cells. (E) Box plots showing the abundance of some of the significantly altered metabolites in normal and MTIF3 knockout cells in normal and glucose-restricted conditions. Statistical analysis was performed using two-way analysis of variance (ANOVA) test, p values are presented in each graph.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | MTIF3 | UCSC Genome Browser | GRCh38/hg38 | |
| Cell line (Homo sapiens) | hWAs | Tseng laboratory at Joslin Diabetes Center | Xue et al., 2015 | |
| Cell line (Homo sapiens) | hWAs-iCas9 | This paper | Cell line maintained at Lund University Diabetes Center | |
| Antibody | Anti-MTIF3 (rabbit polyclonal antibody) | Proteintech | Cat: 14219-1-AP | WB (1:2000) |
| Antibody | Anti-OXPHOS antibody cocktail (mouse polyclonal antibody) | Thermo Fisher Scientific | Cat: 45-8199 | WB (1:1000) |
| Antibody | Anti-FABP4 (rabbit polyclonal antibody) | Cell Signalling Technology | Cat: 12589 | WB (1:1000) |
| Antibody | Anti-ACC rabbit polyclonal antibody | Cell Signalling Technology | Cat: 12589 | WB (1:1000) |
| Antibody | Anti-FAS rabbit polyclonal antibody | Cell Signalling Technology | Cat: 12589 | WB (1:1000) |
| Antibody | Anti-ATP8 (rabbit polyclonal antibody) | Proteintech | Cat: 26723-1-AP | WB (1:2000) |
| Antibody | Anti-ND2 (rabbit polyclonal antibody) | Proteintech | Cat: 19704-1-AP | WB (1:2000) |
| Antibody | Anti-CYTB (rabbit polyclonal antibody) | Proteintech | Cat: 55090-1-AP | WB (1:2000) |
| Antibody | Anti-β-Actin (rabbit polyclonal antibody) | Cell Signaling Technology | Cat: #4967 | WB (1:10,000) |
| Antibody | Anti-GAPDH (rabbit polyclonal antibody) | Abcam | Cat: ab37168 | WB (1:10,000) |
| Antibody | Anti-rabbit IgG, HRP-linked Antibody (goat polyclonal antibody) | Cell Signaling Technology | Cat: #7074 | WB (1:10,000) |
| Antibody | Anti-mouse IgG, HRP-linked Antibody (horse polyclonal antibody) | Cell Signaling Technology | Cat: #7076 | WB (1:10,000) |
| Recombinant DNA reagent | Super PiggyBac transposase (plasmid) | System Biosciences | PB210PA-1 | |
| Recombinant DNA reagent | pGL4.23 vectors | Promega | E8411 | |
| Recombinant DNA reagent | pGL4.75 CMV-Renilla reporter vectors | Promega | E6931 | |
| Recombinant DNA reagent | pPB-rtTA-hCas9-puro-PB plasmid | doi:10.1038/nprot.2016.152 | ||
| Sequence-based reagent | PCR primer (Forward) for rs67785913 genotyping | IDT | 5′–3′: GATTTGCAGGTGAGCAGACA | |
| Sequence-based reagent | PCR primer (Reverse) for rs67785913 genotyping | IDT | 5′–3′: ACTTGGAAATGGCCAAGATG | |
| Sequence-based reagent | sgRNA for rs67785913 editing | IDT | Spacer sequence: 5′-TTCAATAAGAAATTCCTCAA-3′ | |
| Sequence-based reagent | sgRNA for rs67785913 editing | IDT | Spacer sequence: 5′-GAAGAAAAAGGGGGGACACG-3′ | |
| Sequence-based reagent | Donor template for rs67785913 editing | IDT | ssDNA sequence: 5′’TGTGGACTCGCAGTCTGCCCTTGAGGAATTTCTTATTGAAGAAGAAAAAGAGGGGGGACACGGGGCCCAGACCCCCAGCACCCGGCTTTCGAGCAGGCTC-3′ | |
| Sequence-based reagent | sgRNA against MTIF3 | IDT | Design ID: Hs.Cas9.MTIF3.1.AB | Spacer sequence: 5′-GCAATAGGGGACAA CTGTGC-3′ |
| Sequence-based reagent | Taqman assay for MTIF3 | Thermo Fisher Scientific | Hs00794538_m1 | |
| Sequence-based reagent | Taqman assay for GTF3A | Thermo Fisher Scientific | Hs00157851_m1 | |
| Sequence-based reagent | Taqman assay for ADIPOQ | Thermo Fisher Scientific | Hs00977214_m1 | |
| Sequence-based reagent | Taqman assay for PPARG | Thermo Fisher Scientific | Hs01115513_m1 | |
| Sequence-based reagent | Taqman assay for CEBPA | Thermo Fisher Scientific | Hs00269972_s1 | |
| Sequence-based reagent | Taqman assay for SREBF1 | Thermo Fisher Scientific | Hs02561944_s1 | |
| Sequence-based reagent | Taqman assay for FASN | Thermo Fisher Scientific | Hs01005622_m1 | |
| Sequence-based reagent | Taqman assay for TFAM | Thermo Fisher Scientific | Hs01073348_g1 | |
| Sequence-based reagent | Taqman assay for MT-CO1 | Thermo Fisher Scientific | Hs02596864_g1 | |
| Sequence-based reagent | Taqman assay for PRDM16 | Thermo Fisher Scientific | Hs00223161_m1 | |
| Sequence-based reagent | Taqman assay for TOMM20 | Thermo Fisher Scientific | Hs03276810_g1 | |
| Sequence-based reagent | Taqman assay for CPT1B | Thermo Fisher Scientific | Hs00189258_m1 | |
| Sequence-based reagent | Taqman assay for ACADM | Thermo Fisher Scientific | Hs00936584_m1 | |
| Sequence-based reagent | Taqman assay for ACAT1 | Thermo Fisher Scientific | Hs00608002_m1 | |
| Sequence-based reagent | Taqman assay for ABHD5 | Thermo Fisher Scientific | Hs01104373_m1 | |
| Sequence-based reagent | Taqman assay for PNP1A2 | Thermo Fisher Scientific | Hs00386101_m1 | |
| Sequence-based reagent | Taqman assay for ACACB | Thermo Fisher Scientific | Hs01565914_m1 | |
| Sequence-based reagent | Taqman assay for MT-ND1 | Thermo Fisher Scientific | Hs02596873_s1 | |
| Sequence-based reagent | Taqman assay for MT-ND2 | Thermo Fisher Scientific | Hs02596874_g1 | |
| Sequence-based reagent | Taqman assay for MT-ND3 | Thermo Fisher Scientific | Hs02596875_s1 | |
| Sequence-based reagent | Taqman assay for MT-ND4 | Thermo Fisher Scientific | Hs02596876_g1 | |
| Sequence-based reagent | Taqman assay for MT-CO2 | Thermo Fisher Scientific | Hs02596865_g1 | |
| Sequence-based reagent | Taqman assay for MT-CO3 | Thermo Fisher Scientific | Hs02596866_g1 | |
| Sequence-based reagent | Taqman assay for HPRT-1 | Thermo Fisher Scientific | Hs99999909_m1 | |
| Sequence-based reagent | Taqman assay for TBP | Thermo Fisher Scientific | Hs00427620_m1 | |
| Sequence-based reagent | Taqman assay for RPL13A | Thermo Fisher Scientific | Hs03043885_g1 | |
| Commercial assay or kit | DNeasy Blood and Tissue kit | QIAGEN | 69506 | |
| Commercial assay or kit | Dual-Glo Stop&Glo reagents | Promega | E2920 | |
| Commercial assay or kit | Alt-R S.p. Cas9 D10A Nickase V3 | IDT | 1081058 | |
| Commercial assay or kit | Nucleofector reagent L | Lonza | VCA-1005 | |
| Commercial assay or kit | Alt-R HDR Enhancer V2 | IDT | 10007921 | |
| Commercial assay or kit | QuickExtract DNA Extraction Solution | Lucigen | QE09050 | |
| Commercial assay or kit | Alt-R Genome Editing Detection Kit | IDT | 1075932 | |
| Commercial assay or kit | Mitochondrial isolation kit | Thermo Fisher Scientific | 89874 | |
| Commercial assay or kit | NativePAGE Sample Prep Kit | Invitrogen | BN2008 | |
| Commercial assay or kit | Quant-iT PicoGreen dsDNA Assay Kit | Thermo Fisher Scientific | P7589 | |
| Commercial assay or kit | Triglyceride-Glo Assay kit | Promega | J3161 | |
| Commercial assay or kit | Glycerol-Glo Assay | Promega | J3150 | |
| Chemical compound, drug | Insulin | Sigma-Aldrich | I2643 | |
| Chemical compound, drug | Isoproterenol | Sigma-Aldrich | 1351005 | |
| Chemical compound, drug | Glucose | Angilent | 103577-100 | |
| Chemical compound, drug | Glutamine | Angilent | 103579-100 | |
| Chemical compound, drug | Carnitine | Sigma-Aldrich | C0283 | |
| Chemical compound, drug | Pyridine | Thermo Scientific | 019378.K2 | |
| Chemical compound, drug | N-Methyl-N-(trimethylsilyl) trifluoroacetamide | Thermo Scientific | A13141.22 | |
| Chemical compound, drug | Trimethylsilyl chloride | Thermo Scientific | A12535.30 | |
| Chemical compound, drug | 3-Nitrophenylhydrazine | Sigma-Aldrich | N21804 | |
| Chemical compound, drug | 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride | Thermo Scientific | 22980 | |
| Chemical compound, drug | Formic acid | Fisher Chemical | A117-50 | |
| Chemical compound, drug | Etomoxir | Sigma-Aldrich | E1905 | |
| Chemical compound, drug | Oligomycin | Sigma-Aldrich | O4876 | |
| Chemical compound, drug | FCCP | Sigma-Aldrich | C2920 | |
| Chemical compound, drug | Rotenone | Sigma-Aldrich | R8875 | |
| Chemical compound, drug | Antimycin A | Sigma-Aldrich | A8674 |
Additional files
-
Supplementary file 1
Design of fine mapping luciferase reporter assays, and association of MTIF3 locus with adiposity traits in UK Biobank.
(a) Thirty-one SNPs in tight linkage disequilibrium (r2 ≥ 0.8) with the lead variant rs1885988 tiled down into 12 DNA segments of the MTIF3 gene for luciferase reporter assay. To fine map the transcriptional regulatory regions in the MTIF3 locus, we first identified the common genetic variants which were in tight linkage disequilibrium (r2 ≥ 0.8) with the lead variant rs1885988 in HaploReg v4.1. The identified 31 SNPs were tiled down into 12 DNA segments of the MTIF3 gene depending on PCR primer design constraints. (b) SNPs in MTIF3 locus associated with body mass index (BMI), whole-body fat mass and arm fat mass (right). We checked the rapid GWAS analysis results from 337,000 samples in the UK Biobank, which were made available by Benjamin Neale’s lab and visualized in Oxford BIG browser, we found SNPs in MTIF3 locus showed nominal associations with body weight-related traits including BMI, whole-body fat mass and arm fat mass (right).
- https://cdn.elifesciences.org/articles/84168/elife-84168-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84168/elife-84168-mdarchecklist1-v2.pdf





