On the role of nucleotides and lipids in the polymerization of the actin homolog MreB from a Gram-positive bacterium
Figures

Crystal structure of the apo protofilament of MreB from G. stearothermophilus.
(A) Crystal structure of apo MreBGs (PDB ID 7ZPT), colored by subdomains, superimposed on the crystal structure of apo MreBTm (PDB ID 1JCF), in beige. The sequence similarity between the two proteins is 55.8%. Subdomain IA (blue) of MreBGs is formed by residues 1–32, 66–145 and 315–347; subdomain IB (yellow) by residues 33–65; IIA (red) by residues 146–181 and 246–314 and IIB (green) by residues 182–245. Superimposition of the two forms highlights the distinct positions of loops β6-α2 and α2-β7 as well as the movement of domain IB (two-headed arrow) resulting in slightly distinct subunit interaction modes as shown in panel C. (B) Protofilament structure of apo MreBGs. Three subunits of the protofilament formed upon crystal packing are displayed as cartoon and colored by subdomains. The subunit repeat distance is indicated. (C) Close view of the MreBGs intra-protofilament interface. The two subunits are colored by subdomains as in panel A, and shown as cartoons. Residues involved in putative salt bridges (gray dashed lines) are displayed as sticks colored by atom type (N in blue and O in red) and labeled. (D) Close view of the MreBTm intra-protofilament interface (PDB ID 1JCF). The two subunits are colored in beige as in panel A, and shown as cartoons. Residues involved in putative salt bridges (gray dashed lines) are displayed as sticks colored by atom type (N in blue and O in red) and labeled.
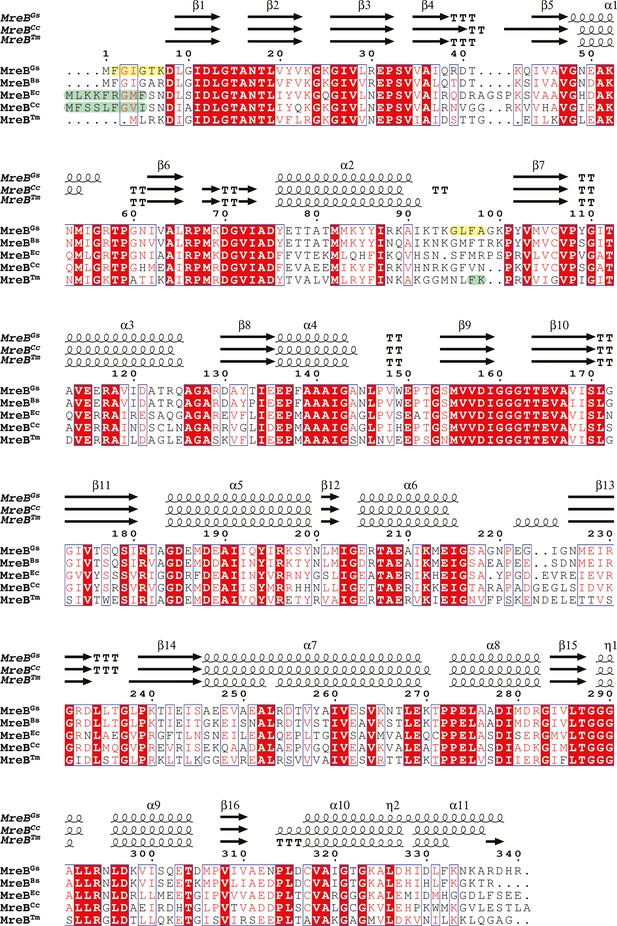
Multiple sequence alignment of MreB proteins from several bacteria.
The sequence of G. stearothermophilus (MreBGs) was aligned using Clustal-Ω at PRABI against the homologous MreB sequences of the Gram-positive bacterium B. subtilis (MreBBs, GenBank ID ATA60829.1) and the Gram-negative bacteria E. coli (MreBEc, GenBank ID P_417717), C. crescentus (MreBCc, GenBank ID YP_002516985.1) and T. maritima (MreBTm, GenBank ID AAD35673.1). Sequence numbering is relative to MreBGs. Secondary structure information extracted from the crystal structures of MreBGs (PDB ID 7ZPT), MreBCc (PDB ID 4CZM) and MreBTm (PDB ID IJCE) are indicated above the sequences using ESPript (Robert and Gouet, 2014). Beta strands are numbered β1 to β16, alpha helices α1 to α11, and 310 helices η1 to η2, according to the MreBGs structure. α- and β-turns are depicted as TTT and TT, respectively. Blue boxes indicate homologous regions, similar residues are indicated in red and identical residues in white on red background. The residues of the amino-terminus and α2β7 hydrophobic loops deleted in the mutants ΔNter and ΔGLFA of MreBGs are highlighted in yellow. Amphipathic helices in MreBEc and MreBCc and the hydrophobic loop in MreBTm are highlighted in green.
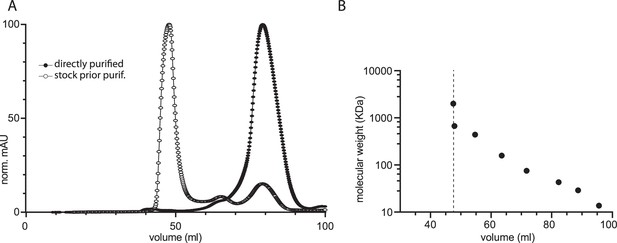
Calibration curve and typical size exclusion chromatography profiles of MreB.
(A) Typical size exclusion chromatography elution profiles of MreBGs. MreBGs (wild-type) was loaded on a HiLoad 16/600 Superdex 200 pg (GE healthcare) size exclusion column immediately after elution from a Nickel-NTA affinity purification column (filled circles) or after a subsequent 4 °C overnight incubation (empty circles). When size exclusion chromatography was performed using freshly purified protein from the affinity column, MreBGs (37.36 kDa) eluted mainly as a single peak corresponding to the monomeric form of the protein according to the calibration of the column (B). In contrast, overnight conservation of the eluate at 4 °C before loading it onto the Superdex column led to the irreversible formation of high molecular weight assemblies (aggregates) eluting at the dead volume of the column. (B) Calibration curve for the HiLoad 16/600 Superdex 200 pg (GE healthcare) size exclusion column performed with gel filtration calibration kits HMW and LMW (GE healthcare). The void volume, determined with Blue Dextran 2000, corresponds to an elution volume of ~47.5 ml (dashed line).
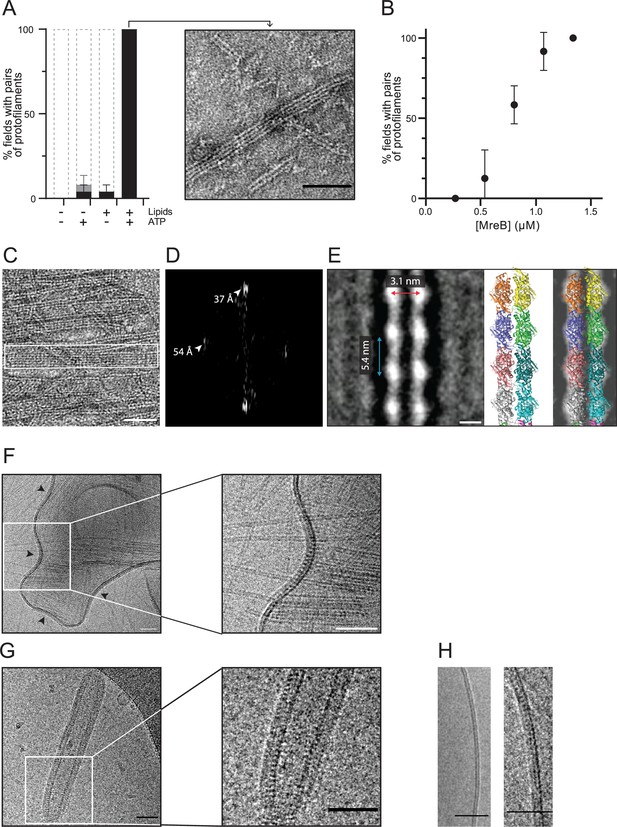
MreBGs forms double protofilaments in the presence of ATP and lipids.
(A) Polymerization of MreBGs into pairs of protofilaments depends on the presence of lipids and ATP. MreBGs was set to polymerize in standard conditions in the presence or absence of ATP and lipid total extract from E. coli. Polymer formation is expressed as percent of fields containing high (black) or low (grey) density of polymers (see Figure 2—figure supplement 1 for details of the quantification method of MreB polymers on TEM grids). Values are average of two independent experiments. Error bars are standard deviations. Inset shows an example of a field of dual protofilaments on a negative stained TEM image. Scale bar, 50 nm. (B) Polymer formation as a function of MreBGs concentration. MreBGs was set to polymerize in standard conditions at a concentration ranging from 0.27 to 1.34 µM (0.01–0.05 mg/mL). Values are the average of two independent experiments. Error bars are standard deviations. (C, D) MreBGs polymers assemble into sheets. (C) EM image of MreBGs set to polymerize in standard conditions. Scale bar, 50 nm. Fourier transform (D) was obtained from the area indicated by a white box in (C) and revealed a longitudinal subunit repeat of the filaments of 54 Å and a lateral spacing of ~37 Å (arrowheads). (E) (Left) 2D averaging of images of negatively stained dual protofilaments of MreBGs from 1 554 individual particles. Scale bar, 3 nm. Two copies of the atomic structure of the protofilaments found in the MreBGs crystals shown to scale (Middle, for illustration the two protofilaments are displayed arbitrarily in an antiparallel conformation but could fit in a parallel conformation as well) and docked into the 2D averaged EM image (Right). (F, G) MreBGs polymers assemble on lipid bilayers and distort liposomes as shown by cryo-electron microscopy (cryo-EM). Cryo-EM micrographs of liposomes (0.37 mg/mL) made from E. coli lipid total extracts incubated with purified MreBGs (1.34 µM; 0.05 mg/mL) in the presence of ATP (2 mM), and low (F, 100 mM) or high (G, 500 mM) concentration of KCl. Arrowheads point to MreB accumulations. Scale bars, 50 nm. (H). Cryo-EM micrographs showing the cross-section of the membrane of liposomes in the absence (Left) and in the presence (Right) of ATP-bound MreBGs at 500 mM KCl. Scale bars, 50 nm.
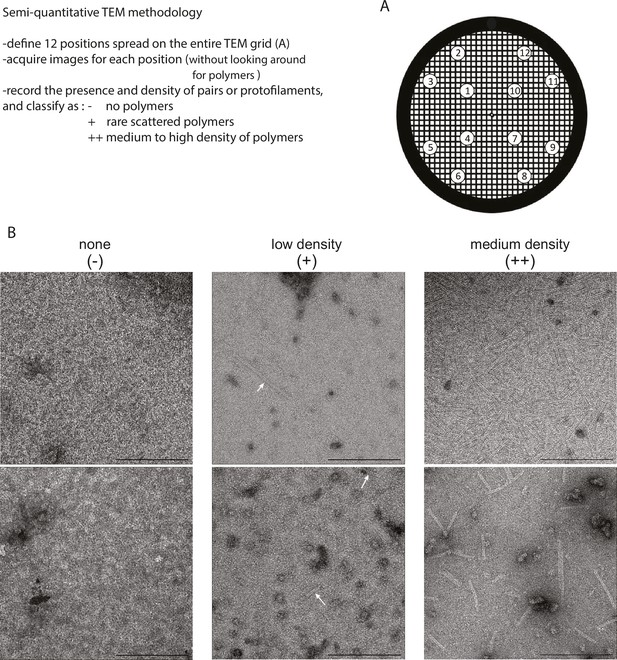
Workflow for quantification of MreB polymers on TEM grids.
(A) Schematic drawing of a 300 mesh EM grid displaying 12 imaging localization widespread on the observation field. (B) EM images of typical fields of view presenting no polymers (Left, ‘-‘), low density (Middle, ‘+’), or medium density (Right, ‘++’) of dual filament polymers of MreB. Scale bars, 200 nm.
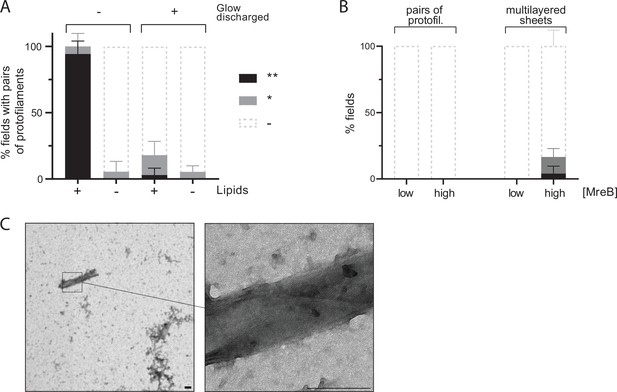
MreB presents limited capacity to form polymers in solution.
(A) Quantification of pairs of protofilaments of MreBGs shows limited polymerization in solution and in the absence of lipids. MreBGs was set to polymerize in standard conditions (ATP and 100 mM KCl) in the presence or absence of lipid total extract from E. coli. Following polymerization, samples were mounted onto untreated (-) or glow-discharged (+) grids favoring binding of hydrophobic or hydrophilic molecules respectively. Polymer formation is expressed as percent of fields containing high (black) or low (grey) density of polymers. Values are average of three independent experiments. Error bars are standard deviations. (B) Quantification of polymers of MreBGs shows limiting polymerization in solution regardless of the concentration of the protein. MreBGs was set to polymerize in standard conditions at low (1.34 µM) or high (6.7 µM) protein concentration in the absence of lipids, and samples were mounted on glow-discharged grids. MreB forming isolated pairs of protofilaments (Left) or large multilayered sheets (Right) were quantified separately. Polymeric structures are expressed as percent of fields containing high (black) or low (grey) density of polymers. Values are average of two independent experiments. Error bars are standard deviations. (C) Example of multilayered sheets of MreB. High levels of MreBGs (6.7 µM) were set to polymerize in the absence of lipids and in the presence of ATP. The crisscrossed pattern suggests that the sheet forms a tubular structure. Disorganized assemblies are visible in the bottom right corner of the zoomed-out image. Scale bars, 200 nm.
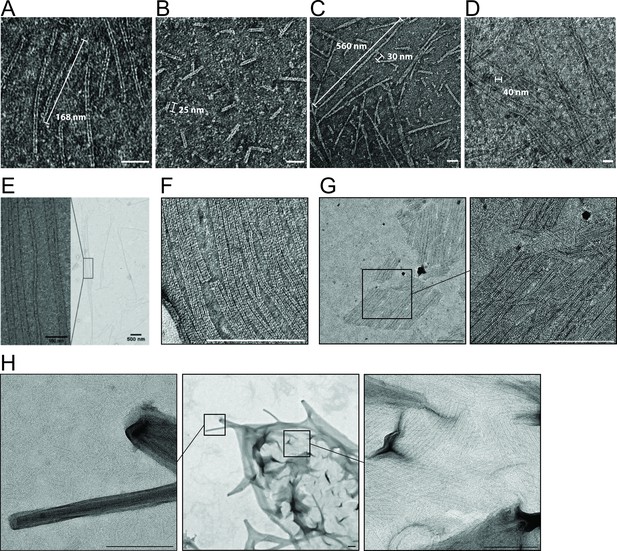
MreBGs polymers display a broad range of lengths and widths.
(A–D) Dual protofilaments of MreBGs observed on various fields of a single EM grid. Example of fields containing exclusively medium size polymers (>100 nm) (A); exclusively short polymers (<50 nm) (B); a mix of medium (some bundling) and short polymers (C), and a mix of long (>1 μm) and short polymers (D). In D, the long polymers are extending beyond the edges of the field of view. Scale bars, 50 nm. (E, F) MreBGs polymers can form filaments extending over several μm. Scale bar on F, 200 nm. (G) MreBGs polymers associate laterally to form short sheets of limited widths. Scale bars, 200 nm. (H) MreBGs can also form complex multilayered sheet-like structures. Scale bars, 200 nm.
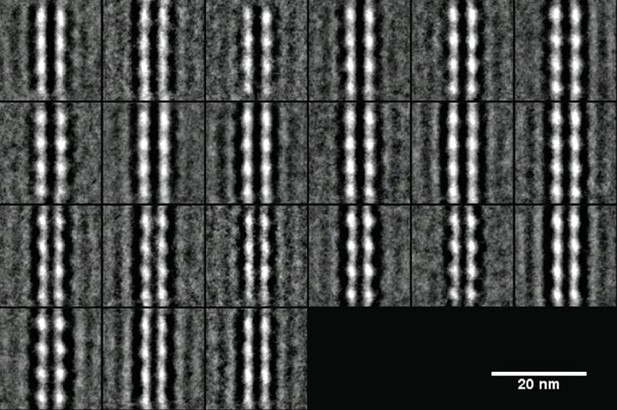
2D averaging of negatively stained images of MreBGs dual protofilaments showing the symmetrical arrangement of monomers.
Displayed are the 21 classes of images generated by 2D image processing (alignment and classification from 1 554 individual raw images). Scale bar, 20 nm.
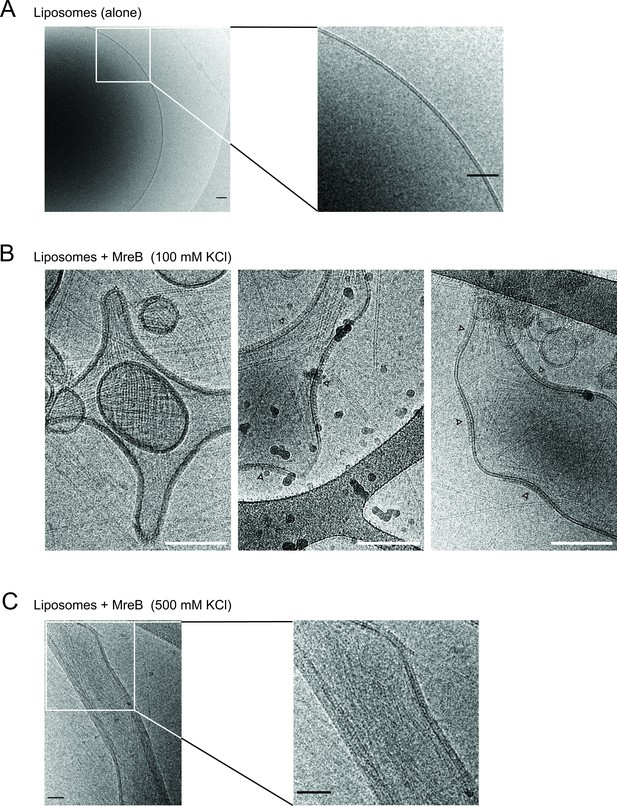
MreBGs polymers coat and distort liposomes.
Cryo-EM micrographs of 0.37 mg/mL liposomes made from E. coli lipid total extract, alone (A) or mixed with 1.34 µM (0,05 mg/mL) purified MreBGs in the presence of 2 mM ATP and 100 mM (B) or 500 mM (C) KCl. MreBGs extensively coated the liposomes and deformed them into negatively curved (B) or faceted (C) vesicles. White arrowheads point to MreB accumulations. White scale bars, 100 nm, black scale bars, 50 nm.
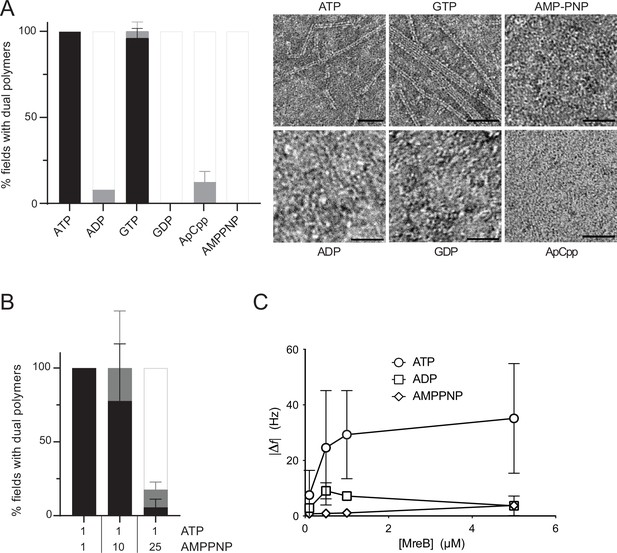
Double protofilaments of Geobacillus MreB efficiently form in the presence of hydrolysable nucleotides.
(A) ATP and GTP promote efficient assembly of MreBGs polymers on a lipid surface. MreB (1.34 µM; 0,05 mg/mL) was incubated in the presence of either ATP, ADP, GTP, GDP, or the non-hydrolysable AMP-PNP or ApCpp (2 mM), on a lipid monolayer. (Left) Quantification of MreBGs pairs of filaments. Polymer formation is expressed as percent of fields containing high (black), low (grey) density of polymers, or no polymers. Values are averages of at least two independent experiments. Error bars are standard deviations. (Right) representative TEM images. Scale bars, 50 nm. (B) AMP-PNP-MreBGs does not form double filaments on a lipid monolayer. AMP-PNP competes with ATP for binding to MreBGs, preventing polymerization. MreBGs was set to polymerize in standard conditions except that 2 mM ATP was replaced by a mix of ATP and AMP-PNP at the indicated concentrations (in mM). Polymer formation was quantified as in (A). Values are average of three independent experiments. Error bars are standard deviations. (C) Adsorption of MreBGs to a supported lipid bilayer (SLB) requires ATP. Frequency changes (|Δf|) in QCM-D experiments measured with varying amount (0.1–5 µM) of MreBGs on SLBs made of DOPC:DOPG 80:20 and in the presence of 2 mM of either ATP, ADP, or AMP-PNP. Values are an average of four independent experiments.
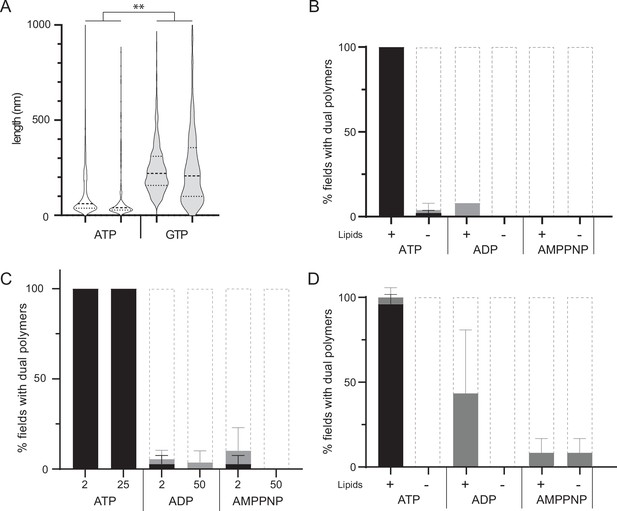
Effect of nucleotides, lipids and protein concentration on the polymerization of MreB.
(A) Size distribution of MreBGs double filaments set to polymerize in the presence of ATP or GTP (2 mM) and 500 mM KCl. Negative stained EM micrographs were analyzed using FIJI and the length of filaments < 1 µm were individually measured. Values are distributions of length of at least 800 filaments per condition from two independent experiments. Median (dashed lines) and quartiles (dotted line) are displayed. The difference between the two conditions is significantly different in a nested T-test (p-value = 0.006). (B) Quantification of MreBGs pairs of protofilaments. MreB was set to polymerize in standard conditions (Tris 20 mM pH7, KCl 100 mM, MgCl2 5 mM, MreB 1.3 µM) in the absence (-) and in the presence (+) of E. coli total lipid extract, and in the presence of either ATP, ADP, or AMP-PNP (2 mM). Polymer formation is expressed as percent of fields containing high (black) or low (grey) density of polymers, or no polymers. Values are average of at least two independent experiments. Error bars are standard deviations. (C) Quantification of MreBGs polymer formation in the presence of high concentrations of nucleotides. MreBGs (1.34 µM) was set to polymerize in the presence of ATP (2 and 25 mM), ADP (2 and 50 mM) or AMP-PNP (2 and 50 mM) in otherwise standard polymerization conditions. Values are average of at least two independent experiments. Error bars are standard deviations. (D) Quantification of MreBGs pairs of protofilaments formed at high MreB concentration. MreB was set to polymerize as in (B) except that concentration was 6.7 µM. Values are average of at least two independent experiments. Error bars are standard deviations.
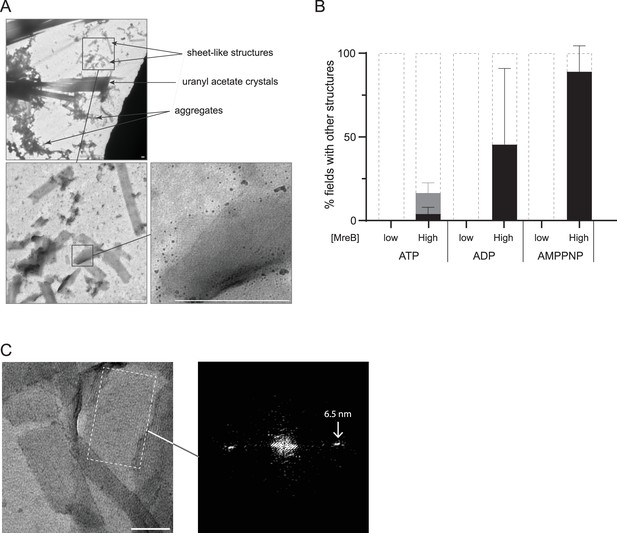
Sheet-like structures formed by MreB in solution.
MreB at high concentration (6.7 µm; 0.25 mg/ml) was set to polymerize in solution in the presence of 2 mM of various nucleotides (ATP, ADP, or AMP-PNP). (A). Negatively stained EM images of typical large sheet-like structures found in solution among aggregates and acetate uranyl crystals. The close zoomed-in shows regular arrangement compatible with MreB sheets of polymers. Scale bars, 300 nm. (B) Quantification of these structures formed in the presence of the different nucleotides. Values are average of two independent experiments. Structure formation is expressed as percent of fields containing high (black) or low (grey) density of structures, or no structures. Error bars are standard deviations. (C) Fourier transform (right panel) of the selected area (dashed rectangle) form the EM image (left panel) containing a large sheet of MreBGs reveals a 6.5 nm repeat between protofilaments in solution.
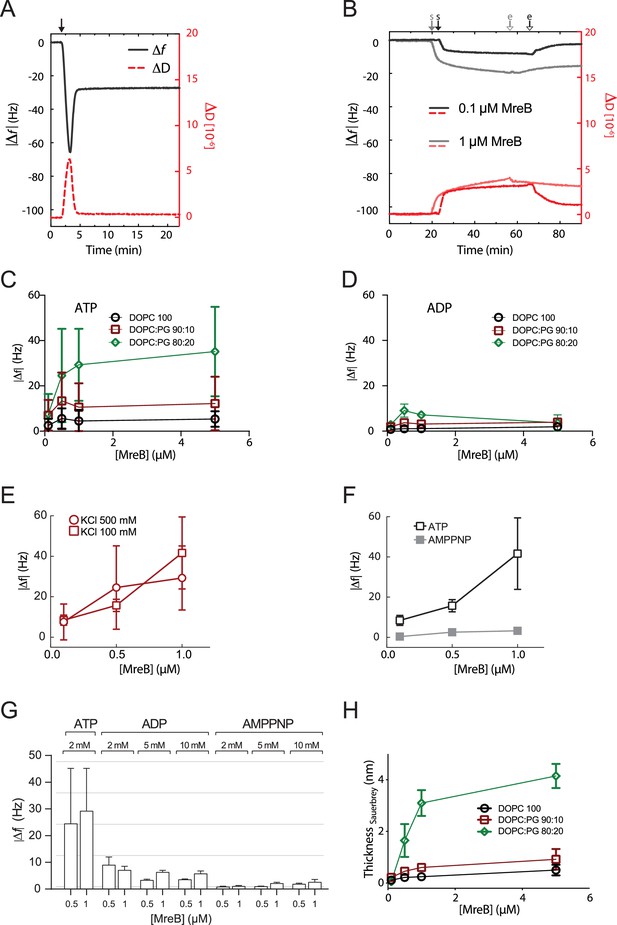
QCM-D experiments of MreBGs adsorption on supported lipid bilayers.
(A) Lipid bilayer formation on crystal with SiO2 layers. Supported lipid bilayers (SLBs) are formed by spontaneous rupture of adsorbed liposomes as indicated by frequency shifts (Δf, black solid lines) and dissipation shifts (ΔD, red dotted lines). Exemplarily shown is the formation of DOPC:DOPG 90:10 SLB from DOPC:DOPG 90:10 liposomes. The solid black arrow indicates the addition of liposomes to the SiO2 surface. (B) Subsequently, 1 µM (light colors) or 0.1 µM (dark colors) wild-type MreBGs are added to SLBs. The closed and open arrows indicate the start (s) and end (e) of the protein addition (followed by rinsing with polymerization buffer), respectively. (C, D) MreBGs binds to SLBs of different lipid compositions (SLBs from DOPC:DOPG: 80:20, green; 90:10, red; 100:0, black) in the presence of ATP (C) but not in the presence of ADP (D). Error bars are standard deviations of n=3. (E) MreBGs binds to SLBs (DOPC:DOPG 80:20) similarly in the presence of 100 mM (square) or 500 mM (circle) KCl. Error bars are standard deviations of n=4. (F) MreBGs binds to SLBs (DOPC:DOPG 80:20) in the presence of ATP but not AMP-PNP at low ionic strength (100 mM KCl). Error bars are standard deviations of n=4. (G) High concentrations of ADP and AMP-PNP do not support adsorption of MreBGs to SLBs. Frequency changes were measured for the binding of purified MreBGs to SLBs in QCM-D experiments. Incubations were performed in polymerization buffer containing 2 mM ATP or the indicated concentrations of ADP or AMP-PNP. SLBs consisted of DOPC:DOPG 80:20. Values are averages of at least two independent experiments. Error bars represent standard deviations. (H) Thickness of the MreB protein layer on the SLBs calculated with the Sauerbrey equation. Error bars are standard deviations of n=3.
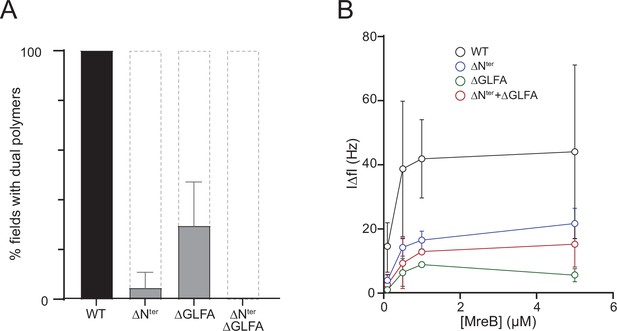
in theThe N-terminus and the α2β7 hydrophobic loop of MreBGs promote membrane binding and polymerization on a lipid surface.
(A) Both the hydrophobic α2-β7 loop and the N-terminus sequence of MreBGs are required for efficient polymerization on a lipid monolayer. Frequency and density of polymer formation in high salt (500 mM KCl) polymerization conditions, observed on negatively stained TEM images for the wild type (WT) and the mutants of the α2-β7 loop (ΔGLFA), the N-terminus (ΔNter) or both domains (ΔNter+ ΔGLFA) of MreBGs. Images were categorized based on the absence or the presence of low or high density of polymers. Values are the average of two independent experiments. Error bars are standard deviations. (B) The α2-β7 loop and the N-terminus domains of MreBGs enhance its adsorption to supported lipid bilayers. Frequency change (IΔfI) measured for the binding of various concentrations (0.1–5 µM) of purified wild-type (WT) and mutant forms of MreBGs to SLBs. Incubations were performed in polymerization buffer containing 500 mM KCl and 2 mM ATP. SLBs contained an 80:20 molecular ratio of DOPC:DOPG. Values are an average of at least two independent experiments.
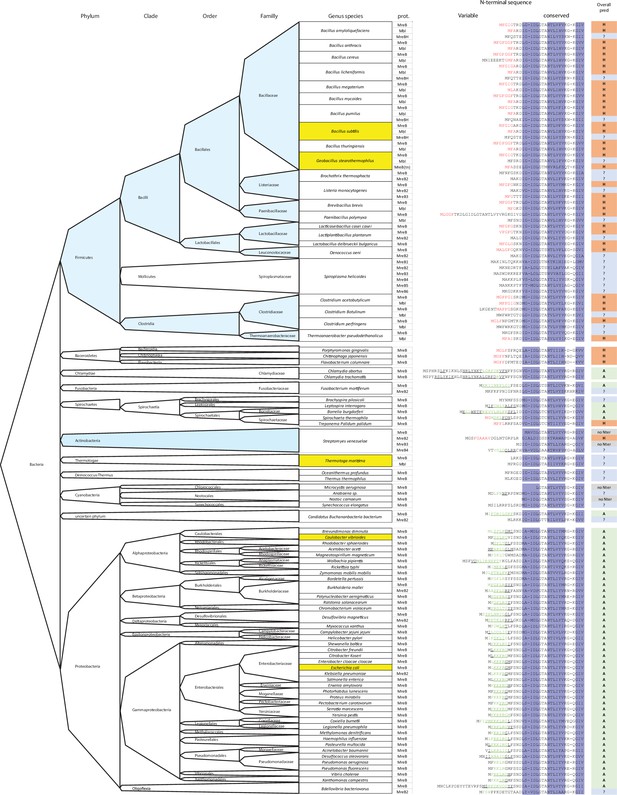
Distribution of N-terminal amphipathic helices and hydrophobic sequences on MreBs proteins in the bacterial kingdom.
N-terminal sequences of MreB proteins from selected species across the bacterial kingdom were aligned using Clustal-Ω. The N-terminal sequences were analyzed for the presence of a putative α-helix (underscore) and/or amphipaticity (green) using the Amphipaseek tool at Prabi (https://npsa-prabi.ibcp.fr/), and for the presence of hydrophobic sequences (red). Dark blue columns mark the β-sheets 1, 2, and 3 according to MreBGs structure (Figure 1—figure supplement 1). The prediction for putative anchoring structures is summarized in the right column: A (green), amphipathic helix; H (red), hydrophobic sequence; ? (blue), unknown. Species of interest aligned in Figure 1—figure supplement 1 are highlighted in yellow. Gram-positive bacteria (with low and high GC %) are colored in light blue.
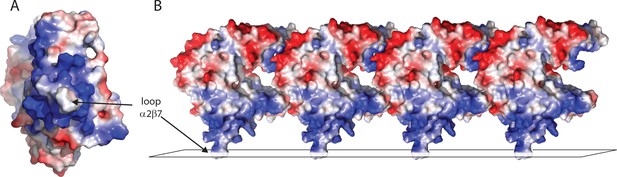
The protruding hydrophobic subdomain IA of MreBGs is surrounded by a positively charged cluster.
(A). Electrostatic surface potential of an MreBGS monomer (PDB ID 7ZPT), facing the domains I (IA, down and IB, up) side. The view corresponds to a 90° rotation along a vertical axis compared to the monomer shown in Figure 1A. Positively charged regions are shown in blue, negatively charged areas in red and hydrophobic patches in white. (B). Electrostatic surface potential of an MreBGS protofilament. The view corresponds to a 90° clockwise rotation compared to the protein in Figure 1A. The loop α2-β7 is shown embedded into a lipid layer (black parallelogram).
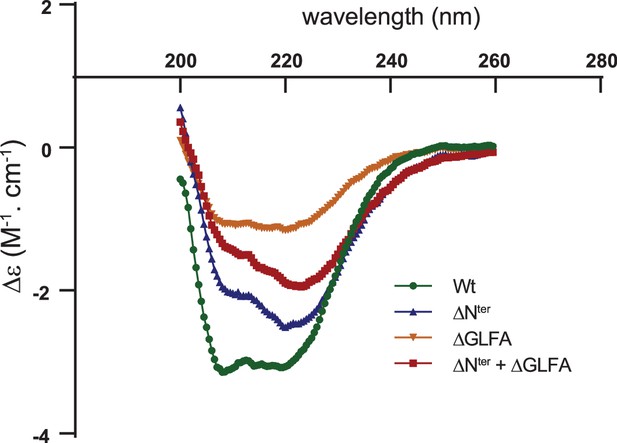
Circular dichroism (CD) spectra showing similar folding of the wild-type and the deletion mutants of the α2-β7 loop (ΔGLFA), the N-terminus (ΔNter) or both domains (ΔNter + ΔGLFA) of recombinant MreBGs.
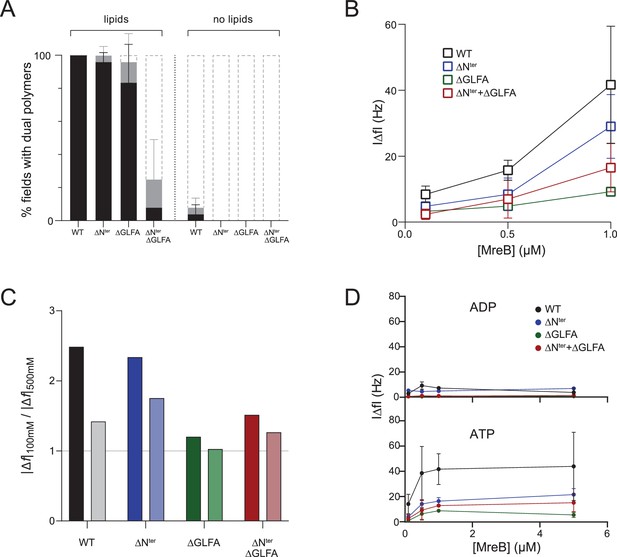
The amino-terminal sequence, the GLFA residues of the α2-β7 hydrophobic loop and electrostatic interactions mediate binding of MreB to a lipid surface.
(A) Quantification of dual protofilament formation by MreBGs wild type (WT) and the mutants of the N-terminus (ΔNter), the α2-β7 loop (ΔGLFA), or both domains (ΔNter+ ΔGLFA), in the presence of lipids and in solution. MreBGs was set to polymerize in standard polymerization conditions (ATP 2 mM, KCl 100 mM) with or without lipid total extract from E. coli. Following polymerization, samples incubated with or without lipids were mounted onto untreated or glow-discharged grids, respectively. Polymer formation is expressed as percent of fields containing high (black) or low (grey) density of polymers, or no polymers. Values are average of two independent experiments. Error bars are standard deviations. (B,C,D) Adsorption of the wild-type (WT, black) and the mutants of the N-terminus (ΔNter; blue), the α2-β7 (ΔGLFA, green), or both domains (ΔNter+ ΔGLFA; red) of MreBGs to supported lipid bilayers made of DOPC:DOPG 80:20. (B) Frequency changes (|Δf|) in QCM-D experiments measured with varying amounts (0.1–1 µM) of each MreBGs in the presence of 2 mM ATP and 100 mM KCl. Error bars are standard deviations of n=2. (C) Ratio of frequency changes (|Δf|) in QCM-D experiments measured in the presence of low (100 mM KCl) and high (500 mM KCl) ionic strength, and in the presence of 2 mM ATP, 1 µM of each MreBGs and SLBs made of DOPC:DOPG 80:20 (light colors) or 90:10 (dark colors). (D) Frequency changes (|Δf|) in QCM-D experiments measured with varying amounts (0.1–5 µM) of each MreBGs in the presence of 2 mM ATP or ADP, and 500 mM KCl. Error bars are standard deviations of n=2.
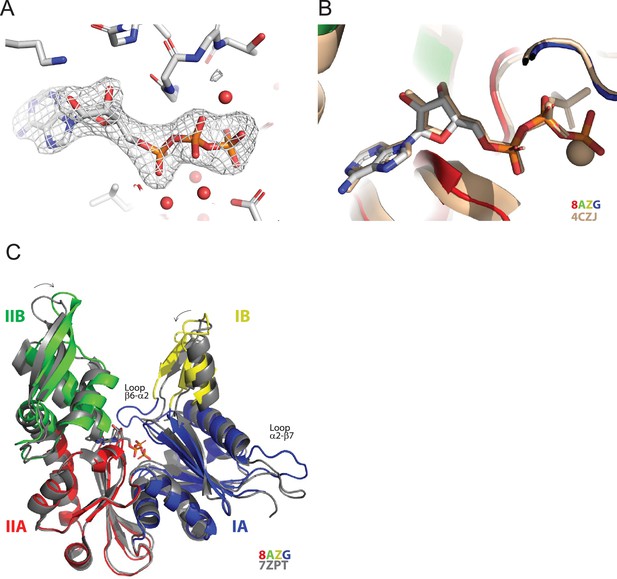
Crystal structure of MreBGs bound to ATP.
(A) Electron density of the ATP molecule bound to MreBGs. The Fo-Fc omit map calculated by omitting the nucleotide from the model is shown as a grey mesh contoured at 4σ. The nucleotide and the surrounding protein residues are shown as sticks colored by atom type (C in grey, N in blue, O in red and P in orange). Water molecules are shown as red spheres. (B) Comparison of the ATP-bound MreBGs with MreBCc bound to AMP-PNP/Mg++. Close view of the superimposed nucleotide-binding sites. MreBGs (PDB ID 8AZG) and MreBCc (PDB ID 4CZJ) are shown as cartoon colored by subdomains and in beige, respectively. The nucleotide molecules are shown as sticks. The bound ATP is colored by atom type and the bound AMP-PNP/Mg++ is shown in beige. (C) Comparison of the ATP-bound form with the apo form of MreBGs. One subunit of ATP-bound MreBGs (PDB ID 8AZG), colored by subdomains, is superimposed with the apo form of the protein (PDB ID 7ZPT), colored in gray. The bound nucleotide is shown as sticks colored by atom type. Loop β6-α2, stabilized by the presence of the bound nucleotide, is labeled, as well as loop α2-β7 and the N-terminus, which display alternative conformations.
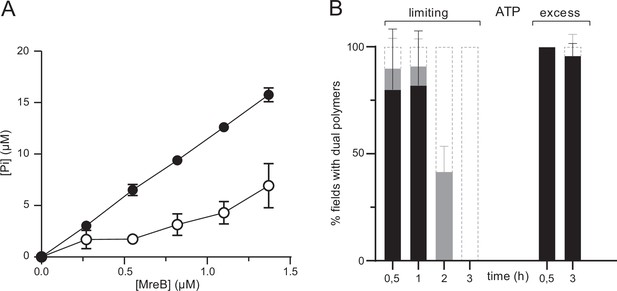
ATPase activity of MreBGs.
(A) The ATPase activity of MreBGs is stimulated in the presence of lipids. ATPase activity, measured by monitoring inorganic phosphate (Pi) release, of MreBGs at different concentrations (0.26–1.34 µM) in the presence of 0.5 mM ATP, 100 mM KCl, and in the presence (filled circles) or absence (empty circles) of 0.05 mg/mL liposomes, after 1 hr incubation at 53 °C. Values are averages of at least two independent experiments. Error bars are standard deviations. (B) Kinetics of MreBGs polymer formation in the presence of limiting ATP. MreBGs was set to polymerize in the presence of standard (2 mM; ‘excess’) or low (13 µM; ‘limiting’) concentration of ATP. Samples were prepared for TEM observation at different incubation times, up to 3 hr. Polymer formation is expressed as percent of fields containing high (black) or low (grey) density of polymers, or no polymers. Values are averages of two independent experiments. Error bars are standard deviations.
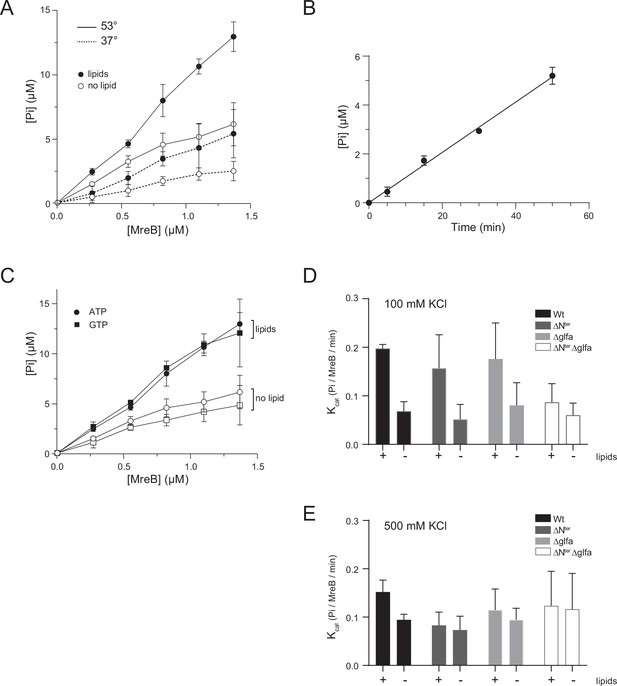
Effects of lipids, temperature and deletions on the hydrolytic activity of MreB.
(A) The ATPase activity of MreBGs is stimulated at high temperature. Release of Pi detected by malachite green assay for a range of MreBGs concentrations (0.26–1.34 µM) in the presence or absence of 0.05 mg/mL liposomes in polymerization buffer (0.5 mM ATP, 100 mM KCl) after 1 h incubation at 53 °C or 37 °C. Error bars are standard deviations of at least two independent measurements. (B) Kinetics of ATP hydrolysis detected via Pi release in the presence of 1.34 µM MreBGs, 0.5 mM ATP and 0.05 mg/mL liposomes at 53 °C. The line is a simple linear regression fit (goodness of fit R²=0.9910). Values are averages of two independent experiments. Error bars are standard deviations. (C) MreB shows a similar hydrolytic activity toward GTP and ATP and is stimulated in the presence of lipids. Release of Pi detected by malachite green assay in the presence of ATP or GTP (0.5 mM), after 1 hr incubation at 53 °C in the presence or absence of 0.05 mg/mL liposomes for a range of MreBGs concentrations (0.27–1.34 µM) at 500 mM KCl. Error bars are standard deviations of at least two independent measurements. (D, E) The ATPase activity of the MreBGs deletion mutants depends on the presence of lipids and KCl concentration. Release of Pi were detected after 1 hr incubation at 53 °C, by malachite green assay for the wild type (Wt, black) and the mutants of the N-terminus (ΔNter; dark grey) the α2-β7 (ΔGLFA, light grey), or both domains (ΔNter+ ΔGLFA; white) of MreBGs, set to polymerize in standard conditions (1.34 µM MreBGs, 0.5 mM ATP) in the presence or absence of 0.05 mg/mL liposomes, at low (100 mM; D) or high (500 mM; E) KCl concentration. Displayed are kcat and error bars are standard deviations of at least two independent measurements.

Model for ATP-driven MreBGs membrane binding and polymerization into pairs of filaments.
ATP hydrolysis stimulates MreBGs adsorption to lipids, possibly by promoting a conformational change that renders the hydrophobic α2-β7 loop and N-terminal protruding region prone for insertion into the membrane, either as a monomeric (1) or a nucleated (2) MreBGs form (dotted lines). Lipid binding would trigger formation of double-protofilaments on the lipid surface, which in turn would promote Pi release. Membrane-associated pairs of filaments may be mostly in the ADP-Pi form, and Pi release may destabilize them and ultimately promote disassembly.
Additional files
-
Supplementary file 1
Data-collection and refinement statistics.
- https://cdn.elifesciences.org/articles/84505/elife-84505-supp1-v2.xlsx
-
Supplementary file 2
List of polymerization conditions assayed.
- https://cdn.elifesciences.org/articles/84505/elife-84505-supp2-v2.xlsx
-
Supplementary file 3
List of proteins used in this study.
- https://cdn.elifesciences.org/articles/84505/elife-84505-supp3-v2.xlsx
-
Supplementary file 4
List of strains used in this study.
- https://cdn.elifesciences.org/articles/84505/elife-84505-supp4-v2.xlsx
-
Supplementary file 5
List of oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/84505/elife-84505-supp5-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84505/elife-84505-mdarchecklist1-v2.docx






