Sexual dimorphic regulation of recombination by the synaptonemal complex in C. elegans
Figures
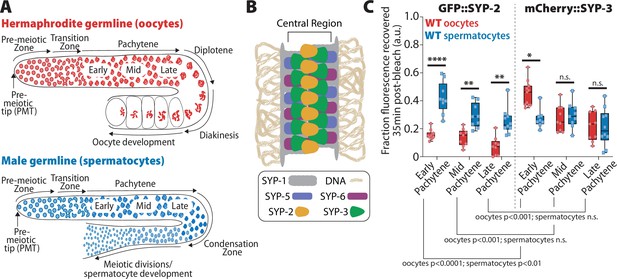
SYP-2 and SYP-3 dynamics are sexually dimorphic.
(A) Diagrams of hermaphrodite (top, red) and male (bottom, blue) germlines with developing oocytes and spermatocytes, respectively. The stages of the germlines are labeled starting at the pre-meiotic tip (PMT) and ending at the meiotic divisions. Nuclei proliferate at the distal end of the germline (pre-meiotic tip) and physically move proximally as they proceed into the stages of meiosis: the transition zone (leptotene/zygotene), pachytene, diplotene, and diakinesis (in spermatocytes diplotene/diakinesis is termed the condensation zone). At the end of prophase I, oocyte nuclei arrest at diakinesis until they are fertilized, but spermatocytes rapidly complete the meiosis I and meiosis II divisions to generate mature sperm. (B) Diagram of the synaptonemal complex showing the positions of SYP-1 (gray), SYP-2 (yellow), SYP-3 (green), SYP-5 (blue-purple), and SYP-6 (red-purple) within the central region of the complex. (C) Quantification of the fraction of fluorescence recovered 35 min after photobleaching a small region of either GFP::SYP-2 (left) or mCherry::SYP-3 (right) in both oocytes (red)and spermatocytes (blue) (a.u. = arbitrary units). All statistics are multiple comparisons Bonferroni–Dunn adjusted Mann–Whitney U test and unless the p-value is indicated the asterisk indicates number of significant digits from p=0.05 (n.s. = not significant). Oocyte data with GFP::SYP-2 is from 10 nuclei (early), 9 nuclei (mid), and 9 nuclei (late), and with mCherry::SYP-3 is from 11 nuclei (early), 9 nuclei (mid), and 9 nuclei (late). Spermatocyte data with GFP::SYP-2 is from 9 nuclei (early), 9 nuclei (mid), and 10 nuclei (late), and with mCherry::SYP-3 is from 8 nuclei (early), 9 nuclei (mid), and 10 nuclei (late).
-
Figure 1—source data 1
The normalized fluorescence recovery for the fluorescent recovery after photobleaching (FRAP) data for each nucleus analyzed with GFP::SYP-2 and mCherry::SYP-3 at early, mid, and late pachytene in both sexes (Figure 1C).
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig1-data1-v2.xlsx
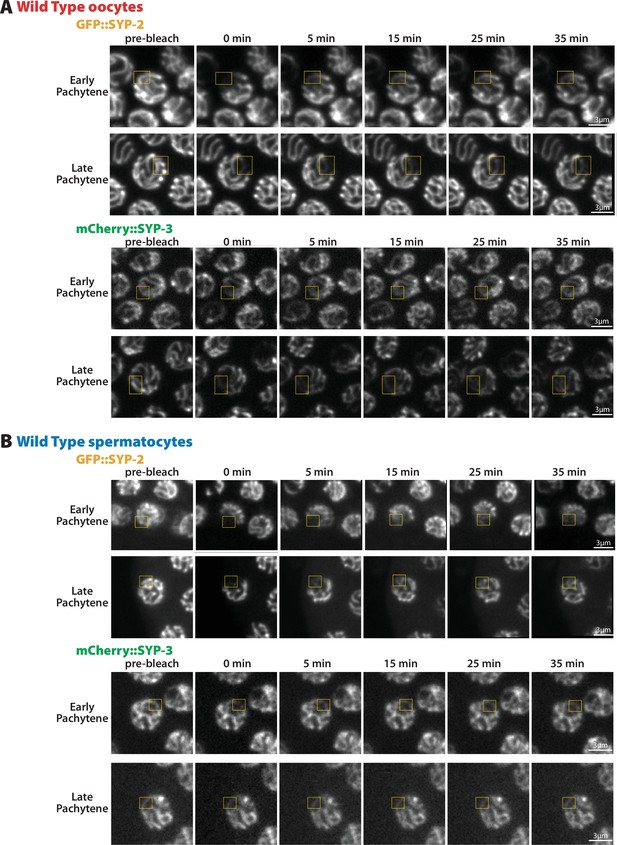
Montages of fluorescent recovery after photobleaching (FRAP) from oocytes and spermatocytes.
Representative image montages of GFP::SYP-2 and mCherry::SYP-3 FRAP from oocytes (A) and spermatocytes (B) in early and late pachytene. Yellow box indicates the bleached region of each nucleus.
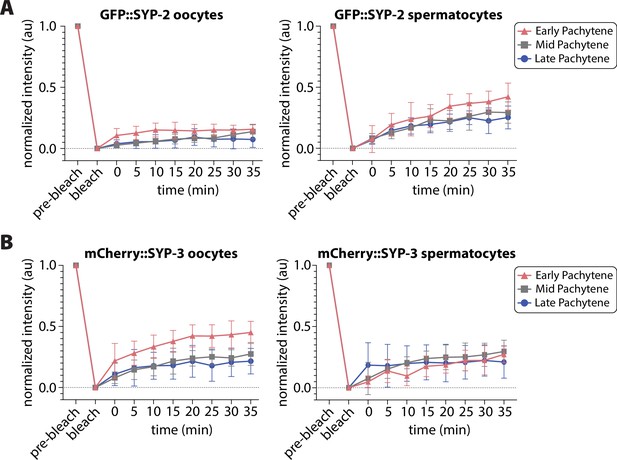
Fluorescent recovery after photobleaching (FRAP) recovery curves from oocytes and spermatocytes.
Normalized recovery curves from FRAP analysis of nuclei with GFP::SYP-2 (A) or mCherry::SYP-3 (B) from oocytes (left plot) and spermatocytes (right plot) (see ‘Methods’). Nuclei were analyzed at early pachytene (pink triangles), mid pachytene (gray squares), and late pachytene (blue circles). Error bars indicate standard deviation.
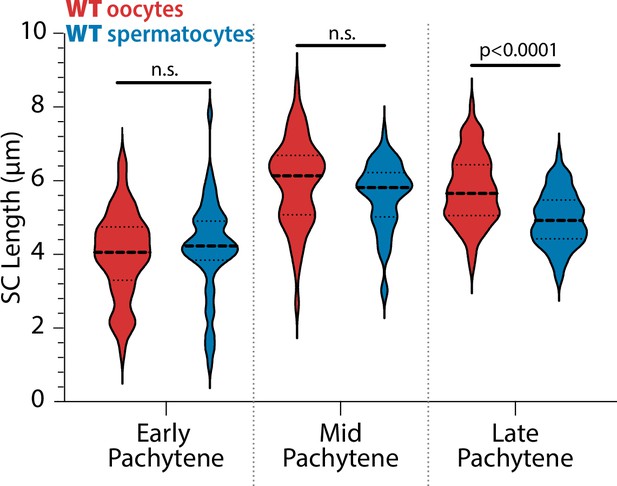
Synaptonemal complex (SC) lengths are not different between the sexes in early and mid pachytene.
Quantification of the measurement of SYP-1 length on each chromosome between oocytes (red) and spermatocytes (blue) throughout pachytene. Only late pachytene display significant differences in SC lengths (Mann–Whitney U test; n.s. = not significant).
-
Figure 1—figure supplement 3—source data 1
The SYP-1 length measurements for each chromosome in wild-type oocytes and spermatocytes.
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig1-figsupp3-data1-v2.xlsx
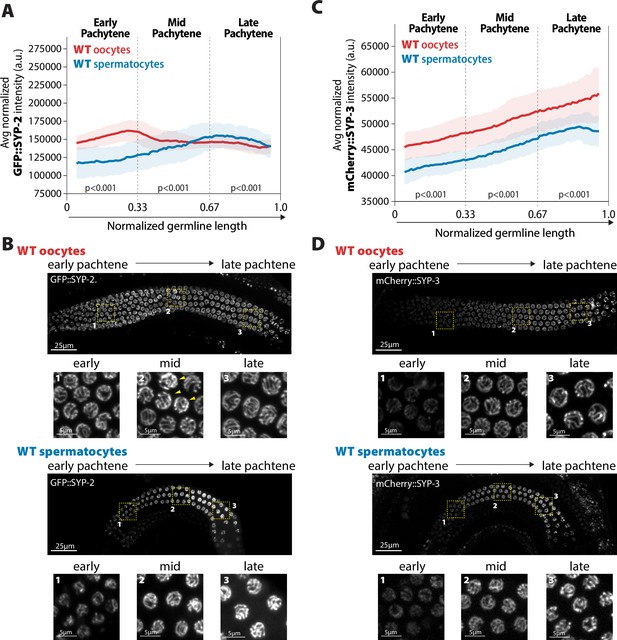
Accumulation of SYP-2 and SYP-3 in the synaptonemal complex (SC) is sexually dimorphic.
(A,C) Quantification of the mean intensity of GFP::SYP-2 (A) or mCherry::SYP-3 (C) per nucleus normalized by the volume of each nucleus (see ‘Methods’) throughout pachytene for wild-type (WT) oocytes (red, pale red band is the standard deviation) and spermatocytes (blue, pale blue band is the standard deviation). p-Values on the plot are comparisons between oocytes and spermatocytes for each region of pachytene using Mann–Whitney U tests. (B, D) Represented images of GFP::SYP-2 (B) or mCherry::SYP-3 (D) in WT hermaphrodite (top, oocytes) and male (bottom, spermatocytes) germlines. Germlines are oriented with the start of pachytene on the left and meiotic progression continues to the right. Yellow boxes identify the regions enlarged in each image below to show representatives of early, mid, and late pachytene. Arrowheads indicate GFP::SYP-2 bright nuclei in mid pachytene. The intensity adjustments are the same for both GFP::SYP-2 germlines and mCherry::SYP-3 germlines, respectively. n values for the number of germlines and nuclei can be found in Figure 2—source data 2.
-
Figure 2—source data 1
Raw sum intensity and normalized sum intensity per nucleus for GFP::SYP-2, mCherry::SYP-3 in both sexes and wild-type genotypes (Figure 2A,C).
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig2-data1-v2.xlsx
-
Figure 2—source data 2
SC intensity n values for nuclei and germlines scored.
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig2-data2-v2.docx
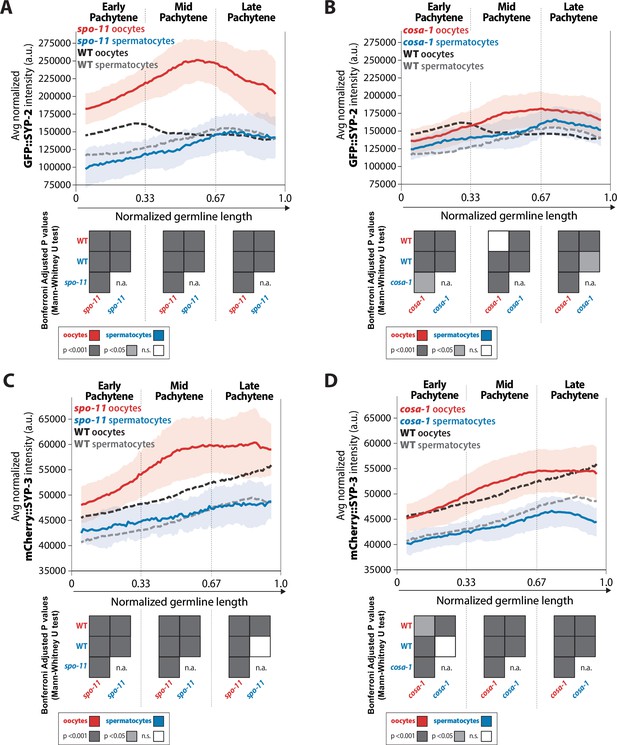
Recombination influences the incorporation of SYP-2 and SYP-3 in the synaptonemal complex (SC) differently in each sex.
(A, B) Quantification of the mean intensity of GFP::SYP-2 per nucleus normalized by the volume of each nucleus (see ‘Methods’) throughout pachytene for spo-11 (A) and cosa-1 (B). (C, D) Quantification of the mean intensity of mCherry::SYP-3 per nucleus normalized by the volume of each nucleus (see ‘Methods’) throughout pachytene for spo-11 (C) and cosa-1 (D). Oocytes are shown in red with the standard deviation shown as a pale red band and spermatocytes are shown in blue with the standard deviation shown as a pale blue band. The mean intensity of GFP::SYP-2 (A, B) or mCherry::SYP-3 (C, D) for wild-type (WT) oocytes (black) and WT spermatocytes (gray) are shown as dashed lines. Heat maps below each pachytene region show the Bonferroni adjusted p-values from Mann–Whitney U tests, with dark gray indicating p<0.001, light gray indicating p<0.05, and white indicating not significant (n.s.). The self-comparison between spermatocyte spo-11 or cosa-1 mutants was not determined (n.a.). n values for the number of germlines and nuclei can be found in Figure 3—source data 2.
-
Figure 3—source data 1
Raw sum intensity and normalized sum intensity per nucleus for GFP::SYP-2 and mCherry::SYP-3 in both sexes and all genotypes analyzed in Figure 3.
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig3-data1-v2.xlsx
-
Figure 3—source data 2
SC intensity n values for nuclei and germlines scored.
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig3-data2-v2.docx
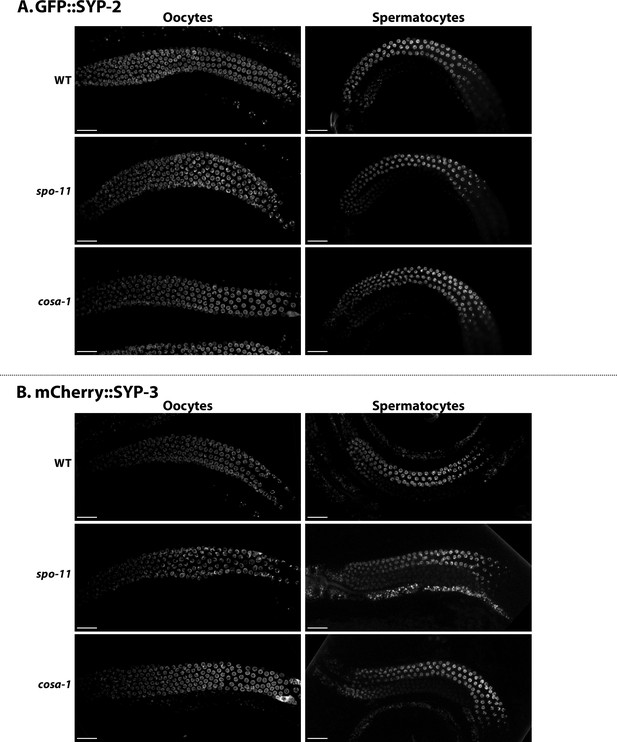
Representative images of spo-11 and cosa-1 germlines with GFP::SYP-2 (A) and mCherry::SYP-3 (B) in oocytes and spermatocytes.
Scale bar represents 25 µm.
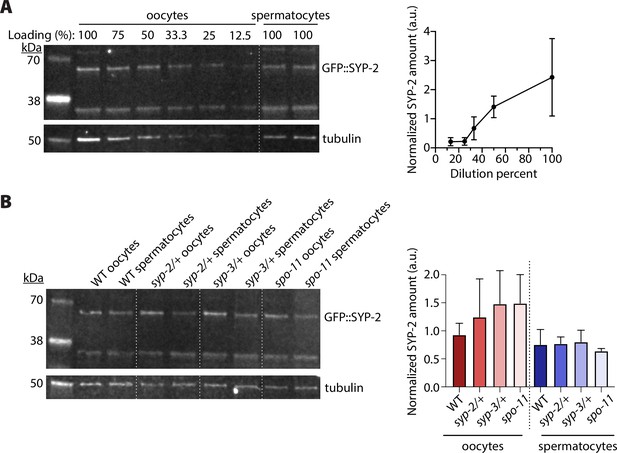
Western blot analysis of SYP-2 protein levels in oocytes and spermatocytes.
(A) Dilution series blot with adult hermaphrodite samples to determine the level of dilution necessary to match the oocyte GFP::SYP-2 amounts to the spermatocyte GFP::SYP-2 amounts (see ‘Methods’). The loading percent above each well is the percent of hermaphrodite sample and diluted with sample buffer prior to loading. 100% is undiluted sample for both hermaphrodite (oocytes) and male (spermatocytes) samples. To the right of the blot is the quantification of the GFP::SYP-2 band normalized to the averaged undiluted spermatocyte SYP-2 amounts. From the plot, we determined that a 40% dilution of the hermaphrodite samples was necessary to be equivalent to an undiluted spermatocyte sample from male worms. (B) Blot of GFP::SYP-2 levels in wild-type (WT) and mutant oocytes and spermatocytes. To the right of the blot is quantification of the GFP::SYP-2 band normalized to tubulin loading control. Three independent western blots were quantified with 100 worm samples loaded per genotype, and the error bars represent the standard deviation.
-
Figure 3—figure supplement 2—source data 1
Original western blot images, raw SYP-2 band intensity measurements, and normalized SYP-2 amounts in both sexes and all genotypes analyzed in this manuscript.
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig3-figsupp2-data1-v2.zip
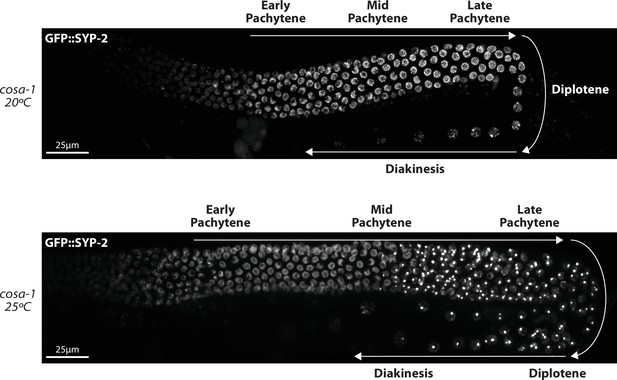
GFP::SYP-2 aggregates in cosa-1 at 25°C during mid and late pachytene.
Representative hermaphrodite germline images of GFP::SYP-2 in cosa-1 mutants from worms kept at 20°C and worms upshifted to 25°C for 18–24 hr prior to imaging. White arrows indicate the direction oocyte nuclei are moving within the germline, and the stages of prophase are labeled.
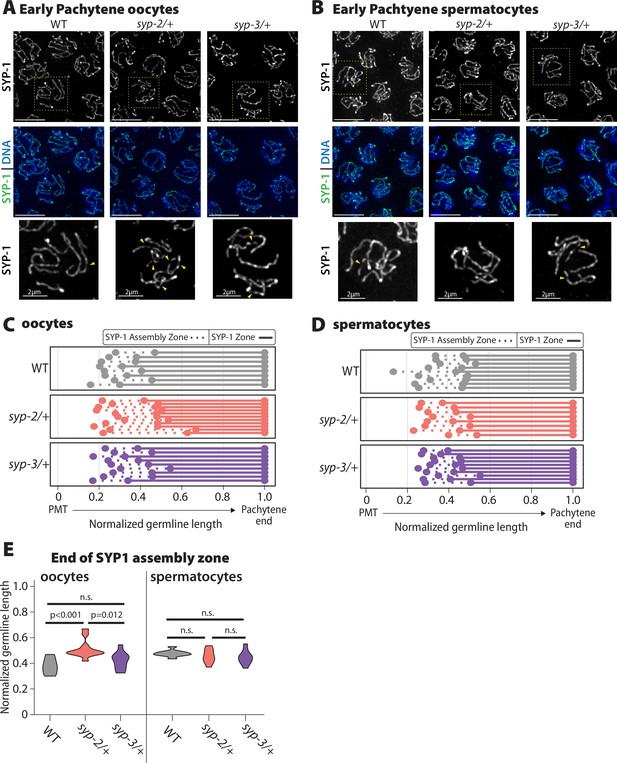
Oocyte SYP-1 assembly is uniquely sensitive to SYP-2 dosage.
(A, B) Representative images early pachytene stained for SYP-1 in oocytes (A) and spermatocytes (B) from wild-type (WT), syp-2/+, and syp-3/+. Yellow dashed box shows the nucleus that is enlarged below each merged image. Yellow arrowheads indicate the regions where the SYP-1 signal is broken/not continuous. Scale bar represents 5 µm (C, D) Measurement of the relative length of the SYP-1 assembly zone in oocytes (C) and spermatocytes (D) from the pre-meiotic tip (PMT) to the end of pachytene in WT (gray), syp-2/+ (pink), and syp-3/+ (purple). Dashed lines represent the SC assembly zone and solid lines represent fully assembled synaptonemal complex (SC). (E) Quantification of the end of the SYP1 assembly zone in WT (gray), syp-2/+ (pink), and syp-3/+ (purple) in oocytes (left) and spermatocytes (right). All statistics are Bonferroni adjusted p values from Mann–Whitney U tests (n.s. = not significant). Oocyte data is from 8 WT germlines, 10 syp-2/+ germlines, and 9 syp-3/+ germlines. Spermatocyte data is from 9 WT germlines, 8 syp-2/+ germlines, and 10 syp-3/+ germlines.
-
Figure 4—source data 1
Distance data from germlines where the length of SYP-1 assembly was determined (Figure 4C–E).
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig4-data1-v2.xlsx
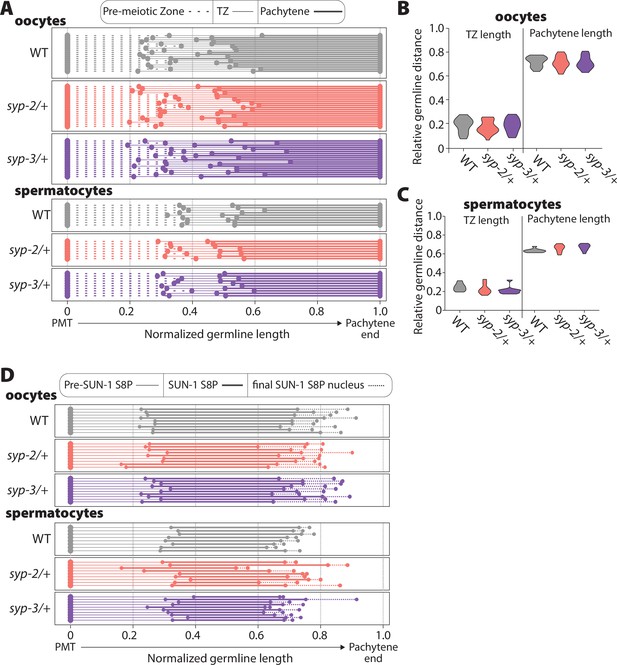
Transition zone length and SUN-1 phosphorylation zone are unaltered by SYP dosage.
(A) Measurement of the pre-meiotic zone length (dashed line), transition zone length (thin solid line), and pachytene length (thick solid line) in wild-type (WT) (gray), syp-2/+ (pink), and syp-3/+ (purple) based on DAPI morphology. All germline lengths have been normalized from the pre-meiotic tip (PMT) to the end of pachytene in both oocytes and spermatocytes. Each line represents an individual germline. Overall, neither syp-2/+ nor syp-3/+ displayed altered transition zone length. (B, C) Violin plots of the transition zone (TZ) length and pachytene length in WT (gray), syp-2/+ (pink), and syp-3/+ (purple) in oocytes (B) and spermatocytes (C). There are no statistical differences in transition zone length or pachytene length (Mann–Whitney U test). Oocyte data is from 18 WT germlines, 19 syp-2/+ germlines, and 18 syp-3/+ germlines. Spermatocyte data is from 9 WT germlines, 8 syp-2/+ germlines, and 10 syp-3/+ germlines. (D) Measurement of SUN-1 S8 phosphorylation (S8P) zone length in WT (gray), syp-2/+ (pink), and syp-3/+ (purple) oocytes and spermatocytes. Thin solid line indicates the distance from PMT to start of SUN-1 S8P; a thicker solid line indicates the distance from start to end of most nuclei with SUN-1 S8P staining; and the dashed line indicates the distance to the final nucleus in the germline with SUN-1 S8P staining (see ‘Methods’). Each line represents an individual germline. Overall, neither syp-2/+ nor syp-3/+ displayed altered the germline region with SUN-1 S8P.
-
Figure 4—figure supplement 1—source data 1
Distance data for the length of meiosis stages and SUN-1 S8P.
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig4-figsupp1-data1-v2.zip
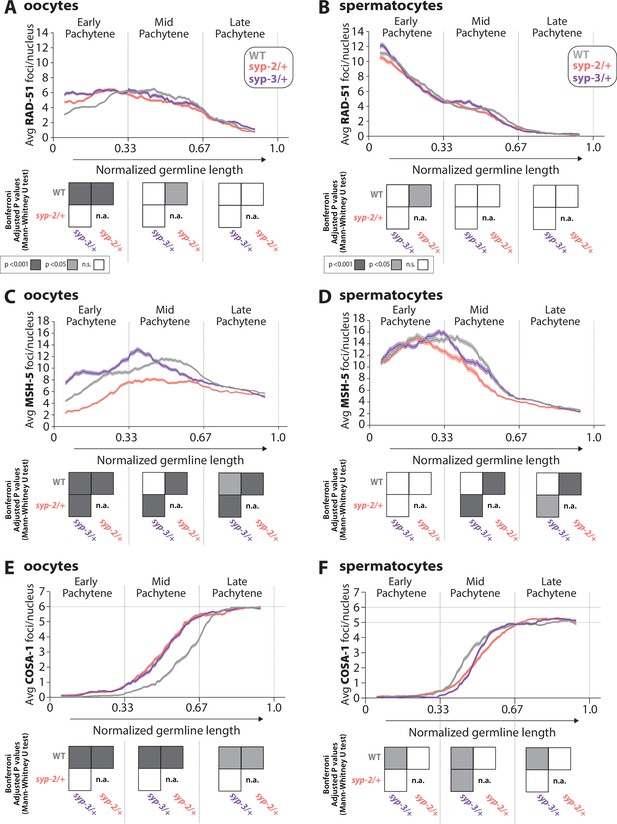
Sex-specific regulation of recombination by SYP-2 and SYP-3 dosage.
(A, B) Quantification of the average number of RAD-51 foci per nucleus throughout pachytene in wild-type (WT, gray), syp-2/+ (pink), and syp-3/+ (purple) from oocytes (A) and spermatocytes (B). (C, D) Quantification of the average number of MSH-5 foci per nucleus throughout pachytene in WT (gray), syp-2/+ (pink), and syp-3/+ (purple) from oocytes (C) and spermatocytes (D). (E, F) Quantification of the average number of COSA-1 foci per nucleus throughout pachytene in WT (gray), syp-2/+ (pink), and syp-3/+ (purple) from oocytes (E) and spermatocytes (F). Heat maps below each pachytene region show the Bonferroni adjusted p-values from Mann–Whitney U tests, with dark gray indicating p<0.001, light gray indicating p<0.05, and white indicating not significant (n.s.). The self-comparison between syp-2/+ was not determined (n.a.). n values for the number of germlines and nuclei can be found in Figure 5—source data 2.
-
Figure 5—source data 1
RAD-51, MSH-5, COSA-1 foci per nucleus counts for wild-type, syp-2/+ (syp2het), and syp-3/+ (syp3het).
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig5-data1-v2.xlsx
-
Figure 5—source data 2
RAD-51, MSH-5, and COSA-1 n values for nuclei and germlines scored.
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig5-data2-v2.docx
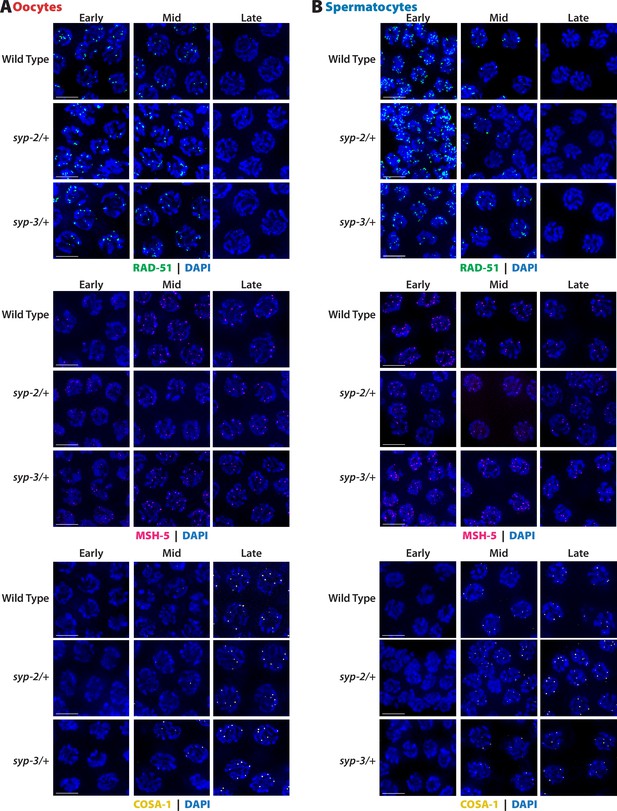
Representative images of the oocytes (A) and spermatocytes (B) quantification in Figure 5.
Scale bar represents 5 µm.
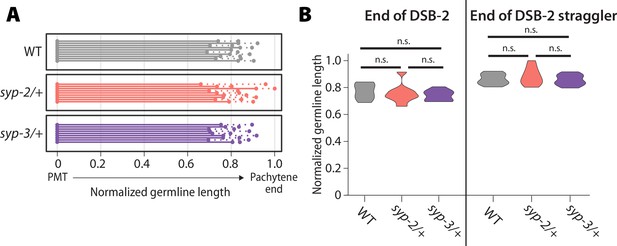
DSB-2 zone is unaltered in oocytes when SYP dosage is reduced.
(A) Measurement of the length of DSB-2 staining (solid line) and the final DSB-2 stained straggler nucleus (dashed line) in wild-type (WT) (gray), syp-2/+ (pink), and syp-3/+ (purple). All germline lengths have been normalized from the pre-meiotic tip (PMT) to the end of pachytene in oocytes. Each line represents an individual germline. Overall, the zone of DSB-2 staining is largely unaltered in syp-2/+ and syp-3/+. (B) Violin plots of the ending of the DSB-2 zone (left) and the ending of the DSB-2 straggler nuclei (right) in WT (gray), syp-2/+ (pink), and syp-3/+ (purple). There are no statistical differences at the end of the DSB-2 zone or the end of the DSB-2 straggler nuclei (Mann–Whitney U test). Data is from 10 WT germlines, 9 syp-2/+ germlines, and 9 syp-3/+ germlines.
-
Figure 5—figure supplement 2—source data 1
Distance data from germlines where the length of DSB-2 zone was determined.
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig5-figsupp2-data1-v2.xlsx
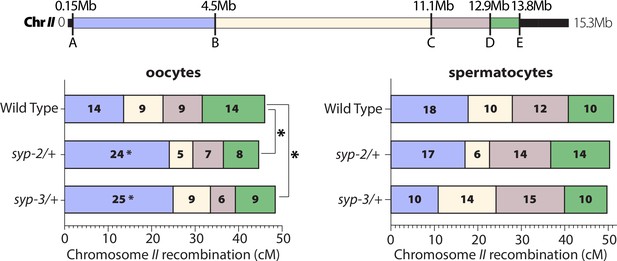
SYP dosage influences the crossover landscape in only oocytes.
Recombination SNP mapping of chromosome II in WT, syp-2/+, and syp-3/+ from oocytes (left) and spermatocytes (right) (see ‘Methods’ for details). A diagram of the 15.3 Mb chromosome II shows the megabase location of each SNP assayed (A–E), and the colored boxes between each SNP show the intervals where crossovers were assessed. The map length (cM) is indicated in each crossover interval. The asterisks next to the map lengths indicate significance based on Fisher’s exact tests compared between the mutants and wild-type (syp2/+ p=0.0449; syp3/+ p=0.0170). The asterisks outside the bars indicate significance based on chi-squared tests between mutants and wild-type (syp2/+ p=0.0343; syp3/+ p=0.0333). The worm counts for these plots can be found in Table 1.
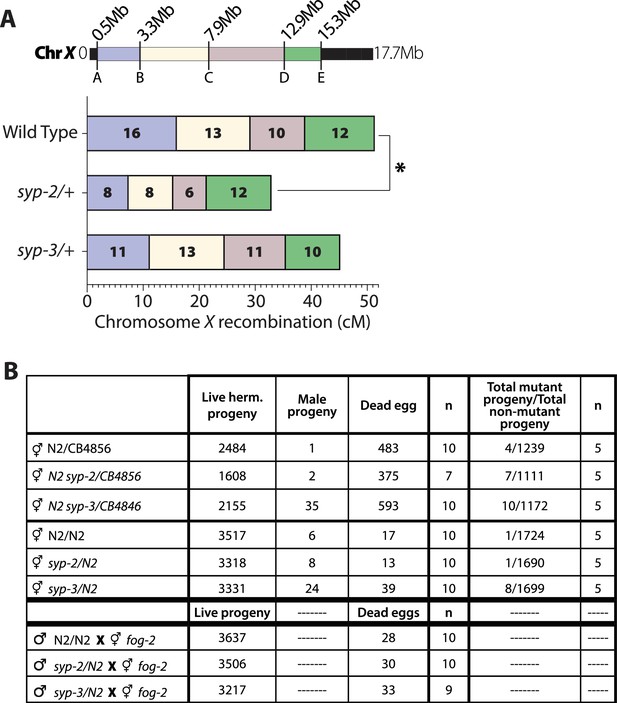
X chromosome may have some chromosome distortion in syp-2/+ oocytes.
(A) Recombination SNP mapping of chromosome X in wild-type, syp-2/+, and syp-3/+ oocytes. A diagram of the 17.7 Mb chromosome X shows the megabase location of each SNP assayed (A–E), and the colored boxes between each SNP show the intervals where crossovers were assessed. The map length (cM) is indicated in each crossover interval. syp-2/+ displays a significant reduction in map length on chromosome X (p=0.0100, chi-squared), while syp-3/+ displays no significant changes to wild-type. The worm counts for each interval are in Figure 6—figure supplement 1—source data 1. (B) Fertility, male progeny, and mutant progeny counts for wild-type, syp-2/+, and syp-3/+ with either all N2 (Bristol) chromosomes or N2/CB4856 (Bristol/Hawaiian). Mutant progeny were scored by displaying dumpy and/or uncoordinated mutant phenotypes. The first n from the left is for the number of worms scored with live hermaphrodite progeny, male progeny, and dead eggs. The second n from the left is for the number of worms scored having mutant progeny.
-
Figure 6—figure supplement 1—source data 1
Worm counts for chromosome X SNP mapping recombination.
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig6-figsupp1-data1-v2.docx
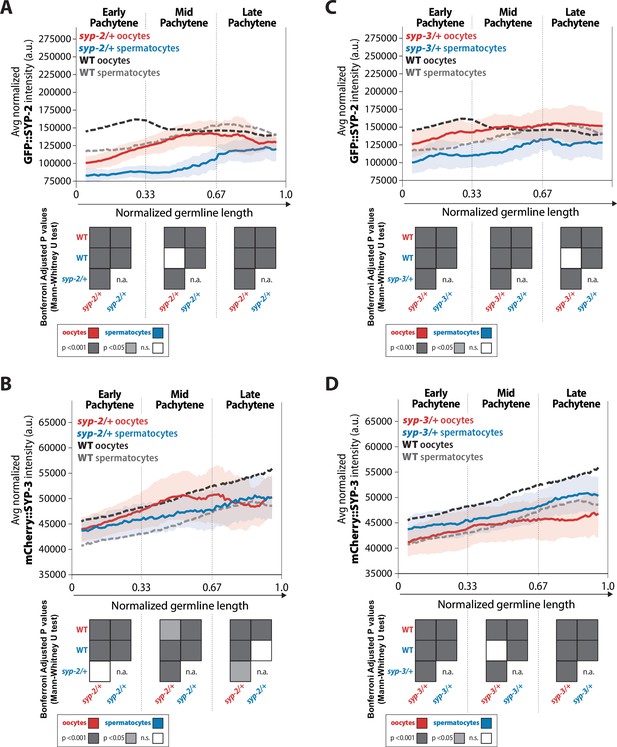
syp-2 and syp-3 gene dosage influence the amount of each SYP loaded within the synaptonemal complex (SC) via sex-specific mechanisms.
(A, B) Quantification of the mean intensity of GFP::SYP-2 (A) and mCherry::SYP-3 (B) per nucleus normalized by the volume of each nucleus (see ‘Methods’) throughout pachytene for syp-2/+. (C, D) Quantification of the mean intensity of GFP::SYP-2 (C) mCherry::SYP-3 (D) per nucleus normalized by the volume of each nucleus throughout pachytene (normalized, see ‘Methods’) for syp-3/+. Oocytes are shown in red with the standard deviation shown as a pale red band, and spermatocytes are shown in blue with the standard deviation shown as a pale blue band. The mean intensity of GFP::SYP-2 (A, B) and mCherry::SYP-3 (C, D) for wild-type (WT) oocytes (black) and WT spermatocytes (gray) are shown as dashed lines. Heat maps below each pachytene region show the Bonferroni adjusted p-values from Mann–Whitney U tests, with dark gray indicating p<0.001, light gray indicating p<0.05, and white indicating not significant (n.s.). The self-comparison between spermatocyte syp-2/+ or syp-3/+ mutants was not determined (n.a.). n values for the number of germlines and nuclei can be found in Figure 7—source data 2.
-
Figure 7—source data 1
Raw sum intensity and normalized sum intensity per nucleus for GFP::SYP-2 and mCherry::SYP-3 in both sexes and all genotypes in Figure 7.
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig7-data1-v2.xlsx
-
Figure 7—source data 2
Synaptonemal complex (SC) intensity n values for nuclei and germlines scored.
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig7-data2-v2.docx
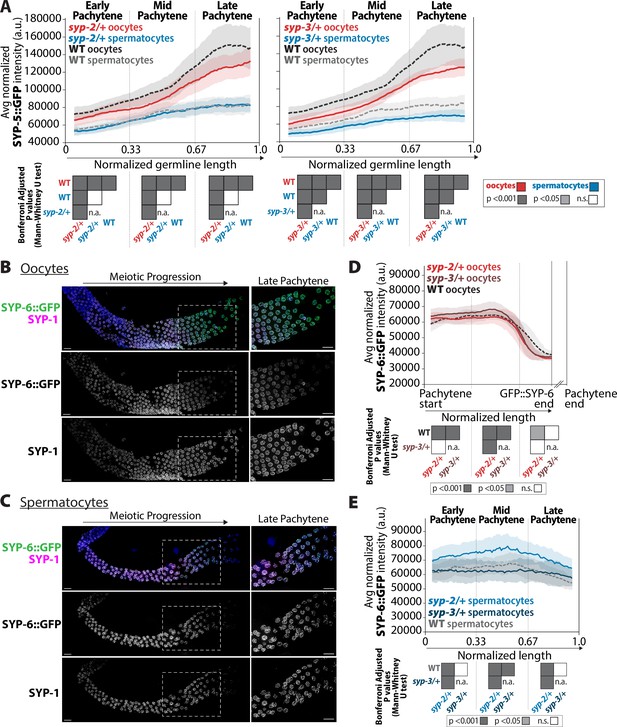
syp-2 and syp-3 gene dosage impacts the composition of SYP-5 and SYP-6 within the synaptonemal complex (SC).
(A) Quantification of the mean intensity of SYP-5::GFP per nucleus normalized by the volume of each nucleus (see ‘Methods’) throughout pachytene for syp-2/+ (left plot) and syp-3/+ (right plot). Mutants are in solid lines with oocytes in red and spermatocytes in blue. Wild-type is in dashed lines with oocytes in black and spermatocytes in gray. The pale shading around each line is the standard deviation. (B, C) Representative images of wild-type germlines stained with SYP-6::GFP (green) and SYP-1 (magenta) in germlines with oocytes (B) and spermatocytes (C). The white dashed box shows the region enlarged in the image on the right. Scale bar represents 10 µm. (D) Quantification of the mean intensity of oocyte SYP-6::GFP per nucleus normalized by the volume of each nucleus throughout pachytene for syp-2/+ (bright red) and syp-3/+ (dark red). Since SYP-6 disassembles prior to the end of pachytene, the germline length is normalized to the germline region with SYP-6::GFP signal starting at the beginning of pachytene (Pachytene start) to the end of the SYP-6::GFP signal (SYP-6::GFP end) (see ‘Methods’). The broken x-axis indicates the unknown distance to the end of pachytene (Pachytene end). Mutants are in solid lines, wild-type oocytes are in a dashed line, and the pale shading around each line represents the standard deviation. (E) Quantification of the mean intensity of spermatocyte SYP-6::GFP per nucleus normalized by the volume of each nucleus (see ‘Methods’) throughout pachytene for syp-2/+ (bright blue) and syp-3/+ (dark blue). Mutants are in solid lines, wild-type spermatocytes are in a dashed line, and the pale shading around each line represents the standard deviation. Heat maps below plots in panels (A), (D), and (E) show the Bonferroni adjusted p-values from Mann–Whitney U tests, with dark gray indicating p<0.001, light gray indicating p<0.05, and white indicating not significant (n.s.). The self-comparison between spermatocyte syp-2/+ or syp-3/+ mutants was not determined (n.a.). n values for the number of germlines and nuclei can be found in Figure 8—source data 2.
-
Figure 8—source data 1
Raw sum intensity and normalized sum intensity per nucleus for SYP-5::GFP and SYP-6::GFP in both sexes and all genotypes in Figure 8.
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig8-data1-v2.xlsx
-
Figure 8—source data 2
Synaptonemal complex (SC) intensity n values for nuclei and germlines scored.
- https://cdn.elifesciences.org/articles/84538/elife-84538-fig8-data2-v2.docx
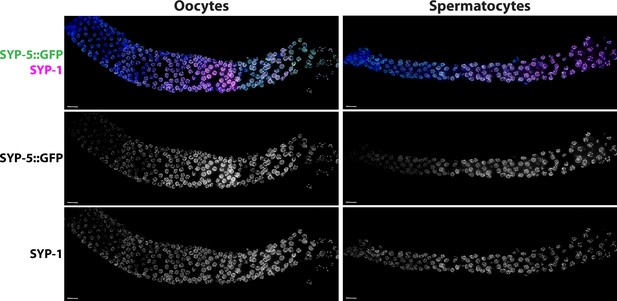
Representative images of wild-type germlines stained with SYP-5::GFP and SYP-1 in oocytes and spermatocytes.
Scale bar represents 10 µm.
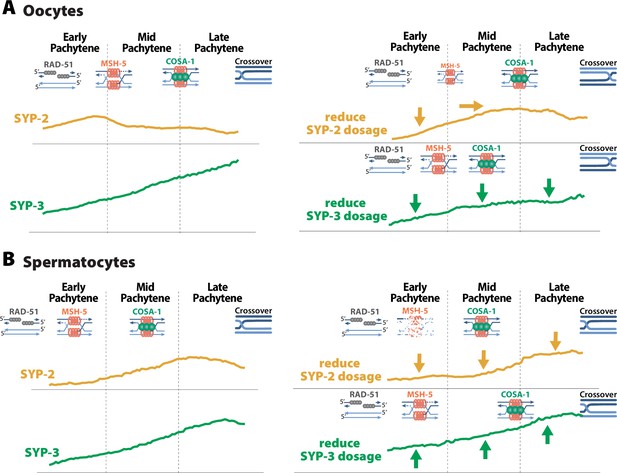
SYP dosage influences the sexually dimorphic regulation of recombination.
(A) In oocytes, SYP-2 amounts with the synaptonemal complex (SC) are critical for the proper formation and/or maintenance of joint molecules stabilized by MSH-5. Reducing the amount of SYP-2 dosage causes decreases in the amount of SYP-2 in early pachytene and shifts the peak amounts of SYP-2 toward mid/late pachytene. This alteration to SYP-2 composition within the SC causes a severe reduction in MSH-5. SYP-3 amounts in the SC are important for the proper timing of recombination. When the dosage of SYP-3 is reduced, SYP-3 composition within the SC is reduced throughout pachytene causing faster resolution of jointed molecules and faster designation of crossovers during pachytene. This ultimately causes changes in the recombination landscape where crossovers are more often positioned near the pairing center. (B) In spermatocytes, SYP-2 amounts are important for the maintenance of MSH-5 stabilized joint molecules. When SYP-2 dosage is reduced, the amount of SYP-2 in the SC is reduced and MSH-5 foci are rapidly lost either because they are resolved quickly or the stability of the joint molecules is compromised. SYP-3 amounts in spermatocytes also influence the timing of recombination, but when SYP-3 dosage is reduced SYP-3 amounts are increased rather than decreased. Thus, elevated SYP-3 levels in spermatocytes cause a delay in crossover designation in spermatocytes.
Tables
Chromosome II SNP mapping recombination.
| Recombinant Intervals | |||||||
|---|---|---|---|---|---|---|---|
| Sex | Genotype | A—B | B—C | C—D | D—E | Non-recombinant | Total worms |
| Oocytes | Wild-type | 26 | 17 | 17 | 27 | 103 | 190 |
| syp-2/+ | 45 | 10 | 13 | 16 | 102 | 186 | |
| syp-3/+ | 44 | 15 | 10 | 16 | 90 | 175 | |
| Spermatocytes | Wild-type | 31 | 17 | 21 | 17 | 79 | 165 |
| syp-2/+ | 29 | 10 | 24 | 23 | 84 | 170 | |
| syp-3/+ | 20 | 25 | 29 | 18 | 92 | 184 | |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Caenorhabditis elegans) | syp-2 | https://wormbase.org/species/c_elegans/gene/WBGene00006376#0-9f-10 | WormBase ID: WBGene00006376 | |
| Gene (C. elegans) | syp-3 | https://wormbase.org/species/c_elegans/gene/WBGene00006377#0-9f-10 | WormBase ID: WBGene00006377 | |
| Gene (C. elegans) | syp-5 | https://wormbase.org/species/c_elegans/gene/WBGene00021832#0-9f-10 | WormBase ID: WBGene00021832 | |
| Gene (C. elegans) | syp-6 | https://wormbase.org/species/c_elegans/gene/WBGene00019002#0-9f-10 | WormBase ID: WBGene00019002 | |
| Gene (C. elegans) | syp-1 | https://wormbase.org/species/c_elegans/gene/WBGene00006375#0-9f-10 | WormBase ID: WBGene00006375 | |
| Gene (C. elegans) | dsb-2 | https://wormbase.org/species/c_elegans/gene/WBGene00194892#0-9f-10 | WormBase ID: WBGene00194892 | |
| Gene (C. elegans) | rad-51 | https://wormbase.org/species/c_elegans/gene/WBGene00004297#0-9f-10 | WormBase ID: WBGene00004297 | |
| Gene (C. elegans) | msh-5 | https://wormbase.org/species/c_elegans/gene/WBGene00003421#0-9f-10 | WormBase ID: WBGene00003421 | |
| Gene (C. elegans) | cosa-1 | https://wormbase.org/species/c_elegans/gene/WBGene00022172#0-9f-10 | WormBase ID: WBGene00022172 | |
| Strain, strain background (C. elegans) | For C. elegans alleles and strain information, see strain table below (‘C. elegans strains, genetics, CRISPR, and culture conditions’) | This paper | See strain table below in ‘C. elegans strains, genetics, CRISPR, and culture conditions’ | |
| Genetic reagent (C. elegans) | For details on CRISPR/Cas9, see ‘C. elegans strains, genetics, CRISPR, and culture conditions’ | This paper | CRISPR/Cas9 transgenics performed by InVivo Biosystems | |
| Antibody | Anti-RAD-51 (chicken polyclonal) | Kurhanewicz et al., 2020; Toraason et al., 2021 | IF (1:1500) | |
| Antibody | Anti-SYP-1 (rabbit polyclonal) | Gift from Nicola Silva lab | IF (1:1000) | |
| Antibody | Anti-DSB-2 (rabbit polyclonal) | Rosu et al., 2013 | IF (1:5000) | |
| Antibody | Anti-OLLAS (rabbit polyclonal) | GenScript | Cat# A01658 | IF (1:1000) |
| Antibody | Anti-SUN-1 S8P (guinea pig polyclonal) | Woglar et al., 2013 | IF (1:700) | |
| Antibody | Anti-Tubulin (mouse monoclonal) | Abcam | Cat# ab7291 | WB (1:1000) |
| Antibody | Alexa Fluor 488 anti-rabbit (goat polyclonal) | Thermo Fisher | Cat# A11034 | IF (1:200) |
| Antibody | Alexa Fluor 488 anti-chicken (goat polyclonal) | Thermo Fisher | Cat# A11039 | IF (1:200) |
| Antibody | Alexa Fluor 555 anti-rabbit (goat polyclonal) | Thermo Fisher | Cat# A21428 | IF (1:200) |
| Antibody | Anti-GFP booster-488 (nanobody) | Chromotek | Cat# gb2AF488-50 | IF (1:200) |
| Antibody | Alexa Fluor 488 anti-guinea pig (goat polyclonal) | Thermo Fisher | Cat# A11073 | IF (1:200) |
| Antibody | IRDye 680 anti-mouse (donkey polyclonal) | LI-COR | Cat# 926-68072 | WB (1:1000) |
| Antibody | IRDye 800CW anti-rabbit (donkey polyclonal) | LI-COR | Cat# 926-32213 | WB (1:1000) |
| Sequence-based reagent | CRISPR primers are in Supplementary file 1 | This paper | PCR primers | Supplementary file 1 |
| Sequence-based reagent | For details on SNP recombination mapping primers, see ‘SNP recombination mapping’ and Supplementary file 2 | This paper | PCR primers | Supplementary file 2 |
| Chemical compound, drug | Serotonin | Sigma-Aldrich | Cat# H7752 | (25 mM) |
| Chemical compound, drug | Tricaine (ethyl 3-aminobenzoate methanesulfonate) | Sigma-Aldrich | Cat# E10521-50G | (0.08% w/v) |
| Chemical compound, drug | Tetramisole hydrochloride | Sigma-Aldrich | Cat# T1512-10G | (0.008% w/v) |
| Chemical compound, drug | Agarose | Invitrogen | Cat# 16500500 | (7–9% w/v) |
| Chemical compound, drug | Naphthaleneacetic acid (K-NAA, auxin) | PhytoTechnology Laboratories | Cat# N610 | (1 mM and 10 mM) |
| Software, algorithm | Whole Gonad Analysis (R script) | Toraason et al., 2021 | https://github.com/libudalab/Gonad-Analysis-Pipeline | |
| Software, algorithm | Prism 10 | GraphPad | https://www.graphpad.com/features | |
| Software, algorithm | Imaris 9 | Oxford Instruments | https://imaris.oxinst.com/products | |
| Software, algorithm | FIJI plug in – ‘stackRegJ’ | https://research.stowers.org/imagejplugins/ | ||
| Software, algorithm | FIJI plug in – Stitcher | Preibisch et al., 2009 | https://imagej.net/plugins/image-stitching | |
| Software, algorithm | FIJI | Schindelin et al., 2012 | https://imagej.net/software/fiji/ | |
| Other | Low fluorescence PVDF membranes | Thermo Fisher | Cat# 22860 | |
| Other | Vectashield | VWR | Cat# 101098-042 | |
| Other | DAPI stain | Invitrogen | Cat# D1306 | (2 µg/mL) |
Additional files
-
Supplementary file 1
CRISPR/Cas9 primer sequences.
- https://cdn.elifesciences.org/articles/84538/elife-84538-supp1-v2.xlsx
-
Supplementary file 2
SNP recombination mapping primer sequences.
- https://cdn.elifesciences.org/articles/84538/elife-84538-supp2-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84538/elife-84538-mdarchecklist1-v2.docx





