Formation and three-dimensional architecture of Leishmania adhesion in the sand fly vector
Figures
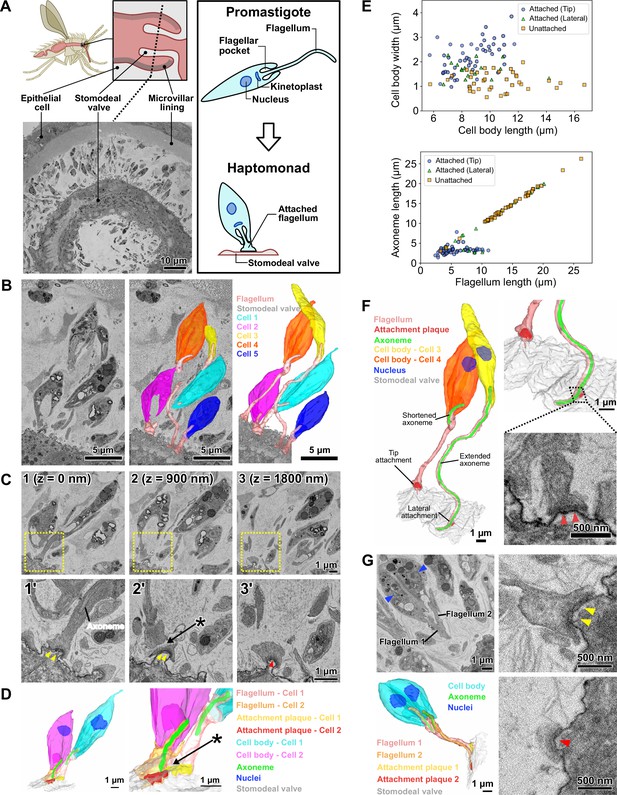
Haptomonads form a dense, multi-layered complex on the sand fly stomodeal valve.
(A) Cartoon of the sand fly digestive tract highlighting the position of the stomodeal valve, the site of differentiation of free-swimming Leishmania promastigotes into haptomonads, which attach to the stomodeal valve surface via a shortened and expanded flagellum (Right black box). SBFSEM slice (bottom left) showing a cross section of the stomodeal valve at the position indicated by the dotted line in the cartoon (Figure 1—video 1). (B) SBFSEM slice and 3D reconstruction showing attached haptomonads in close proximity to each other and attached to the stomodeal valve (grey) by flagella (peach) of different lengths (Figure 1—videos 2–3). (C) Sequential SBFSEM images (every 900 nm) showing details of the attached flagella from cells 1 and 2 in C. Top: lower magnification view showing entire cells. Bottom: higher magnification view of the attachment region indicated by the yellow dotted box in the top image. Both cells were attached to the stomodeal valve by a flagellum containing an attachment plaque (yellow and red arrowheads), but no attachment plaque was formed between neighbouring flagella (asterisk). (D) 3D reconstruction of the inner flagellum structures from cells 1 and 2 in C, showing the attachment plaques and the lack of attachment between flagella (asterisk, as indicated in D). (E) Scatter plots of cell body length against cell body width (Top), and flagellum length against axoneme length (Bottom) of attached cells with the distal flagellar tip attachment (Blue circles; n=50), attached cells with the laterally attached flagellum (Green triangles; n=17), and unattached cells (Orange squares; n=39) in the sand fly. (F) 3D reconstruction (left and top right) showing a slender and wider cell (cell 3 and 4 respectively in C) attached to the stomodeal valve via an attachment plaque formed on the lateral part or the distal tip of the flagellum, respectively. Note: the axoneme extended along the length of the flagellum in cell 3 with the laterally attached flagellum, whereas in the cell 4 with the distal flagellar tip attachment, the axoneme only just extended beyond the end of the cell body. SBFSEM image (bottom right) showing a cross section of an attachment plaque (red arrowheads) formed on the lateral part of the flagellum of cell 3. (G) SBFSEM slices (top left and right) and 3D reconstruction (bottom left) showing a dividing haptomonad with two nuclei (blue arrowheads) and two attached flagella. Top and bottom right: higher magnification views of the attachment plaques in each flagellum (yellow and red arrowheads, corresponding to the colours of the attachment plaques in the 3D reconstruction).
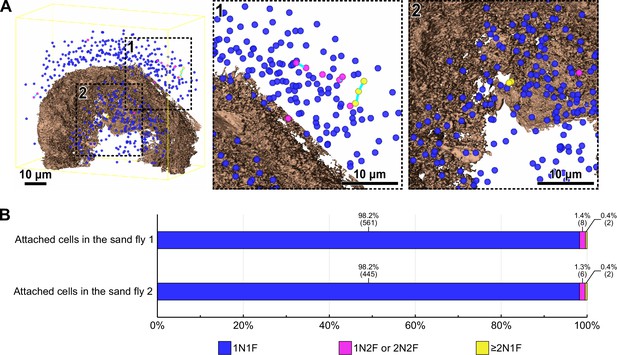
Cell cycle analysis of haptomonads at the sand fly stomodeal valve.
(A) Mapping of an infected stomodeal valve showing the position of the nuclei of different attached cells, colour code according to the cell’s division status (Figure 1—video 4). Whole view (left) and magnified views of the outside (middle; black dotted box 1) and the inside (right; black dotted box 2) of the stomodeal valve. Different coloured spheres indicate nuclei of cells which have one nucleus and one flagellum (1N1F; blue), nuclei of cells which have one or two nucleus and two flagella (1N2F or 2N2F; magenta), and nuclei of cells which have two or more nuclei but in which new flagellum assembly has not occurred (≥2N1 F; yellow). Two or more nuclei in a single cell body are linked by cyan bars. The stomodeal valve is shown in brown. The yellow line indicates the boundary box of the imaged volume. (B) Counts of the nuclei of attached cells present in the volume obtained from two SBFSEM runs. The percentages and number of nuclei counted are shown above the bars.
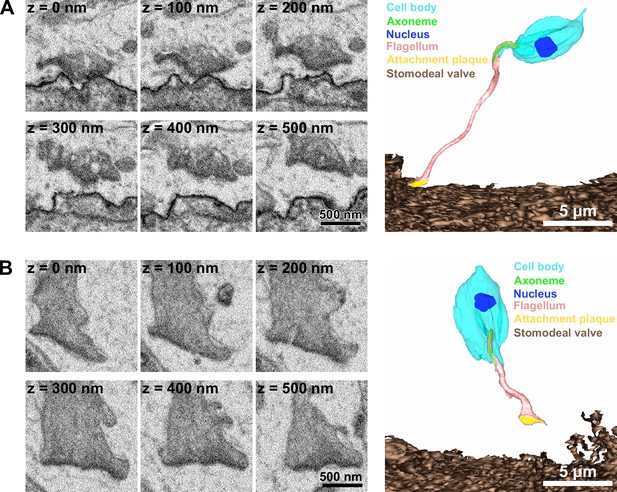
Haptomonads detached from the stomodeal valve.
(A) Sequential SBFSEM images (every 100 nm; left) and 3D reconstruction (right) showing a haptomonad flagellum with part of the attachment plaque detached from the stomodeal valve. (B) Sequential SBFSEM images (every 100 nm; left) and 3D reconstruction showing a haptomonad flagellum completely detached from the stomodeal valve with the attachment plaque left at the tip.
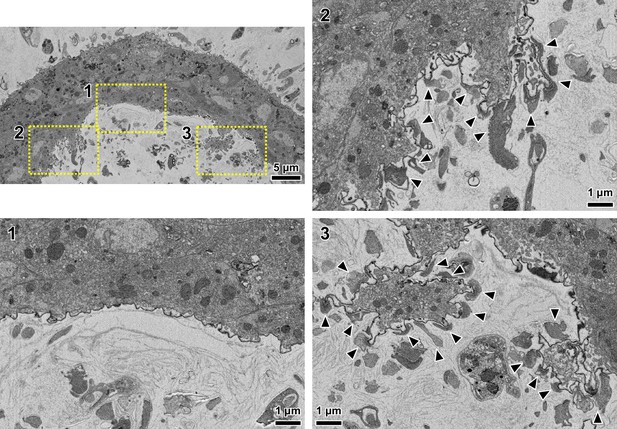
Damage to the stomodeal valve at points of haptomonad attachment.
Top left: lower magnification view showing a SBFSEM cross-section of the stomodeal valve. Bottom left: magnified view of the stomodeal valve surface at the area where no haptomonads were attached and no damage to the stomodeal valve was seen (yellow dotted Box 1 in the top left image). Top and bottom right: magnified views of the stomodeal valve surface at areas where a large number of haptomonads attached (yellow dotted box 2 and 3 in the top left image). Note: compared to the area where there was no haptomonad adhesion, the areas with a large number of haptomonads showed detachment of the cuticle lining indicating potential damage to the stomodeal valve surface. Black arrowheads: areas where the flagellum of a haptomonad is attached on the stomodeal valve.
SBFSEM imaging of haptomonads on the stomodeal valve in the sand fly (whole view).
3D volume taken in 400 cycles of 100 nm cuttings. Scale bar = 10 µm.
SBFSEM imaging of haptomonads on the stomodeal valve in the sand fly (enlarged view).
Scale bar = 5 µm.
3D model generated from the SBFSEM imaging shown in Figure 1—video 2.
Scale bar = 5 µm.
3D model of mapping of an infected stomodeal valve showing the position of the nuclei of different attached cells.
Different coloured spheres indicate nuclei of cells which have one nucleus and one flagellum (1N1F; blue), nuclei of cells which have one or two nucleus and two flagella (1N2F or 2N2F; magenta), and nuclei of cells which have two or more nuclei but in which new flagellum assembly has not occurred (≥2N1 F; yellow). Two or more nuclei in a single cell body are linked by cyan bars. The stomodeal valve is shown in brown. The yellow line indicates the boundary box of the imaged volume. Scale bar = 10 µm.
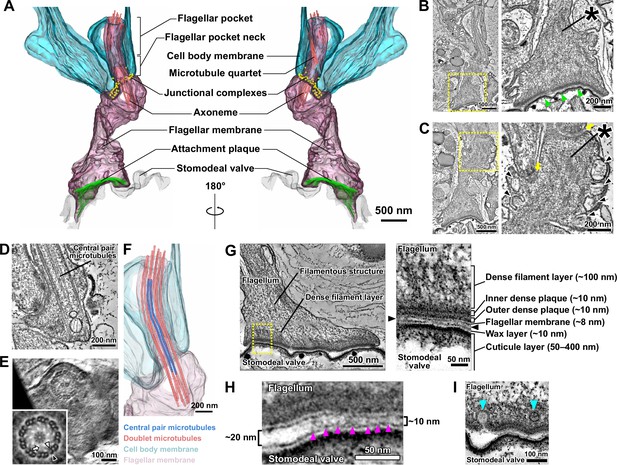
The modified haptomonad flagellum has a highly organised ultrastructure, with multiple discrete elements.
Different serial tomograms of haptomonads in the sand fly, focusing on the attachment region (Figure 2—video 1). (A–C) 3D reconstruction (Figure 2—video 2; A) and slices (B, C) from the serial tomogram shown in Figure 2—video 1. (B) Low- (left) and high- (right) magnification tomogram slices showing the attachment plaque region (green arrowheads), the expanded flagellum attachment zone (FAZ; yellow arrows) found in attached cells, as well as filament bundles (asterisks) and vesicles (black arrowheads) found inside the attached flagellum. (D, E) Slices from a different tomogram showing the shortened haptomonad 9+2 axoneme, in longitudinal section (D) and cross-section (E). The nine-fold Markham rotational averaging of the same axoneme (inset in E) highlights the presence of inner and outer dynein arms (white and black arrowheads, respectively) and radial spokes (white arrow). (F) 3D reconstruction of the shortened 9+2 axoneme shown in D. (G–I) Slices from a higher magnification tomogram of the attachment region. The image on the right in G shows a high-magnification view of the boxed area on the left image, with the distinct ultrastructural elements of the attachment region indicated. The black arrowheads indicate the gap between the flagellum and the stomodeal valve. (H) Magnified view of the gap between the flagellum and the stomodeal valve in a different area of the tomogram. Electron-dense structures (magenta arrowheads) appear to connect the flagellar membrane to the stomodeal valve surface in the narrower (~10 nm wide) regions of the gap, but were absent in the wider (~20 nm wide) region of the gap. (I) Different area of the tomogram showing vesicles (cyan arrowheads) budding from (or fusing with) the flagellar membrane at the attachment interface.
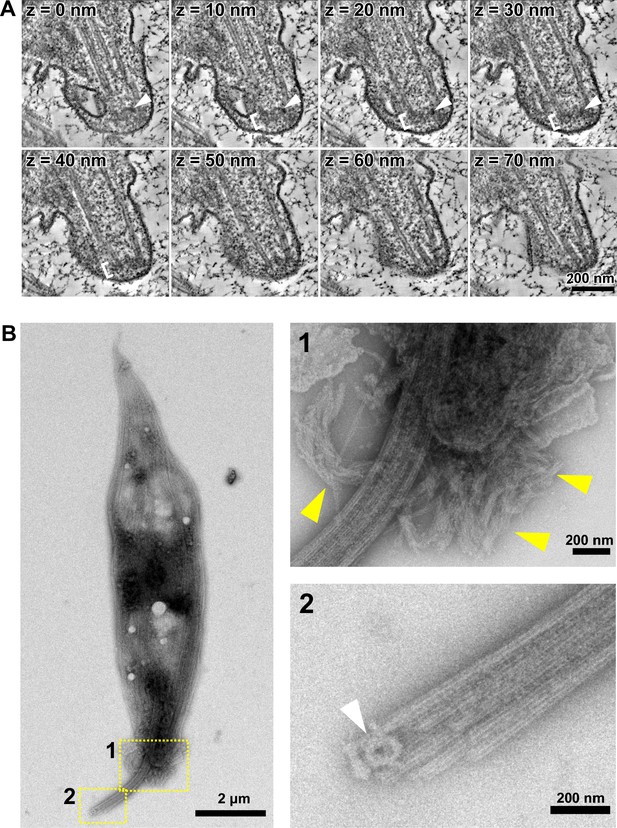
Tip structure of the in vivo haptomonad and in vitro haptomonad-like cell axoneme.
(A) Sequential tomographic slices (every 10 nm) showing the tip structure of the haptomonad axoneme. The order of the slices is indicated at top left of each image. White arrowheads: ring-shaped capping structure. White brackets: amorphous material. (B) Whole mount cytoskeleton of an in vitro haptomonad-like cell attached to a formvar membrane, image by negative staining transmission electron microscopy (left). Top right: higher magnification view of the anterior tip of the cell (yellow dotted box 1) showing filament bundles (yellow arrowheads), likely remnants of filamentous structures seen in the attached flagellum. Bottom right: magnified view of the axoneme tip (yellow dotted box 2), showing the ring-shaped capping structure (white arrowhead).
Serial section electron tomography of a haptomonad on the stomodeal valve in the sand fly.
3D volume reconstructed by joining tomograms from a series of six 150-nm-thick sections. Scale bar = 500 nm.
3D model generated from the serial tomogram shown in Figure 2—video 1.
Scale bar = 500 nm.
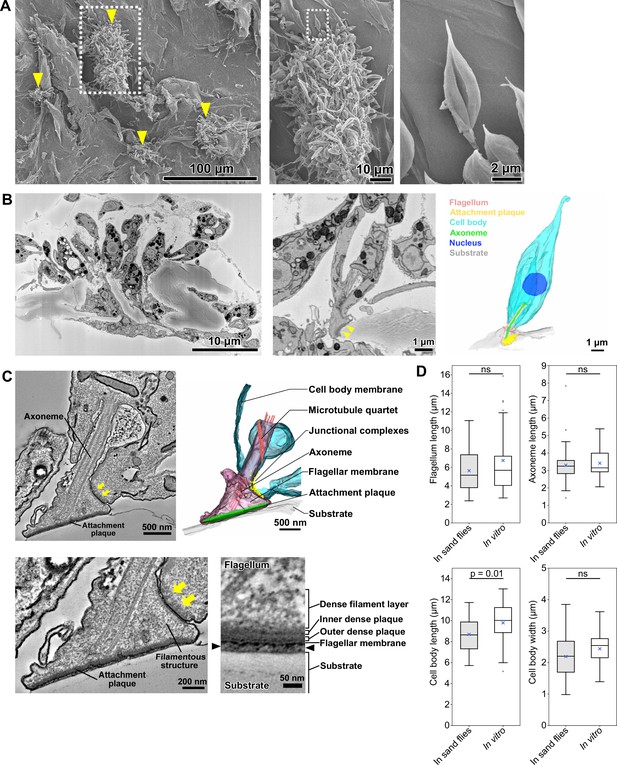
In vitro haptomonad-like cells resemble haptomonads observed in the sand fly.
(A) Scanning electron microscopy images of in vitro haptomonad-like cells on a plastic coverslip. Left: Low-magnification image showing groups of cells (yellow arrowheads) attached to the substrate. Middle: a magnified view of the clump of attached haptomonad-like cells indicated by the white dotted box in the left image. Right: an individual haptomonad-like cell from the attached clump indicated by the white dotted box in the middle image. (B) Low-magnification SBFSEM image of haptomonad-like cells attached to a plastic coverslip in vitro (left; Figure 3—video 1). Magnified SBFSEM image (middle) and 3D reconstruction (right) of an in vitro haptomonad-like cell. SBFSEM slices show the haptomonad-like cell attached to the substrate with an electron-dense attachment plaque (yellow arrowheads). (C) Slices of a serial tomogram and 3D reconstruction (top right) of an in vitro haptomonad-like cell. Top left: tomographic slice through the region of the flagellum attachment to the cell, highlighting the FAZ and the flagellar pocket. Yellow arrows: junctional complexes. Bottom left: high-magnification view of the attachment region and the FAZ. Bottom right: magnified views of the tomogram showing that the attachment plaque observed in vitro has the same distinct ultrastructural elements identified in attachment plaques in vivo (in the sand fly; Figure 2G). Black arrowheads show the gap between the flagellum and the substrate. (D) Box-whisker plots of flagellum (top left), axoneme (top right) and cell body (bottom left) length and cell body width (bottom right) of haptomonads in sand flies and in vitro haptomonad-like cells. Boxes and whiskers indicate the median, upper and lower quartiles and 5th/95th percentiles. Blue crosses and grey dots indicate mean values and outliers, respectively. No significant (ns) morphological differences were found between haptomonads in sand flies and in vitro haptomonad-like cells, except for cell body length. p-Values calculated using two-tailed Welch’s t-test. N=50 (in sand flies) and 30 (in vitro).
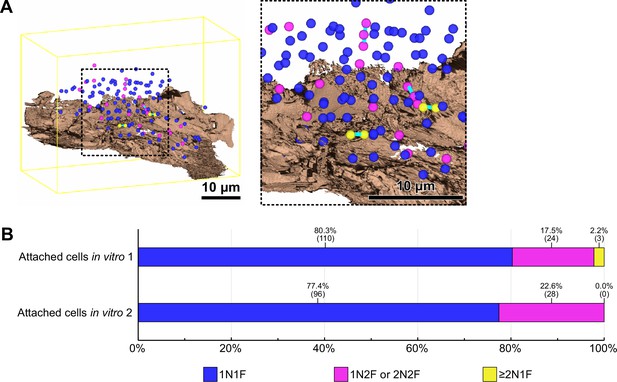
Cell cycle analysis of in vitro haptomonad-like cells.
(A) Mapping of nuclei of in vitro haptomonad-like cells (Figure 3—video 2). Left: whole view. Right: magnified view. Different coloured spheres indicate nuclei of cells which have one nucleus and one flagellum (1N1F; blue), nuclei of cells which have one or two nucleus and two flagella (1N2F or 2N2F; magenta), and nuclei of cells which have two or more nuclei but in which flagellar division was not occurring (≥2N1 F; yellow). Two or more nuclei in a single cell body are linked by cyan bars. The substrate is shown in brown. The yellow line indicates the boundary box of the imaged volume. (B) Counts of the nuclei of cells present in the volume obtained from two SBFSEM runs. The percentages and number of nuclei counted are shown above the bars.
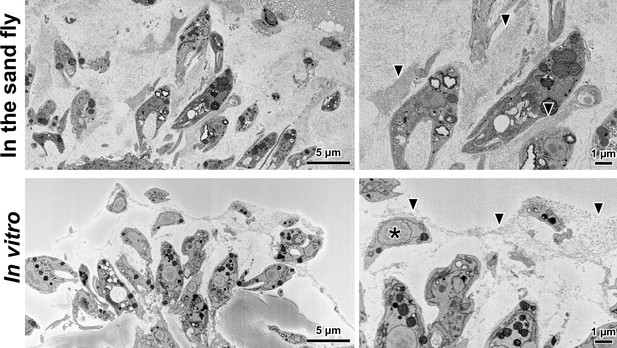
Sand fly haptomonads and in vitro haptomonad-like cells are surrounded by an extracellular filamentous gel-like matrix.
SBFSEM slices of sand fly (top) and in vitro (bottom) cells showing attached cells and unattached promastigotes (asterisk) surrounded by a filamentous matrix (arrowheads).
SBFSEM imaging of in vitro haptomonad-like cells attached to plastic.
3D volume taken in 252 cycles of 100 nm cutting. Scale bar = 10 µm.
3D model of mapping of nuclei of in vitro haptomonad-like cells.
Different coloured spheres indicate nuclei of cells which have one nucleus and one flagellum (1N1F; blue), nuclei of cells which have one or two nucleus and two flagella (1N2F or 2N2F; magenta), and nuclei of cells which have two or more nuclei but in which new flagellum assembly has not occurred (≥2N1 F; yellow). Two or more nuclei in a single cell body are linked by cyan bars. The substrate is shown in brown. The yellow line indicates the boundary box of the imaged volume. Scale bar = 10 µm.
Serial section electron tomography imaging of an in vitro haptomonad-like cell attached to plastic.
3D volume reconstructed by joining tomograms from a series of six 150-nm-thick sections. Scale bar = 500 nm.
3D model generated from the serial tomogram shown in Figure 3—video 3.
Scale bar = 500 nm.
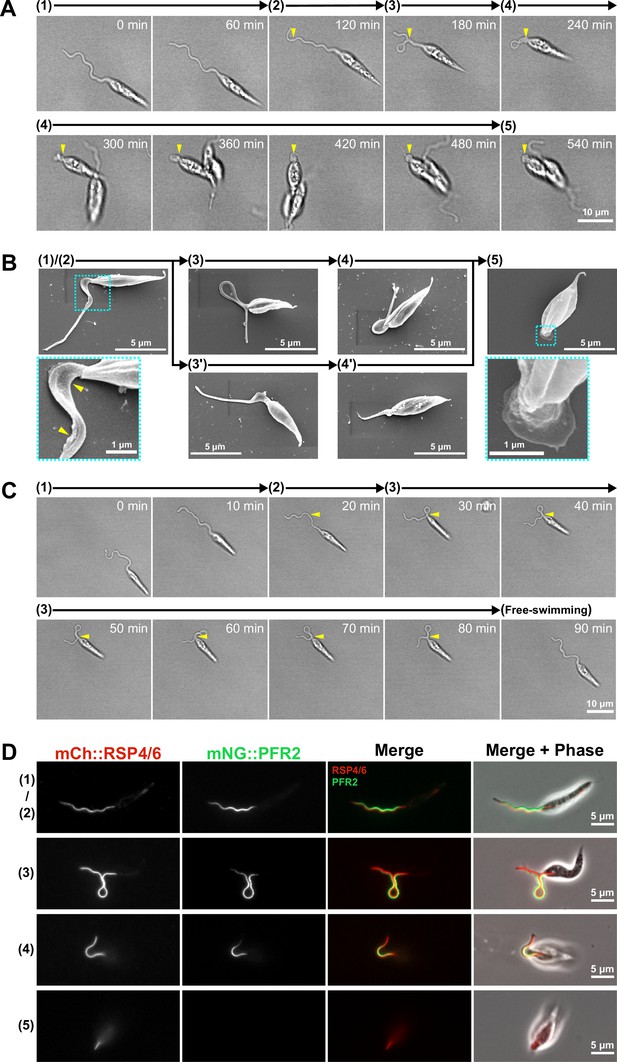
Adhesion of the in vitro haptomonad-like cell has a series of defined sequential steps.
(A) Sequential frames (at 60 min intervals) from a time-lapse video (Figure 4—video 1) of adhesion of a haptomonad-like cell, as viewed by bright field microscopy. The numbers and arrows above the images indicate the 5 different stages of adhesion observed by time-lapse. Yellow arrowheads show the adhesion point. In stage 1, a promastigote cell first probes the substrate surface using the flagellum, and then a small section of the flagellum starts to adhere to the surface (stage 2). In stage 3, the flagellum is more clearly (and widely) adhered to the surface, and then the flagellum disassembles (stage 4). Finally, a clear attachment plaque is observed, representing mature attachment (stage 5). (B) Scanning electron microscopy images of cells in different stages of adhesion, showing that initial adhesion and disassembly of the flagellum can occur with (3 and 4) or without (3’ and 4’) the formation of a loop in the flagellum. The dotted boxes show magnified views of the attachment region in cells at early (1 and 2) and late (5) stages of adhesion. The yellow arrowheads indicate deformation of the flagellar membrane in initial adhesion regions. (C) Sequential frames (at 10 min intervals) from a time-lapse video (Figure 4—video 3) showing that adhesion is reversible at its early stages (stage 1–3). (D) Fluorescent images of promastigotes and haptomonad-like cells expressing an axoneme marker (mCherry::RSP4/6; red) and a PFR marker (mNeonGreen::PFR2; green) showing the disassembly of the PFR and partial disassembly of the axoneme. The stages of adhesion are indicated on the left-hand side of each image row.
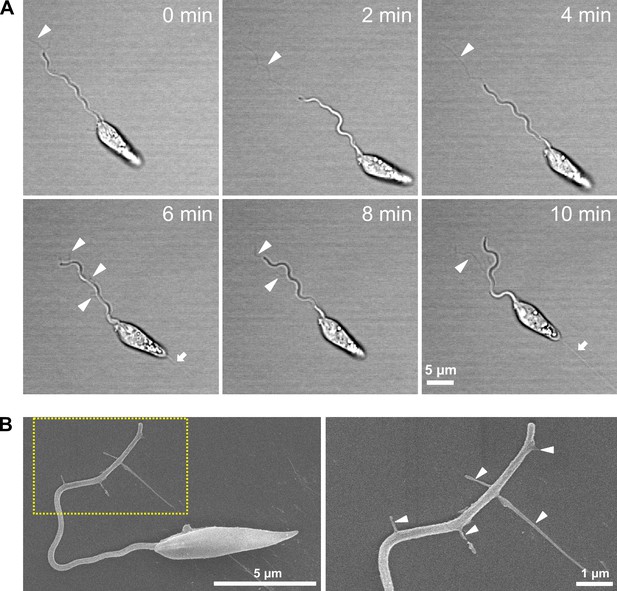
Leishmania explores the surface while releasing membrane streamers from the flagellum.
(A) Sequential frames from a time-lapse video (Figure 4—video 2) of the adhesion of an in vitro haptomonad-like cell (at 2 min intervals), showing membrane streamers from the flagellum (white arrowheads). White arrows indicate membrane streamers emerging from the posterior end of the cell. (B) Scanning electron microscopy images of a cell releasing membrane streamers from the flagellum (left) and magnified view of the flagellar tip (right), showing details of the flagellum membrane streamers (white arrowheads).
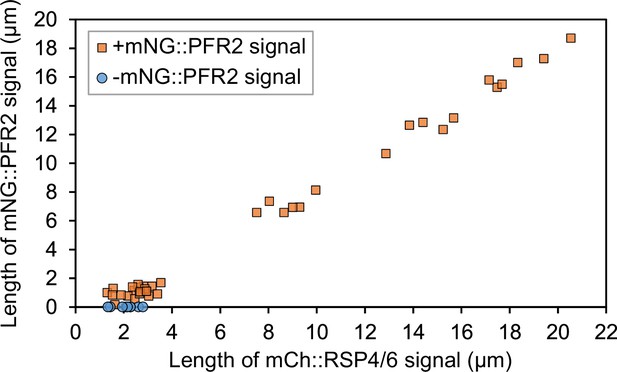
Measurement of mNeonGreen::PFR2 and mCherry::RSP4/6 length in in vitro haptomonad-like cells.
Scatter plot of mCh::RSP4/6 signal length against mNG::PFR2 signal length. The length of the mNG::PFR2 signal of cells whose mNG signal is not visible is indicated as zero, and these points are indicated by blue circles (n=9). Plots for cells with visible mNG::PFR2 signal are indicated by orange squares (n=38).
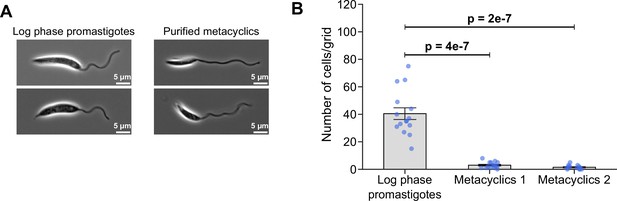
In vitro purified metacyclics do not adhere to glass.
(A) Example images of log phase promastigotes and in vitro purified metacyclics. Note: the metacyclics have a flagellum that is longer than the cell body and a short slender cell body. (B) Comparison of the adhesion ability between log phase promastigotes and in vitro purified metacyclics. Log phase promastigotes and in vitro purified metacyclics (1×106 cells/ml, respectively) were cultured on gridded glass coverslips in a 24-well plate with 1 ml of complete M199 medium for 24 hr at 28 °C with 5% CO2, and the number of attached cells per grid area was quantified in the same method as in the Figure 5C. Data represent mean ± SEM (n=3 independent experiments). For the metacyclics, the quantification was carried out twice using in vitro purified metacyclics from different cultures (metacyclics 1 and 2). The blue dots represent individual measurement from three-independent experiments. p-Values calculated using two-tailed Welch’s t-test.
Time-lapse video of adhesion process of an in vitro haptomonad-like cell on a glass coverslip.
Playback of ~9 hr at 1000 x speed. Scale bar = 10 µm.
Time-lapse video of release of membrane streamers from the flagellum of an in vitro haptomonad-like cell on a glass coverslip.
Playback of ~8 min at 30 x speed. Scale bar = 5 µm.
Time-lapse video of reversible adhesion of an in vitro haptomonad-like cell on a glass coverslip.
Playback of 1.5 hr at 500 x speed. Scale bar = 10 µm.
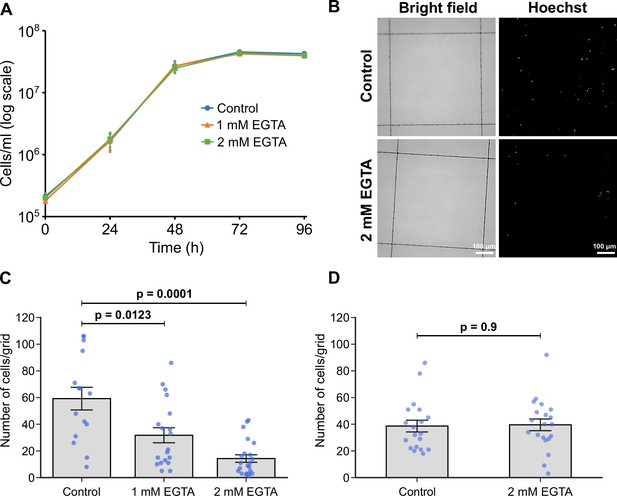
Calcium is necessary for adhesion of in vitro haptomonad-like cells.
(A) Growth curve of cells cultured in control M199 growth medium or M199 with 1 or 2 mM EGTA. Data represent mean ± SD (n=3 independent experiments). (B–D) In vitro analysis of adhesion in the presence of EGTA. Cells were allowed to adhere to gridded coverslips for 24 hr, and subjected to EGTA treatment during (B, C) or after (D) adhesion. (B) Bright field and Hoechst fluorescence images of attached cells after 24 hr of culture in medium with or without 2 mM EGTA. (C) Quantification of the number of attached cells per grid area. Data represent mean ± SEM (n=3 independent experiments). (D) Quantification of the number of attached cells per grid area in attached cultures exposed to 2 mM EGTA (or control medium) for 30 min after 24 hr of adhesion. Data presented mean ± SEM (n=3 independent experiments). In C and D, the blue dots represent individual measurement from three-independent experiments. p-Values calculated using two-tailed Welch’s t-test.
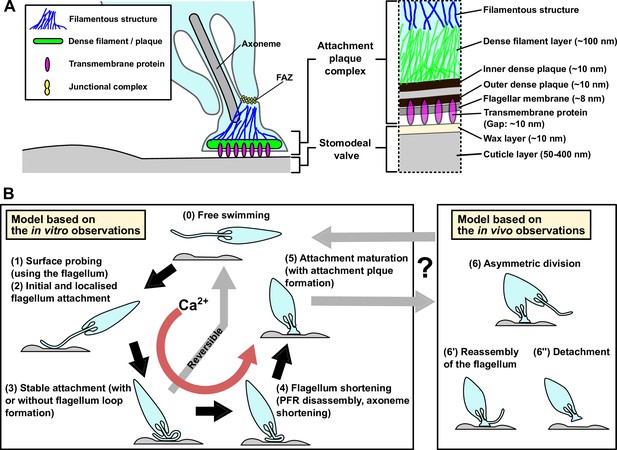
Models of the haptomonad attachment plaque and adhesion process.
(A) Model of the haptomonad attachment plaque. (B) Model of the haptomonad adhesion process based on the in vitro observations. A promastigote cell explores the surface of the substrate using the flagellum (1), and initiates adhesion through the flagellum (2). Subsequently, stable adhesion is established (3), which is often (but not always) associated with the formation of a flagellum loop. Then, the flagellum becomes shorter (partial flagellum disassembly; 4), and finally the adhesion matures, with the formation of an attachment plaque (5). Stage 3 is reversible, as cells can still detach from the substrate before flagellum shortening. Adhesion formation (but not maintenance) is Ca2+-dependent. The box on the right in B shows three different hypotheses for generation of free-swimming promastigotes from haptomonads based on the in vivo observations, including asymmetric division (6), flagellum reassembly followed by detachment (6’), or detachment followed by flagellum reassembly (6”).

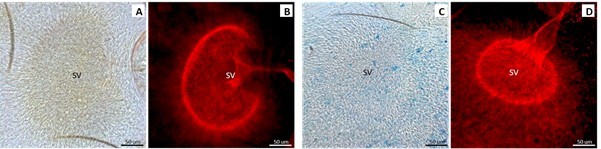
Infection rates and intensities of infections on days 6 and 9 post-infection (PI).
Numbers above the bars indicate the number of dissected females. Levels of Leishmania infections were graded into four categories: negative, light (< 100 parasites/gut), moderate (100-1000 parasites/gut) and heavy (>1000 parasites/gut).
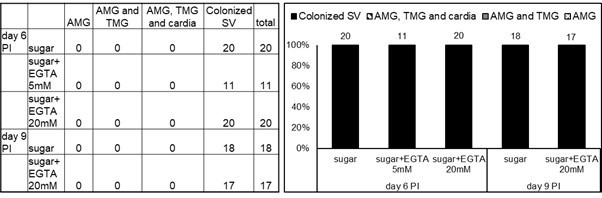
Localization of infections on days 6 and 9 post-infection (PI).
Abdominal midgut (AMG), thoracic midgut (TMG) and stomodeal valve (SV). Numbers above the bars indicate the number of evaluated (positive) females.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Leishmania mexicana) | RSP4/6 | TriTrypDB (http://tritrypdb.org/tritrypdb/) | LmxM.13.0430 | |
| Gene (Leishmania mexicana) | PFR2 | TriTrypDB (http://tritrypdb.org/tritrypdb/) | LmxM.16.1430 | |
| Strain, strain background (Leishmania mexicana) | L. mexicana | Sunter lab stocks | WHO strain MNYC/BZ/1962 /M379 | The identity has been authenticated by genome and mRNA sequencing. The strain was monitored for contamination, including mycoplasma contamination, through DNA staining and microscopy during data capture. |
| Cell line (L. mexicana) | mCherry::RSP4/6, mNeonGreen::PFR2 | Wang et al., 2021 L. mexicana cell line provided by Dr Richard Wheeler (University of Oxford) | NA | The identity has been authenticated by genome sequencing. The cell line was monitored for contamination, including mycoplasma contamination, through DNA staining and microscopy during data capture. |
| Strain, strain background (Lutzomyia longipalpis) | L. longipalpis | Volf and Volfova, 2011 | NA | |
| Software, algorithm | 3dmod | Kremer et al., 1996 | PMID:8742726 | |
| Software, algorithm | IMOD | http://bio3d.colorado.edu/imod | RRID:SCR_003297 | |
| Software, algorithm | SerialEM | Mastronarde, 2005 | PMID:16182563 | |
| Software, algorithm | Fiji | Schindelin et al., 2012 | RRID: SCR_002285 | |
| Software, algorithm | Excel | https://microsoft.com/mac/excel | RRID:SCR_016137 | |
| Software, algorithm | python | https://www.python.org/ | RRID:SCR_008394 | |
| Software, algorithm | matplotlib | Hunter, 2007 | RRID:SCR_008624 | |
| Other | Gridded glass coverslips grid-500 | iBidi | Cat. #: 10816 | In vitro haptomonad-like cell preparation |
| Other | Thermanox plastic coverslips | Nalgene Nunc International | Cat. #: 174950 | In vitro haptomonad-like cell preparation |
| Other | µ-dish 35 mm, high grid-500 glass bottom | iBidi | Cat. #: 81168 | In vitro haptomonad-like cell preparation |