Neural mechanisms of parasite-induced summiting behavior in ‘zombie’ Drosophila
Figures
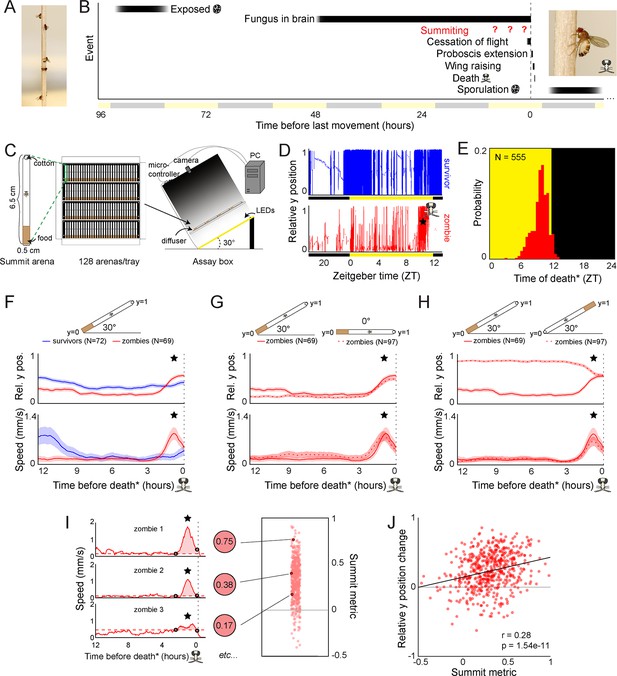
Behavioral signature of E.muscae-induced summiting in wild-type flies.
(A) E. muscae-killed fruit flies that summited on a wooden dowel prior to death. (B) Timeline of events relative to an E. musace-infected fly’s last movement (dashed line). See (Elya et al., 2018; Krasnoff et al., 1995). (C) Summiting assay schematic. (D) Example y position data for a typical survivor fly (top) and zombie (bottom). X-axis is Zeitgeber time (ZT), hours since lights were turned on. The fly ‘skull’ indicates the manually-annotated time of zombie death (see Methods). Black and yellow bars indicate the state of visible illumination. (E) Distribution of time of death for Canton-S flies killed by E. muscae. Background color indicates the state of visible illumination. (F) Mean y position (middle) and mean speed (bottom) of survivor flies (blue) and zombie flies (red) housed in arenas angled at 30° with food at the bottom (schematic at top) during the 12 hr preceding the time of death. Here and in all other panels, shaded regions are +/− 1 standard error of the mean. Time of death for zombies was manually determined as the time of the last movement from the y position trace. Survivors did not die but were assigned fictive times of death from the distribution of zombie death times for comparability (see Methods). (G) As in (F), but comparing zombies in standard arenas (30° with respect to gravity, same data as (F); solid lines) to zombies in flat arenas (0°; dashed lines). (H) As in (F) and (G), but comparing zombies in standard arenas (food at the bottom, same data as (F); solid lines) to zombies in arenas with food at the top (dashed lines). (I) Speed versus time for three examples Canton-S zombies (left) and their corresponding summit metrics (middle) outlined in black (right) amidst all Canton-S summit metrics (N=555, right). Black circles denote the window of summiting behavior as determined from the mean behavior of Canton-S zombie flies. Dashed red line indicates the mean speed in the hour preceding summiting (baseline speed). Summit metric is calculated as the integral of speed minus baseline in the summiting window (shaded region). (J) Relative y position change versus summit metric for Canton-S zombies (N=555). Points are individual flies. Linear regression line in black; Pearson’s correlation r & p-value (upper left).
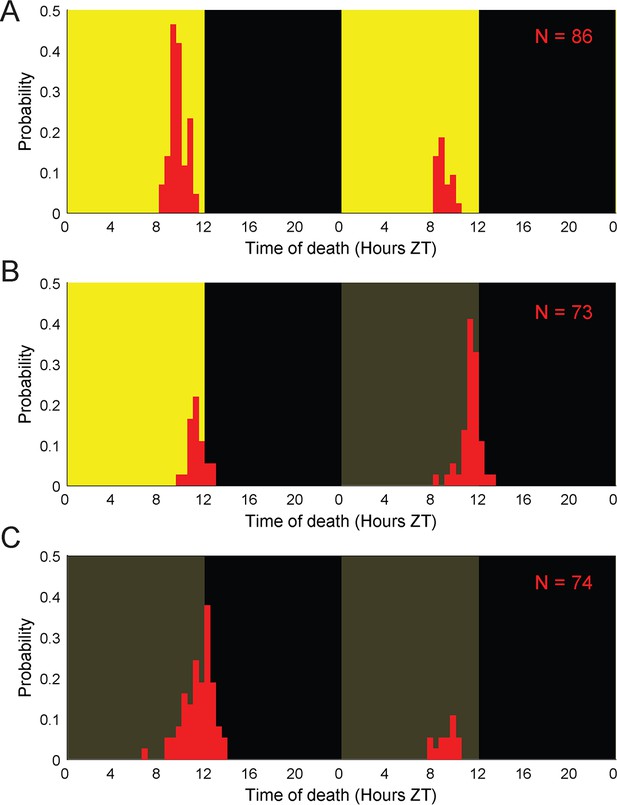
E.muscae-infected flies die at specific times of day in the absence of proximal lighting cues.
Distributions of time of death for Canton-S flies killed by E. muscae with (A) continuous L:D cues, (B) L:D cues terminating after 96 hr, or (C) L:D cues terminating after 72 hr. Background color indicates state of visible illumination (black or gray = no light; yellow = light; gray = entrainment conditions). Time of death for zombies was manually determined as the time of the last movement from the y position trace. Number of total cadavers observed in each panel is indicated in top right.
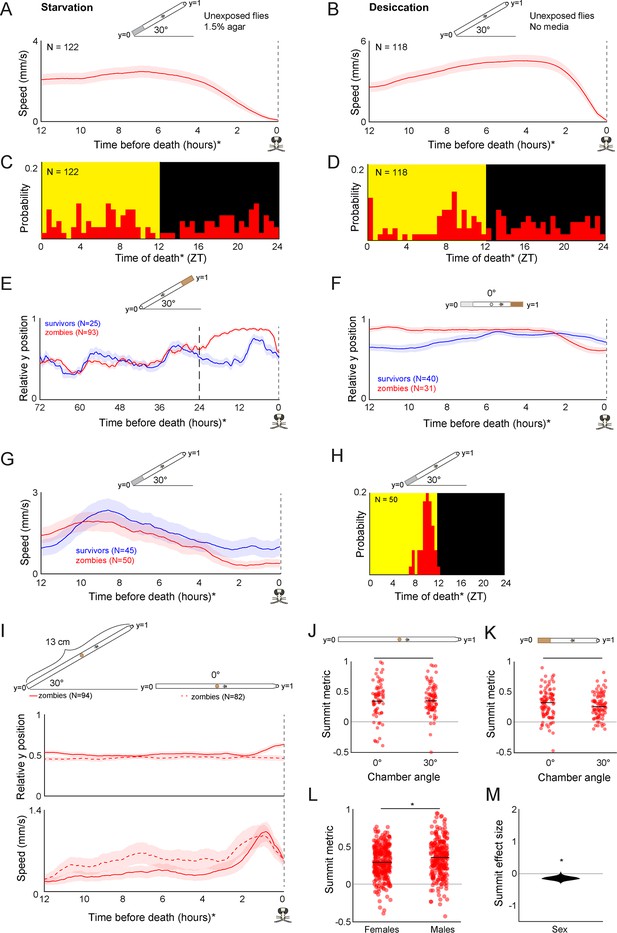
Additional features of summiting behavior in the custom behavior assay.
(A) Unexposed, wild-type flies that die of starvation do not exhibit a burst in activity beginning 2.5 hr prior to their last movement, and (C) do not exhibit specific circadian timing of death. Flies in this experiment were provided with non-nutritive agar instead of food at position y = 0. (B) Unexposed, wild-type flies that die of desiccation do not exhibit a burst in locomotor activity beginning 2.5 hr prior to their last movement, and (D) do not exhibit specific circadian timing of death. Flies in this experiment were provided food nor agar at position y = 0. (E) Mean y position of zombie flies diverges from survivors beginning around 24 hr prior to death when nutritive food is positioned at y = 1. (F) Compared to survivors, zombie flies prefer nutritive (5% sucrose, 1.5% agar) over non-nutritive (0% sucrose, 1.5% agar) media in their final 12 hours of life prior to the onset of summiting at 2.5 hours before death. (G) Zombie flies housed on non-nutritive food for their final 24 hr do not exhibit summiting behavior and (H) show typical timing of last death (compare to Figure 1E). (I) When housed in tall arenas (13 cm) with food placed in the middle, zombie flies tend to move slightly upward when the arena is tilted at 30°, but not 0°. Zombies in arenas at both angles exhibit a burst of speed prior to death (bottom). (J, K) Summit metric (see Figure 1I) does not vary for zombies housed in chambers at 0° versus 30° in tall or standard chambers. (L) Summit metric for female versus male Canton-S zombies. (M) Distribution of estimated effect size of sex on summiting. For (A, B, E-K), diagrams depicting behavior arena setup are shown above corresponding plots. For L and M, behavior arenas were in standard configuration (as in Figure 1F)* = p<0.05; **=p<0.01; *** p<0.001 by two-tailed t-test.
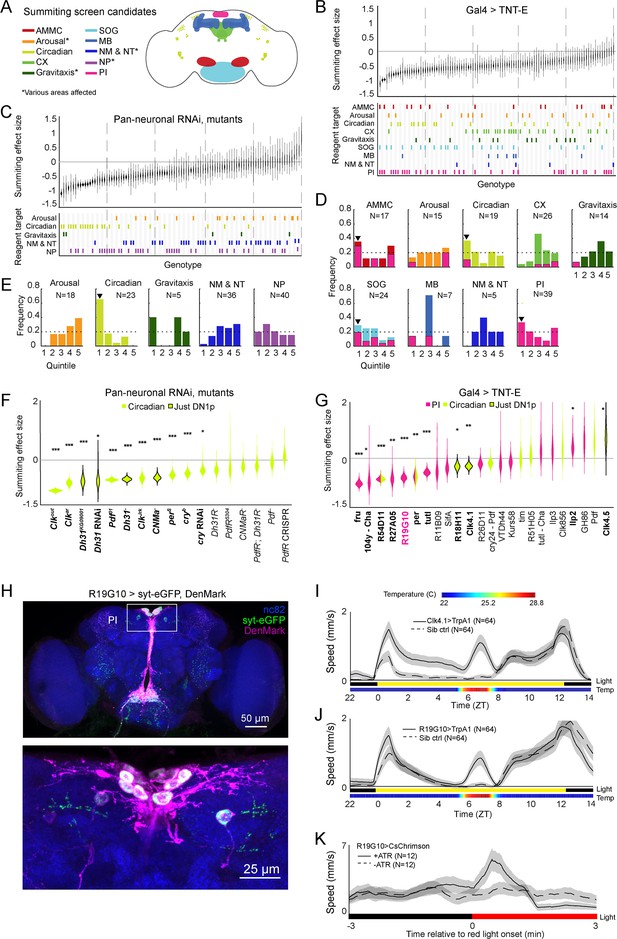
Identification of host circuits and genetic components involved in summiting behavior.
(A) Regions and pathways targeted in the candidate screen. AMMC = antennal mechanosensory and motor center; CX = central complex; SOG = subesophageal ganglion; MB = mushroom body; NM & NT = neuromodulator or neurotransmitter; NP = neuropeptide; PI = pars intercerebralis. (B and C) Effects of neuronal disruption (B; 12<N<111, median N=35) or gene knockdown or mutagenesis (C; 10<N<182, median N=46) on summiting. Above: Summiting effect size estimate distributions as estimated by bootstrapping. Experimental groups are ordered by mean effect (negative to positive). Below: gene function and brain region annotations associated with each screened reagent. See Supplementary file 1 for genotype and annotation details. Solid gray line indicates an effect size of zero. Dashed vertical lines separate ranked data into quintiles. (D and E) Frequency of annotations by quintile for (B) and (C), respectively. The number of lines screened (N) is indicated for each annotation. Dashed line indicates the frequency of annotation expected from a null, uniform distribution. Black arrowheads highlight annotations that are overrepresented in the first quintile. For (D), pink overlays indicate the portion of line annotations that are co-annotated for expression in the PI. (F and G) Summiting effect size estimate distributions of disrupting specific circadian genes (F; 19<N<182, median N=62) or circadian and/or PI neurons (G; 11<N<111, median N=46) compared to genotype-matched controls. Lines are ordered by effect size. Pink indicates Gal4 expression in the PI, lime circadian Gal4 lines and genes, and black outlines expression only in DN1ps. Asterisks indicate statistically significant effects on summiting behavior by a two-tailed t-test (*=p<0.05; **=p<0.01; *** p<0.001). R19G10 is highlighted in pink to emphasize its subsequent use as the main PI reagent. See Supplementary file 2 for genotypes and matched controls. (H) Maximum z-projections of brains showing pre- (synaptotagmin; syt-eGFP) and post- (DenMark) synaptic compartments of R19G10 neurons. Bruchpilot (nc-82) staining (blue) visualizes neuropil. Above: brain imaged from anterior. Below: another brain, imaged from the posterior. (I and J) Mean speed of unexposed flies vs time for Clk4.1>TrpA1 and R19G10>TrpA1 genotypes and sibling controls, respectively. Shaded regions are +/− 1 standard error of the mean. Bars along the x-axis indicate the state of visible illumination (above) and temperature (below). (K) Red light onset-triggered mean speed across flies of unexposed R19G10>CsChrimson flies versus time. All trans retinal (ATR) indicates control flies not fed CsChrimson cofactor. Shaded regions are +/− 1 standard error of the mean. Bar along the x-axis indicates lighting conditions (black: darkness, red: red-light illumination).

Additional experiments assessing summiting after clock neuron and R19G10 disruption.
(A–C) Plots visualizing summiting for manipulations targeting R19G10 and DN1p neurons. Top: Mean zombie baseline-corrected speed versus time. Shaded area is +/− 1 standard error. Bottom: Individual SM (circles) and median SM (line). *=p<0.05; **=p<0.01; ***p<0.001 by two-tailed t-test. Clk4.1 drives expression in 8–10 and R18H11 drives expression in 7–8 of DN1p neurons per hemisphere, respectively (Kunst et al., 2014; Zhang et al., 2010). (D) Left: Summiting effect size estimate distributions for Gal4/effector combinations targeting clock neurons. Sample size of experimental animals is in black, and controls in gray. Right: Diagrams depicting target cells of tested reagents following Shafer et al., 2022. Violin plot outline colors match diagrams. *=p<0.05; **=p<0.01; ***p<0.001 by two-tailed t-test. (E) Confocal micrographs demonstrating ablation of Pdf-expressing neurons in Pdf-Gal4>hid animals. (F) Summiting effect size estimate distributions when targeting components of the circadian locomotor output pathway identified by Cavanaugh et al., 2014. *=p<0.05; **=p<0.01; ***p<0.001 by two-tailed t-test.

Additional experiments assessing the sufficiency of DN1p and R19G10 neuron activation for increased locomotion.
(A–D) Mean speed of unexposed flies with thermogenetically activated DN1p or R19G10 neurons (solid line) versus sibling controls (dashed line) separated by sex. The presence or absence of light is indicated below the horizontal axis with a yellow or black bar, respectively. The measured temperature of the room is shown as a heat map below the light cue indicator. Shaded regions are +/− 1 standard error. For A and B, Clk4.1-Gal4 was used to drive expression in DN1ps. (E and F) Kernel density estimates of the distribution of log-speed for R19G10 >CsChrimson flies with (F) or without (E) of dietary all trans retinal (ATR) in the presence of absence of 5 Hz pulsed red light stimulus (OFF and ON, respectively). A bimodal distribution of speeds is observed regardless of ATR and light treatment. The lower peak (centered around 0) corresponds to non-walking and the higher peak (centered around 6mm/s) corresponds to walking flies. (G) Mean speed across flies of unexposed R19G10>CsChrimson flies before and after the second presentation of red light in the experiment described in Figure 2K. ATR+ flies show a smaller increase in locomotion upon red light stimulus, consistent with CsChrimson depolarization block or adaptation within the stimulated circuit.
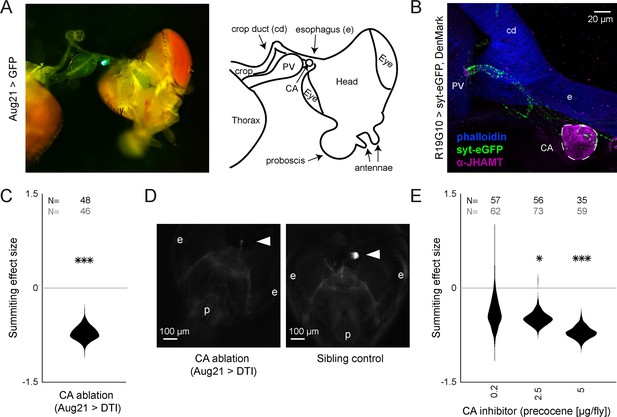
R19G10 (PI-CA) neurons project to the corpora allata, which are required for summiting behavior.
(A) Left: Composite micrograph of dissected Aug21>GFP fly, showing GFP fluorescence in the corpora allata (CA) overlaid on bright field image. Right: Diagram of A with anatomical features labeled. PV = proventriculus. (B) Representative confocal micrograph of immunostained RC from an R19G10>syt-eGFP, DenMark fly. Synaptic terminals are visible as green puncta, including in the CA. Magenta is anti-JHAMT and marks the CA. Blue phalloidin counterstain marks actin. Labels as in A. (C) Summiting effect size estimate distribution of ablating the CA with diphtheria toxin (DTI). Effect size is calculated relative to effector-less sibling controls. (D) Representative micrographs of CA-ablated and effector-less, sibling, temperature-matched control flies (additional examples in Figure 3—figure supplement 1D). White arrows indicate the expected location of CA. e = eye, p = proboscis. (E) Summiting effect size estimate distributions of various concentrations of the CA-ablating drug precocene. Effect size is calculated relative to vehicle (acetone) control. For (C and E), effect sizes were estimated as in Figure 2; asterisks indicate statistically significant effects (*=p<0.05; **=p<0.01; ***p<0.001) by two-tailed t-test. Sample sizes of experimental and control experiments are given in black and gray, respectively.

Supporting data for juvenile hormone involvement in summiting.
(A) Summiting effect size estimate distribution for Akh- mutants, showing no effect compared to controls. Sample size of experimental animals is in black, and controls in gray. *=p<0.05; **=p<0.01; ***p<0.001 by two-tailed t-test. (B) As in (A) for the ablation of the corpora allata (CA) using NiPP1 effector (per Yamamoto et al., 2013). A significant reduction in summiting is seen compared to controls. (C) Confocal micrographs of the anterior foregut and retrocerebral complexes from Aug21>GFP, NiPP1 flies, and Aug21>GFP sibling controls. Green channel is GFP and magenta anti-actin phalloidin counterstain. Arrowheads indicate the observed or expected location of CA. Scale bar is 100 microns. CAs in Aug21>GFP, NiPP1 animals ranged from intact to absent. (D) Compound epifluorescence micrographs of the head, anterior foregut, and retrocerebral complexes of tub-Gal80, Aug21> GFP, DTI flies. In animals in which DTI expression was induced by thermal inactivation of the Gal80 repressor, only one sample showed potential residual CA; all other animals lacked CA.CA were observed in all controls. e = eye, p = proboscis.

Additional experiments examining juvenile hormone involvement in summiting.
(A) Synthesis pathway for juvenile hormone. Enzymes catalyzing each step are shown at right. HMG-CoA reductase (blue) is the target of the drug fluvastatin. (B) Summiting effect size estimate distribution for E. muscae-exposed flies fed with fluvastatin (250 µg/well) starting 72 hr after exposure to E. muscae. *=p<0.05; **=p<0.01; ***p<0.001 by two-tailed t-test. Sample size of experimental animals is in black, and controls in gray. (C) Times of death for flies fed fluvastatin 72 hr after exposure and killed by E. muscae (i.e. with sporulation). (D) Stacked bar plots of survival outcomes for fluvastatin-treated flies and controls, at 72 hr and 24 hr after E. muscae exposure. Flies were manually assessed as alive or dead at the end of behavior tracking; dead flies were further classified as having sporulated (Sp+) or not (Sp−). E. muscae-exposed N = 91–108; unexposed N=20–37. (E) Times of death for flies fed fluvastatin 24 hr after exposure, which showed no signs of sporulation and died roughly uniformly throughout the day. (F) Stacked bar plots of survival outcomes for flies treated topically with the indicated amount of precocene or vehicle control (acetone) with or without exposure to E. muscae. E. muscae-exposed N = 97–100; unexposed N = 28–31. (G) Times of death for flies exposed to E. muscae and treated with three concentrations of precocene or acetone at 72 hr after exposure. (H) Summiting effect size estimate distributions for topical application of the JH analog methoprene at two different doses. This had no effect compared to vehicle (acetone) controls. (I) Summiting effect size estimate distributions for co-applied methoprene or dietary pyriproxyfen. These treatments did not rescue summiting deficits induced by precocene.
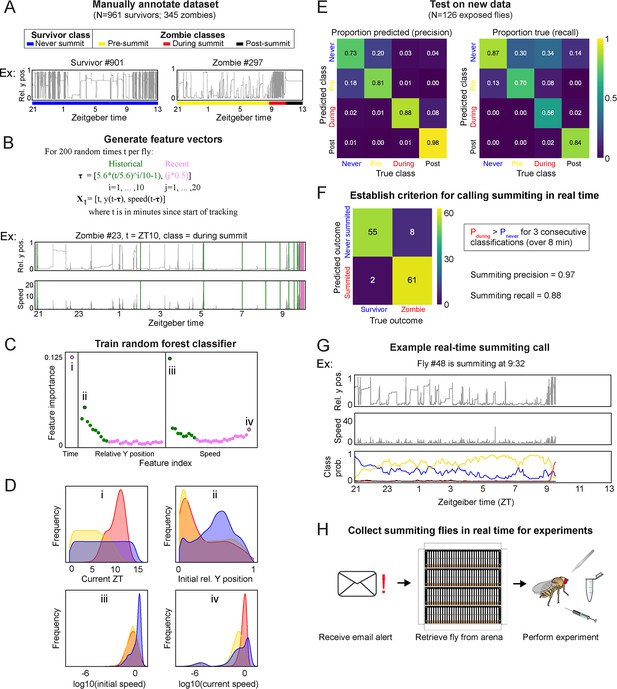
A random-forest classifier (RFC) for identifying summiting flies in real-time.
(A) Top: classes learned by the classifier for zombies were pre-summiting=prior to the onset of summiting (yellow), during summiting = after the onset of summiting but before the time of death (red), and post-summiting=after the time of death (black). For survivors, there was one class, never-summiting (blue). Bottom: annotations of these classes on example y position trajectories from a survivor (left) and zombie (right). (B) Feature vectors (Xt) generated for 200 random time points (t) for each fly. Vertical green and pink lines in the example trajectory below indicate the historical (green) and recent (pink) values selected for the feature vector. (C) Feature importance for classification of the 61 input variables. Roman numerals correspond to plots in subsequent panels. (D) Distributions of important feature variables, visualized with kernel density estimation, across never summiting (blue), pre-summiting (yellow), and summiting (red) classes within the training dataset. (E) Confusion matrices for precision (left) and recall (right) performance of the classifier on the test dataset. (F) Confusion matrix for the survivor and zombie outcomes after implementing the real-time zombie-calling criterion. (G) Example real-time behavior and class probability trajectories for a zombie fly, ending on the frame when it was called as a zombie. (H) Summarized experimental workflow using the real-time classifier.

Development of a real-time random forest classifier for summiting behavior.
(A) Confusion matrices calculated using the validation dataset (25% of the total ground truth data). (B) Example classifications of a summiting (zombie) and a non-summiting (survivor) fly over an entire behavior-tracking experiment. Top and middle plots for each example are the fly’s y position and speed; bottom plot is the class probabilities for the fly at each timepoint. In the summiting example, the fly is consistently classified as pre-summiting (i.e. will become a zombie before the next occurrence of the sunset) starting as early as ZT22 the evening prior to death. This fly was classified as summiting from approximately ZT9.25 to ZT10.75 the following day, and post-summiting (i.e. dead) ZT11 onward. In contrast, the non-summiting example fly was consistently classified as never-summiting (i.e. would live through the next sunset) for the duration of the experiment.
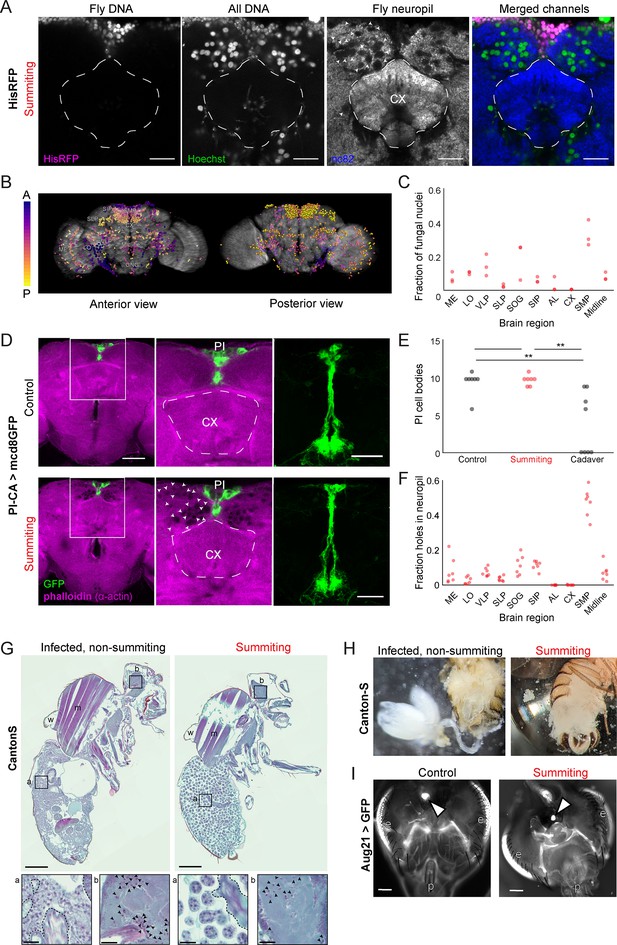
E.muscae densely occupies the superior medial protocerebrum (SMP) during summiting without apparent degradation of pars intercerebralis to corpora allata (PI-CA) neurons or corpora allata (CA).
(A) Confocal micrographs of the superior medial protocerebrum (SMP) from summiting His-RFP fly. Non-fly nuclei (Hoechst+, HisRFP−) are large compared to fly neuronal nuclei (Hoechst+, HisRFP+) and sit in ‘holes’ in the neuropil visible in the nc82 counterstain channel. Scale bar is 20 microns. (B) Whole brain invasion pattern of E. muscae (same brain as A). Nuclei are colored according to depth from anterior (A) to posterior (P). (C) Distribution of fungal nuclei across brain regions (N=3). AL = antennal lobe, SIP = superior intermediate protocerebrum, SLP = superior lateral protocerebrum, CX = central complex, VLP = ventrolateral protocerebrum, SOG = subesophageal ganglion, LO = lobula, ME = medulla, midline = cells along the midline of the brain not in any other region. (D) Confocal micrographs of PI-CA neurons (green) and phalloidin counterstain (magenta) in control and summiting flies. Left: sagittal planes of the central brain. Holes are apparent (in the phalloidin channel) in the SMP of the summiting brain, marked by arrowheads in one hemisphere. Holes are absent in CX of summiting brains and all control brain regions. Middle: Inset from the left. Right: Maximum z-projections of GFP channel from full brain z-stacks. PI-CA morphology appears the same in summiting and control brains. Scale bars are 50 microns. (E) Counts of PI-CA cell bodies in control (unexposed), summiting, or recently-killed (cadaver) PI-CA >mcd8 GFP flies (** indicates p<0.01 by a two-tailed t-test). (F) Distribution of ‘holes’ across brain regions. Abbreviations as in C. (G) Safranin and fast green stained sections of paraffin-embedded Canton-S flies. Left: Infected, non-summiting fly (96 hr after exposure to fungus). Right: summiting, E. muscae-infected fly. a=abdomen, b=brain, w=wing, m=muscle. Scale bars are 200 microns. Insets of the abdomen and brain are shown for each fly below (scale bars are 25 microns). Host tissues are outlined in dashed black; black arrowheads indicate fungal nuclei. (H) Micrographs of dissected abdomens of 96-hour post-exposure non-summiting (left) and summiting (right) female flies. Gut and reproductive organs are still present in the non-summiting fly, but are absent in the summiting fly. Clumps of spherical fungal cells are visible in the dissection saline of summiting but not non-summiting fly. (I) Fluorescence images of dissected Aug21 >GFP flies. White arrowheads indicate CA. p=proboscis, e=eyes. Scale bars are 100 microns. Additional examples are available in Figure 5—figure supplement 1F.
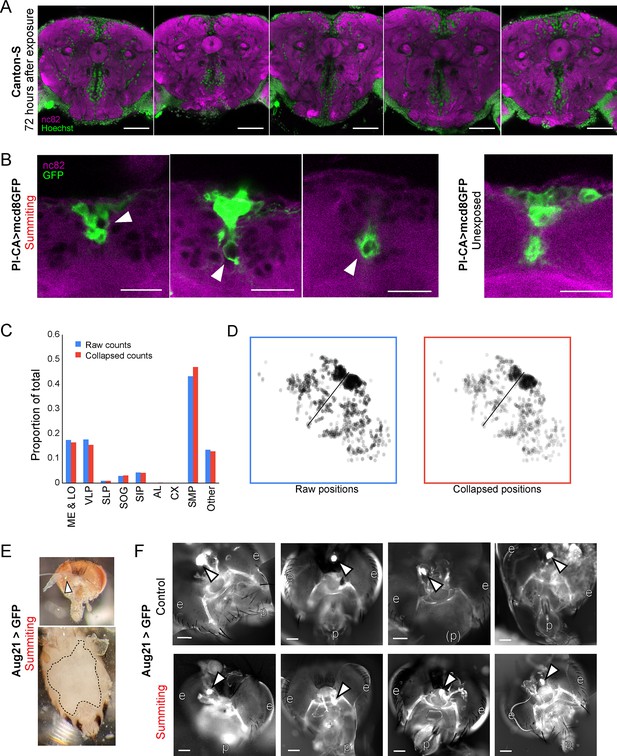
Supporting data for host morphology during E.muscae infection.
(A) Confocal micrographs of five infected brains 72 hr after E. muscae exposure. Green is Hoechst and marks all nuclei. Magenta is the synaptic marker nc82. Fungal cells are present in the SMP and absent from the central complex (CX). Scale bar is 50 microns. (B) Confocal micrographs if PI-CA>mcd8GFP (green) brains during summiting and unexposed to E muscae. White arrowheads indicate apparent displacement of pars intercerebralis to corpora allata (PI-CA) processes by fungal cells. Scale bar=25 microns. (C) Distribution of E. muscae nuclei across brain regions from a whole-brain confocal volume (2-micron z-step), using two counting methods. Counting every nucleus seen in a single z-slice as one cell (‘Raw counts’), and merging cell bodies if they have the same x- and y-position (to within 2 µm) and appear within 10 microns of each other along the z-axis (‘Collapsed counts’) produces very similar distribution estimates (N=1,426 nuclei). (D) Z-projections of nuclei as counted by the Raw and Collapsed methods. (E) Micrographs of dissected summiting Aug21>GFP fly. Top: Head (brightfield overlaid with GFP channel). White arrow points to the corpora allata (CA). Bottom: Abdomen (brightfield) with ventral cuticle peeled back. Dashed line indicates the edges of the fly’s remaining cuticle. Only fungal tissues and no host organs are visible inside the dashed line. (F) Additional whole-mount preparations of Aug21>GFP heads and retrocerebral complexes in control (non-summiting) and summiting flies, as in Figure 5. White arrows indicate CA; p=proboscis, e=eyes. Scale bars each 100 microns.
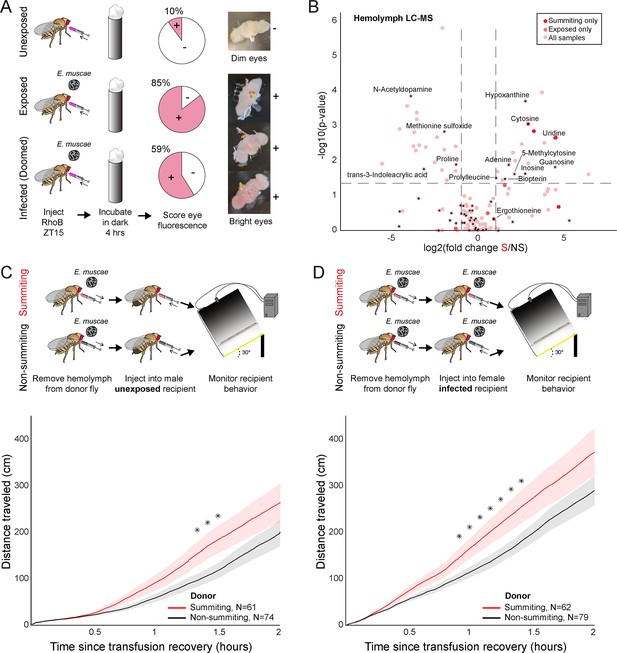
Hemolymph of summiting flies has a distinct metabolome and induces locomotion.
(A) Blood-brain barrier (BBB) permeability of E. muscae exposed (96 hr) or unexposed flies assessed as the portion of flies with eye fluorescence after Rhodamine B (RhoB) injection (N=40–50 per group). Infected (doomed) flies are exposed flies with fungal growth visible by the eye through the abdominal cuticle, all of whom would go on to summit within 22 hr. Bright-eyed flies (+) had visible RhoB uptake. Representative brains from dim and bright-eyed flies are shown at right. (B) Volcano plot of hemolymph metabolites detected by LC-MS mass spectrometry in summiting (S) versus exposed, non-summiting (NS) flies. Putative identifications are given for selected compounds. See Supplementary file 3 for compound abundances and statistical details. (C and D) Total distance traveled versus time for flies receiving a transfusion of hemolymph from summiting donors. Diagrams at the top indicate the hemolymph transfusion experiment configuration. Shaded areas indicate +/− 1 standard error. Asterisks indicate p-values <0.05 for two-tailed t-tests performed at each timepoint.
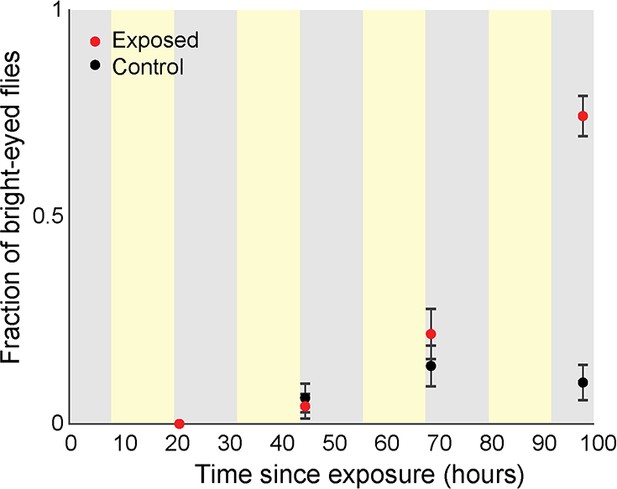
Blood-brain permeability as a function of time since exposure.
Flies were injected with 1.44 mg/mL Rhodamine B (RhoB) and then allowed to incubate for four hours prior to scoring eye fluorescence. Experimenters were blind to treatment type. Error bars are the estimated standard error of the binomial proportion. Background shading indicates entrained lighting conditions experienced by all flies (yellow = lights on, gray = lights off).
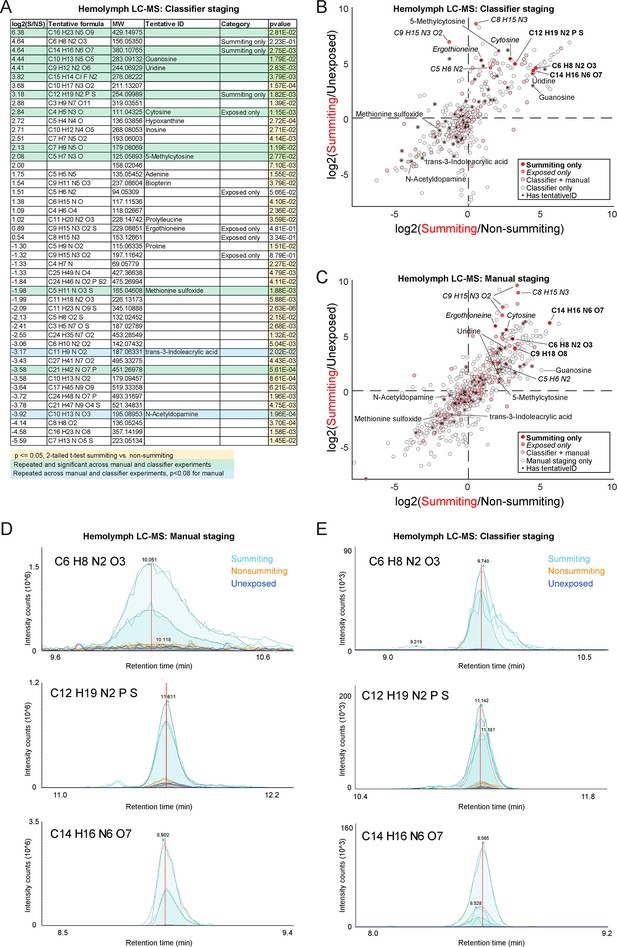
Metabolomics of summiting flies.
(A) Compounds of interest from classifier-staged metabolomics experiment. Compounds are included if at least one of three conditions is met: (1) they are significantly different between summiting and non-summiting, exposed animals, and exhibit at least a twofold change between these treatments, (2) they were only detected in summiting flies (‘Summiting only’), or (3) they were only detected in exposed flies (‘Exposed only’; summiting or non-summiting), but not unexposed flies. p-values <0.05 are highlighted in yellow. Compounds highlighted in green were observed to be significantly different between summiting and non-summiting flies in both the manual and classifier-based experiments (p< = 0.05 by a two-tailed t-test); compounds in blue were significant in the classifier-staged experiment and fell just short of significant (p<0.08) in the manual experiment. (B) Scatter plot of the log2-fold-change of compound abundance in summiting compared to unexposed flies versus the log2-fold-change of compound abundance in summiting compared to exposed, non-summiting flies. Data from the classifier-staged experiment. Compounds in the darkest red were observed only to have chromatogram peaks above baseline (1e3 intensity counts) in summiting samples and not in any other samples. Compounds in medium red were only observed to have real peaks in exposed (summiting or non-summiting) but not unexposed samples across both experiments. Compounds in light red were observed in all experimental groups of both experiments (molecular weight within five parts per million between experiments). Compounds in white were observed in the classifier-based experiment but were not observed in the manually-staged experiment (absolute difference in molecular weight greater than five ppm). Compounds that were observed in both experiments, showed significant differences between summiting and non-summiting flies, and were tentatively identified, are labeled with the compound name or molecular formula. (C) As in (B), for the manually staged experiment. (D,E) Raw chromatogram peaks for three compounds assessed to only be present in summiting flies in manually-staged (D) and classifier-staged (E) metabolomics experiments. Different peaks of the same color are biological replicates. Putative formulas as determined from the measured mass given in the upper left corner for each plot.
-
Figure 6—figure supplement 2—source data 1
Compounds over- or under-abundant in the hemolymph of summiting flies from classifier-staged metabolomics experiment.
- https://cdn.elifesciences.org/articles/85410/elife-85410-fig6-figsupp2-data1-v2.xlsx
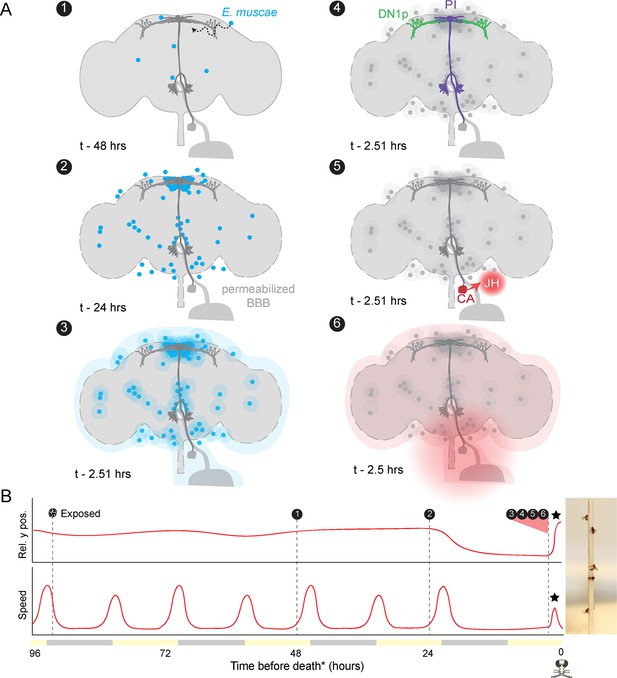
Proposed sequence of E.muscae-induced summiting mechanisms in zombie flies.
(A) Events in the host brain leading to E. muscae-induced summiting. (1) E. muscae cells are present in the brain as soon as 48 hr prior to death (Elya et al., 2018). (2) By 24 hr prior to death, the fungus is present at a high density in the superior medial protocerebrum (SMP). This corresponds to the ‘infected (doomed)’ status of flies in Figure 6. (3) E. muscae alters the hemolymph (perhaps by secreting compounds, as depicted here) to trigger the onset of summiting behavior. (4) Hemolymph-borne factors alter the activity of the circadian network/DN1p and pars intercerebralis to corpora allata (PI-CA) neurons. (5) Juvenile hormone (JH) is released from the corpora allata (CA) following changes in PI-CA activity. (6) Increased JH levels drive an increase in locomotion. The dashed outline of the brain becomes more prominent between steps 1 and 3 to reflect an increase in blood-brain barrier (BBB) permeability over these timepoints. (B) Left: Timeline of events depicted in (A) overlaid on cartoon plot of average relative y position (above) and speed (below) for zombie flies. Summiting is indicated by a black star; death (time of the last movement) is indicated by a fly ‘skull.’ Right: Zombie flies summited on a wooden dowel.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| antibody | anti-Chicken-AF488 (goat polyclonal) | Thermo Fisher | Cat#: A-11039, RRID:AB_2534096 | IF(1:800) |
| antibody | anti-dsRed (rabbit polyclonal) | Takara Bio | Cat#: 632496, RRID:AB_10013483 | IF:(250) |
| antibody | anti-GFP (chicken polyclonal) | Aves Labs | Cat#: GFP-1020, RRID:AB_10000240 | IF(1:4000) |
| antibody | anti-Guinea Pig-AF568 (goat polyclonal) | Thermo Fisher | Cat#: A-11075, RRID:AB_2534119 | IF(1:400) |
| antibody | anti-JHAMT (guinea pig polyclonal) | Niwa et al., 2008 | IF(1:1000) | |
| antibody | anti-Mouse-Cy5 (goat polyclonal) | Millipore | Cat#: AP500S, RRID:AB_805361 | IF(1:400) |
| antibody | anti-nc82 (mouse monoclonal) | Iowa Developmental Studies Hybridoma Bank | Cat#: nc82, RRID:AB_2314866 | IF(1:40) |
| antibody | anti-Rabbit-AF568 (goat polyclonal) | Thermo Fisher | Cat# A-11011, RRID:AB_143157 | IF(1:250) |
| genetic reagent (D. melanogaster) | 104y-Gal4 | Bloomington Drosophila Stock Center | BDSC:81014 | |
| genetic reagent (D. melanogaster) | 104y-Gal4; Cha-Gal80 | Derived from BDSC:81014 & Cha-Gal80 | ||
| genetic reagent (D. melanogaster) | acj6- | Bloomington Drosophila Stock Center | BDSC:30025 | |
| genetic reagent (D. melanogaster) | acj6-Gal4 | Bloomington Drosophila Stock Center | BDSC:30025 | |
| genetic reagent (D. melanogaster) | Akh- | Bloomington Drosophila Stock Center | BDSC:84448 | |
| genetic reagent (D. melanogaster) | AstC- | Bloomington Drosophila Stock Center | BDSC:84453 | |
| genetic reagent (D. melanogaster) | Bl/CyO; tub-Gal80(ts) | Kristin Scott (McGuire et al., 2004) | ||
| genetic reagent (D. melanogaster) | C(1)Dxyfv(X^X)/Y; Aug21-Gal4, UAS-GFP/CyO | Rochele Yamamoto (Yamamoto et al., 2013) | ||
| genetic reagent (D. melanogaster) | c17-Gal4 | Bloomington Drosophila Stock Center | BDSC:39690 | |
| genetic reagent (D. melanogaster) | c41-Gal4 | Bloomington Drosophila Stock Center | BDSC:30834 | |
| genetic reagent (D. melanogaster) | c708a-Gal4 | Bloomington Drosophila Stock Center | BDSC:50743 | |
| genetic reagent (D. melanogaster) | Canton-S | Liming Wang | ||
| genetic reagent (D. melanogaster) | CCha1- | Bloomington Drosophila Stock Center | BDSC:84458 | |
| genetic reagent (D. melanogaster) | CCKR-17D1- | Bloomington Drosophila Stock Center | BDSC:84462 | |
| genetic reagent (D. melanogaster) | CCLKR-17D3- | Bloomington Drosophila Stock Center | BDSC:84463 | |
| genetic reagent (D. melanogaster) | Cha-Gal80/TM3, Sb | Toshihiro Kitamoto (Kitamoto, 2002) | ||
| genetic reagent (D. melanogaster) | Clk4.1-Gal4 | Bloomington Drosophila Stock Center | BDSC:36316 | |
| genetic reagent (D. melanogaster) | Clk4.5-Gal4 | Bloomington Drosophila Stock Center | BDSC:37526 | |
| genetic reagent (D. melanogaster) | Clk856-Gal4/CyO; 911-QF, QUAS-FLP/TM6, Sb | David Cavanaugh (Nettnin et al., 2021) | ||
| genetic reagent (D. melanogaster) | Clk856-Gal4/CyO; MKRS/TM6B | Daniel Cavanaugh (Gummadova et al., 2009) | ||
| genetic reagent (D. melanogaster) | Clkar | Bloomington Drosophila Stock Center | BDSC:24513 | |
| genetic reagent (D. melanogaster) | ClkJrk | Bloomington Drosophila Stock Center | BDSC:24515 | |
| genetic reagent (D. melanogaster) | Clout | Bloomington Drosophila Stock Center | BDSC:56754 | |
| genetic reagent (D. melanogaster) | CNMa- | Bloomington Drosophila Stock Center | BDSC:84485 | |
| genetic reagent (D. melanogaster) | CNMaR- | Bloomington Drosophila Stock Center | BDSC:84486 | |
| genetic reagent (D. melanogaster) | cry-Gal4.Z16 | Bloomington Drosophila Stock Center | BDSC:24514 | |
| genetic reagent (D. melanogaster) | cry-Gal4.Z24 | Bloomington Drosophila Stock Center | BDSC:24774 | |
| genetic reagent (D. melanogaster) | cry02 | Bloomington Drosophila Stock Center | BDSC:86267 | |
| genetic reagent (D. melanogaster) | cryb | Bloomington Drosophila Stock Center | BDSC:80921 | |
| genetic reagent (D. melanogaster) | cyc01 | Bloomington Drosophila Stock Center | BDSC:80929 | |
| genetic reagent (D. melanogaster) | DAT- | Bloomington Drosophila Stock Center | BDSC:25547 | |
| genetic reagent (D. melanogaster) | Dh31- | Bloomington Drosophila Stock Center | BDSC:84490 | |
| genetic reagent (D. melanogaster) | Dh31KG09001 | Bloomington Drosophila Stock Center | BDSC:16474 | |
| genetic reagent (D. melanogaster) | DH31R- | Bloomington Drosophila Stock Center | BDSC:84491 | |
| genetic reagent (D. melanogaster) | disco1 | Bloomington Drosophila Stock Center | BDSC:5682 | |
| genetic reagent (D. melanogaster) | DNc01 | Janelia Research Center | JRC:SS04161 | |
| genetic reagent (D. melanogaster) | DNc02 | Janelia Research Center | JRC:SS02395 | |
| genetic reagent (D. melanogaster) | DNp01 | Janelia Research Center | JRC:SS00726 | |
| genetic reagent (D. melanogaster) | DNp01 | Janelia Research Center | JRC:SS00727 | |
| genetic reagent (D. melanogaster) | DNp01 | Janelia Research Center | JRC:SS02299 | |
| genetic reagent (D. melanogaster) | Dsk- | Bloomington Drosophila Stock Center | BDSC:84497 | |
| genetic reagent (D. melanogaster) | elav-Gal4; UAS-Dcr2 | Bloomington Drosophila Stock Center | BDSC:25750 | |
| genetic reagent (D. melanogaster) | forS | Bloomington Drosophila Stock Center | BDSC:76120 | |
| genetic reagent (D. melanogaster) | fru-Gal4 | Bloomington Drosophila Stock Center | BDSC:30027 | |
| genetic reagent (D. melanogaster) | GH86-Gal4 | Bloomington Drosophila Stock Center | BDSC:36339 | |
| genetic reagent (D. melanogaster) | gl60j | Bloomington Drosophila Stock Center | BDSC:509 | |
| genetic reagent (D. melanogaster) | GLSNP3375-Gal4 | Kyoto Drosophila Stock Center | KDSC:104479 | |
| genetic reagent (D. melanogaster) | His-RFP | Bloomington Drosophila Stock Center | BDSC:23651 | |
| genetic reagent (D. melanogaster) | Hug-Gal4 | Bloomington Drosophila Stock Center | BDSC:58769 | |
| genetic reagent (D. melanogaster) | iav-Gal4 | Bloomington Drosophila Stock Center | BDSC:52273 | |
| genetic reagent (D. melanogaster) | Ilp1-Gal4 | Bloomington Drosophila Stock Center | BDSC:66005 | |
| genetic reagent (D. melanogaster) | Ilp2-Gal4 | Bloomington Drosophila Stock Center | BDSC:37516 | |
| genetic reagent (D. melanogaster) | Ilp3-Gal4 | Bloomington Drosophila Stock Center | BDSC:52660 | |
| genetic reagent (D. melanogaster) | Ilp5-Gal4 | Bloomington Drosophila Stock Center | BDSC:66008 | |
| genetic reagent (D. melanogaster) | JO-ACE-Gal4 | Kyoto Drosophila Stock Center | KDSC:113902 | |
| genetic reagent (D. melanogaster) | JO-CE-Gal4 | Kyoto Drosophila Stock Center | KDSC:113878 | |
| genetic reagent (D. melanogaster) | JO15-Gal4 | Bloomington Drosophila Stock Center | BDSC:6753 | |
| genetic reagent (D. melanogaster) | Kurs58-Gal4 | Bloomington Drosophila Stock Center | BDSC:80985 | |
| genetic reagent (D. melanogaster) | MB010B-Gal4 | Janelia Research Center | JRC:MB010B | |
| genetic reagent (D. melanogaster) | Mmp2NP0509-Gal4 | Kyoto Drosophila Stock Center | KDSC:103625 | |
| genetic reagent (D. melanogaster) | nan-Gal4 | Bloomington Drosophila Stock Center | BDSC:24903 | |
| genetic reagent (D. melanogaster) | nan36a | Kristin Scott (Kim et al., 2003) | ||
| genetic reagent (D. melanogaster) | NPF- | Bloomington Drosophila Stock Center | BDSC:84549 | |
| genetic reagent (D. melanogaster) | Oamb- | Bloomington Drosophila Stock Center | BDSC:22758 | |
| genetic reagent (D. melanogaster) | OctBeta1R- | Bloomington Drosophila Stock Center | BDSC:18589 | |
| genetic reagent (D. melanogaster) | Octbeta2R- | Bloomington Drosophila Stock Center | BDSC:18896 | |
| genetic reagent (D. melanogaster) | OctBeta3R- | Bloomington Drosophila Stock Center | BDSC:24819 | |
| genetic reagent (D. melanogaster) | Pdf- | Bloomington Drosophila Stock Center | BDSC:84561 | |
| genetic reagent (D. melanogaster) | Pdf-Gal4 | Bloomington Drosophila Stock Center | BDSC:6899 | |
| genetic reagent (D. melanogaster) | Pdf-Gal80, cry24-Gal4 | Bloomington Drosophila Stock Center | BDSC:80940 | |
| genetic reagent (D. melanogaster) | Pdf01 | Bloomington Drosophila Stock Center | BDSC:26654 | |
| genetic reagent (D. melanogaster) | PdfR- | Bloomington Drosophila Stock Center | BDSC:84705 | |
| genetic reagent (D. melanogaster) | PdfR-; DH31R- | Derived from BDSC:84705 & BDSC:84491 | ||
| genetic reagent (D. melanogaster) | PdfR-Gal4 | Bloomington Drosophila Stock Center | BDSC:68215 | |
| genetic reagent (D. melanogaster) | PdfR5304 | Bloomington Drosophila Stock Center | BDSC:33068 | |
| genetic reagent (D. melanogaster) | per-Gal4 | Bloomington Drosophila Stock Center | BDSC:7127 | |
| genetic reagent (D. melanogaster) | per01 | Bloomington Drosophila Stock Center | BDSC:80928 | |
| genetic reagent (D. melanogaster) | per30 | Bloomington Drosophila Stock Center | BDSC:63136 | |
| genetic reagent (D. melanogaster) | perS | Bloomington Drosophila Stock Center | BDSC:80919 | |
| genetic reagent (D. melanogaster) | ple-Gal4 | Bloomington Drosophila Stock Center | BDSC:8848 | |
| genetic reagent (D. melanogaster) | Procc04750 | Bloomington Drosophila Stock Center | BDSC:11587 | |
| genetic reagent (D. melanogaster) | ProcMI06590 | Bloomington Drosophila Stock Center | BDSC:42407 | |
| genetic reagent (D. melanogaster) | ProcRMB00909 | Bloomington Drosophila Stock Center | BDSC:22930 | |
| genetic reagent (D. melanogaster) | R10F08-Gal4 | Bloomington Drosophila Stock Center | BDSC:48441 | |
| genetic reagent (D. melanogaster) | R10H10-Gal4 | Bloomington Drosophila Stock Center | BDSC:48445 | |
| genetic reagent (D. melanogaster) | R11B09-Gal4 | Bloomington Drosophila Stock Center | BDSC:48288 | |
| genetic reagent (D. melanogaster) | R11C01-Gal4 | Bloomington Drosophila Stock Center | BDSC:49240 | |
| genetic reagent (D. melanogaster) | R14F05-Gal4 | Bloomington Drosophila Stock Center | BDSC:49257 | |
| genetic reagent (D. melanogaster) | R16C05-Gal4 | Bloomington Drosophila Stock Center | BDSC:48718 | |
| genetic reagent (D. melanogaster) | R18H11-Gal4 | Bloomington Drosophila Stock Center | BDSC:48832 | |
| genetic reagent (D. melanogaster) | R19B09-Gal4 | Bloomington Drosophila Stock Center | BDSC:48840 | |
| genetic reagent (D. melanogaster) | R19G10-Gal4 | Bloomington Drosophila Stock Center | BDSC:47887 | |
| genetic reagent (D. melanogaster) | R20A02-Gal4 | Bloomington Drosophila Stock Center | BDSC:48870 | |
| genetic reagent (D. melanogaster) | R20E05-Gal4 | Bloomington Drosophila Stock Center | BDSC:48898 | |
| genetic reagent (D. melanogaster) | R21H04-Gal4 | Bloomington Drosophila Stock Center | BDSC:48958 | |
| genetic reagent (D. melanogaster) | R23E10-Gal4 | Bloomington Drosophila Stock Center | BDSC:49032 | |
| genetic reagent (D. melanogaster) | R25G04-Gal4 | Bloomington Drosophila Stock Center | BDSC:49136 | |
| genetic reagent (D. melanogaster) | R26D11-Gal4 | Bloomington Drosophila Stock Center | BDSC:49323 | |
| genetic reagent (D. melanogaster) | R27A05-Gal4 | Bloomington Drosophila Stock Center | BDSC:49208 | |
| genetic reagent (D. melanogaster) | R30G08-Gal4 | Bloomington Drosophila Stock Center | BDSC:48101 | |
| genetic reagent (D. melanogaster) | R32G08-Gal4 | Bloomington Drosophila Stock Center | BDSC:49729 | |
| genetic reagent (D. melanogaster) | R32H03-Gal4 | Bloomington Drosophila Stock Center | BDSC:49733 | |
| genetic reagent (D. melanogaster) | R34C05-Gal4 | Bloomington Drosophila Stock Center | BDSC:49778 | |
| genetic reagent (D. melanogaster) | R43D05-Gal4 | Bloomington Drosophila Stock Center | BDSC:41259 | |
| genetic reagent (D. melanogaster) | R44B02-Gal4 | Bloomington Drosophila Stock Center | BDSC:50199 | |
| genetic reagent (D. melanogaster) | R45B03-Gal4 | Bloomington Drosophila Stock Center | BDSC:50221 | |
| genetic reagent (D. melanogaster) | R46E11-Gal4 | Bloomington Drosophila Stock Center | BDSC:50272 | |
| genetic reagent (D. melanogaster) | R47A08-Gal4 | Bloomington Drosophila Stock Center | BDSC:50288 | |
| genetic reagent (D. melanogaster) | R50C11-Gal4 | Bloomington Drosophila Stock Center | BDSC:38742 | |
| genetic reagent (D. melanogaster) | R50H05-Gal4 | Bloomington Drosophila Stock Center | BDSC:38764 | |
| genetic reagent (D. melanogaster) | R51H05-Gal4 | Bloomington Drosophila Stock Center | BDSC:41275 | |
| genetic reagent (D. melanogaster) | R54D11-Gal4 | Bloomington Drosophila Stock Center | BDSC:41279 | |
| genetic reagent (D. melanogaster) | R57C10-Gal4 | Bloomington Drosophila Stock Center | BDSC:39171 | |
| genetic reagent (D. melanogaster) | R57F07-Gal4 | Bloomington Drosophila Stock Center | BDSC:46389 | |
| genetic reagent (D. melanogaster) | R61G12-Gal4 | Bloomington Drosophila Stock Center | BDSC:41286 | |
| genetic reagent (D. melanogaster) | R64C04-Gal4 | Bloomington Drosophila Stock Center | BDSC:39296 | |
| genetic reagent (D. melanogaster) | R64C10-Gal4 | Bloomington Drosophila Stock Center | BDSC:39301 | |
| genetic reagent (D. melanogaster) | R65C07-Gal4 | Bloomington Drosophila Stock Center | BDSC:39344 | |
| genetic reagent (D. melanogaster) | R65C11-Gal4 | Bloomington Drosophila Stock Center | BDSC:39347 | |
| genetic reagent (D. melanogaster) | R66B05-Gal4 | Bloomington Drosophila Stock Center | BDSC:39389 | |
| genetic reagent (D. melanogaster) | R70F10-Gal4 | Bloomington Drosophila Stock Center | BDSC:39545 | |
| genetic reagent (D. melanogaster) | R70G01-Gal4 | Bloomington Drosophila Stock Center | BDSC:39546 | |
| genetic reagent (D. melanogaster) | R78G02-Gal4 | Bloomington Drosophila Stock Center | BDSC:40010 | |
| genetic reagent (D. melanogaster) | R85A11-Gal4 | Bloomington Drosophila Stock Center | BDSC:40415 | |
| genetic reagent (D. melanogaster) | R86H08-Gal4 | Bloomington Drosophila Stock Center | BDSC:40471 | |
| genetic reagent (D. melanogaster) | R91A01-Gal4 | Bloomington Drosophila Stock Center | BDSC:40569 | |
| genetic reagent (D. melanogaster) | R95E11-Gal4 | Bloomington Drosophila Stock Center | BDSC:40711 | |
| genetic reagent (D. melanogaster) | RNAi-acj6 | Bloomington Drosophila Stock Center | BDSC:29335 | |
| genetic reagent (D. melanogaster) | RNAi-Akh | Bloomington Drosophila Stock Center | BDSC:27031 | |
| genetic reagent (D. melanogaster) | RNAi-Cry | Bloomington Drosophila Stock Center | BDSC:51033 | |
| genetic reagent (D. melanogaster) | RNAi-Crz | Bloomington Drosophila Stock Center | BDSC:25999 | |
| genetic reagent (D. melanogaster) | RNAi-Crz | Bloomington Drosophila Stock Center | BDSC:26017 | |
| genetic reagent (D. melanogaster) | RNAi-CrzR | Bloomington Drosophila Stock Center | BDSC:42751 | |
| genetic reagent (D. melanogaster) | RNAi-DAT | Bloomington Drosophila Stock Center | BDSC:31256 | |
| genetic reagent (D. melanogaster) | RNAi-DAT | Bloomington Drosophila Stock Center | BDSC:50619 | |
| genetic reagent (D. melanogaster) | RNAi-DDC | Bloomington Drosophila Stock Center | BDSC:27030 | |
| genetic reagent (D. melanogaster) | RNAi-DDC | Bloomington Drosophila Stock Center | BDSC:51462 | |
| genetic reagent (D. melanogaster) | RNAi-Dh31 | Bloomington Drosophila Stock Center | BDSC:41957 | |
| genetic reagent (D. melanogaster) | RNAi-Dh44 | Bloomington Drosophila Stock Center | BDSC:25804 | |
| genetic reagent (D. melanogaster) | RNAi-for | Bloomington Drosophila Stock Center | BDSC:21592 | |
| genetic reagent (D. melanogaster) | RNAi-for | Bloomington Drosophila Stock Center | BDSC:31698 | |
| genetic reagent (D. melanogaster) | RNAi-Lk | Bloomington Drosophila Stock Center | BDSC:25936 | |
| genetic reagent (D. melanogaster) | RNAi-LkR | Bloomington Drosophila Stock Center | BDSC:25836 | |
| genetic reagent (D. melanogaster) | RNAi-Nplp2 | Bloomington Drosophila Stock Center | BDSC:53967 | |
| genetic reagent (D. melanogaster) | RNAi-Nplp2 | Bloomington Drosophila Stock Center | BDSC:54041 | |
| genetic reagent (D. melanogaster) | RNAi-Oamb | Bloomington Drosophila Stock Center | BDSC:31171 | |
| genetic reagent (D. melanogaster) | RNAi-Oamb | Bloomington Drosophila Stock Center | BDSC:31233 | |
| genetic reagent (D. melanogaster) | RNAi-Oct-Tyr | Bloomington Drosophila Stock Center | BDSC:28332 | |
| genetic reagent (D. melanogaster) | RNAi-OctAlpha2R | Bloomington Drosophila Stock Center | BDSC:50678 | |
| genetic reagent (D. melanogaster) | RNAi-OctBeta1R | Bloomington Drosophila Stock Center | BDSC:31106 | |
| genetic reagent (D. melanogaster) | RNAi-OctBeta1R | Bloomington Drosophila Stock Center | BDSC:31107 | |
| genetic reagent (D. melanogaster) | RNAi-OctBeta1R | Bloomington Drosophila Stock Center | BDSC:50701 | |
| genetic reagent (D. melanogaster) | RNAi-OctBeta1R | Bloomington Drosophila Stock Center | BDSC:58179 | |
| genetic reagent (D. melanogaster) | RNAi-OctBeta2R | Bloomington Drosophila Stock Center | BDSC:34673 | |
| genetic reagent (D. melanogaster) | RNAi-OctBeta2R | Bloomington Drosophila Stock Center | BDSC:50580 | |
| genetic reagent (D. melanogaster) | RNAi-OctBeta3R | Bloomington Drosophila Stock Center | BDSC:31108 | |
| genetic reagent (D. melanogaster) | RNAi-Pdf | Bloomington Drosophila Stock Center | BDSC:25802 | |
| genetic reagent (D. melanogaster) | RNAi-ple | Bloomington Drosophila Stock Center | BDSC:25796 | |
| genetic reagent (D. melanogaster) | RNAi-ple | Bloomington Drosophila Stock Center | BDSC:65875 | |
| genetic reagent (D. melanogaster) | RNAi-ple | Bloomington Drosophila Stock Center | BDSC:76062 | |
| genetic reagent (D. melanogaster) | RNAi-ple | Bloomington Drosophila Stock Center | BDSC:76069 | |
| genetic reagent (D. melanogaster) | RNAi-ppk25 | Bloomington Drosophila Stock Center | BDSC:27088 | |
| genetic reagent (D. melanogaster) | RNAi-ProcR | Bloomington Drosophila Stock Center | BDSC:29414 | |
| genetic reagent (D. melanogaster) | RNAi-ProcR | Bloomington Drosophila Stock Center | BDSC:29570 | |
| genetic reagent (D. melanogaster) | RNAi-ptp69D | Bloomington Drosophila Stock Center | BDSC:29462 | |
| genetic reagent (D. melanogaster) | RNAi-ShakB | Bloomington Drosophila Stock Center | BDSC:27292 | |
| genetic reagent (D. melanogaster) | RNAi-SifA | Bloomington Drosophila Stock Center | BDSC:29428 | |
| genetic reagent (D. melanogaster) | RNAi-SifA | Bloomington Drosophila Stock Center | BDSC:60484 | |
| genetic reagent (D. melanogaster) | RNAi-Tbh | Bloomington Drosophila Stock Center | BDSC:27667 | |
| genetic reagent (D. melanogaster) | RNAi-Tbh | Bloomington Drosophila Stock Center | BDSC:67968 | |
| genetic reagent (D. melanogaster) | RNAi-Tdc2 | Bloomington Drosophila Stock Center | BDSC:25871 | |
| genetic reagent (D. melanogaster) | RNAi-Tk | Bloomington Drosophila Stock Center | BDSC:25800 | |
| genetic reagent (D. melanogaster) | RNAi-TkR86C | Bloomington Drosophila Stock Center | BDSC:31884 | |
| genetic reagent (D. melanogaster) | RNAi-TkR99D | Bloomington Drosophila Stock Center | BDSC:27513 | |
| genetic reagent (D. melanogaster) | RNAi-trh | Bloomington Drosophila Stock Center | BDSC:25842 | |
| genetic reagent (D. melanogaster) | RNAi-tutl | Bloomington Drosophila Stock Center | BDSC:54850 | |
| genetic reagent (D. melanogaster) | RNAi-TyrR | Bloomington Drosophila Stock Center | BDSC:25857 | |
| genetic reagent (D. melanogaster) | RNAi-TyrR | Bloomington Drosophila Stock Center | BDSC:57296 | |
| genetic reagent (D. melanogaster) | RNAi-TyrRII | Bloomington Drosophila Stock Center | BDSC:27670 | |
| genetic reagent (D. melanogaster) | RNAi-TyrRII | Bloomington Drosophila Stock Center | BDSC:64964 | |
| genetic reagent (D. melanogaster) | ry506 | Bloomington Drosophila Stock Center | BDSC:225 | |
| genetic reagent (D. melanogaster) | RyaR- | Bloomington Drosophila Stock Center | BDSC:84571 | |
| genetic reagent (D. melanogaster) | shakB-Gal4 | Bloomington Drosophila Stock Center | BDSC:51633 | |
| genetic reagent (D. melanogaster) | SifA-Gal4 | Bloomington Drosophila Stock Center | BDSC:84690 | |
| genetic reagent (D. melanogaster) | sNPF- | Bloomington Drosophila Stock Center | BDSC:84574 | |
| genetic reagent (D. melanogaster) | SS00078-Gal4 | Janelia Research Center | JRC:SS00078 | |
| genetic reagent (D. melanogaster) | SS00090-Gal4 | Janelia Research Center | JRC:SS00090 | |
| genetic reagent (D. melanogaster) | SS00097-Gal4 | Janelia Research Center | JRC:SS00097 | |
| genetic reagent (D. melanogaster) | SS00117-Gal4 | Janelia Research Center | JRC:SS00117 | |
| genetic reagent (D. melanogaster) | SS01566-Gal4 | Janelia Research Center | JRC:SS01566 | |
| genetic reagent (D. melanogaster) | SS02214-Gal4 | Janelia Research Center | JRC:SS02214 | |
| genetic reagent (D. melanogaster) | SS02216-Gal4 | Janelia Research Center | JRC:SS02216 | |
| genetic reagent (D. melanogaster) | SS02255-Gal4 | Janelia Research Center | JRC:SS02255 | |
| genetic reagent (D. melanogaster) | SS02391-Gal4 | Janelia Research Center | JRC:SS02391 | |
| genetic reagent (D. melanogaster) | SS27853-Gal4 | Janelia Research Center | JRC:SS27853 | |
| genetic reagent (D. melanogaster) | SS50464-Gal4 | Janelia Research Center | JRC:SS50464 | |
| genetic reagent (D. melanogaster) | SS52578-Gal4 | Janelia Research Center | JRC:SS52578 | |
| genetic reagent (D. melanogaster) | Tbh- | Bloomington Drosophila Stock Center | BDSC:56660 | |
| genetic reagent (D. melanogaster) | Tdc-Gal4 | Bloomington Drosophila Stock Center | BDSC:9313 | |
| genetic reagent (D. melanogaster) | tim-Gal4 | Bloomington Drosophila Stock Center | BDSC:80941 | |
| genetic reagent (D. melanogaster) | Trh- | Bloomington Drosophila Stock Center | BDSC:10531 | |
| genetic reagent (D. melanogaster) | Trh-Gal4 | Bloomington Drosophila Stock Center | BDSC:38388 | |
| genetic reagent (D. melanogaster) | Trh-Gal4 | Bloomington Drosophila Stock Center | BDSC:38389 | |
| genetic reagent (D. melanogaster) | tutl-Gal4 | Bloomington Drosophila Stock Center | BDSC:63344 | |
| genetic reagent (D. melanogaster) | tutl-Gal4/CyO;Cha-Gal80 | Derived from BDSC:63344 and Cha-Gal80 | ||
| genetic reagent (D. melanogaster) | tutl1/CyO | Kendal Broadie (Bodily et al., 2001) | ||
| genetic reagent (D. melanogaster) | TyrR- | Bloomington Drosophila Stock Center | BDSC:27797 | |
| genetic reagent (D. melanogaster) | TyrRII- | Bloomington Drosophila Stock Center | BDSC:23837 | |
| genetic reagent (D. melanogaster) | UAS-CsChrimson | Bloomington Drosophila Stock Center; Klapoetke et al., 2014 | BDSC:55135 | |
| genetic reagent (D. melanogaster) | UAS-DTI | Bloomington Drosophila Stock Center | BDSC:25039 | |
| genetic reagent (D. melanogaster) | UAS-eGFP-Kir2.1.FRT.mCherry | David Anderson; Watanabe et al., 2017 | ||
| genetic reagent (D. melanogaster) | UAS-hid | Bloomington Drosophila Stock Center | BDSC:65403 | |
| genetic reagent (D. melanogaster) | UAS-Kir2.1 | Jess Kanwal; Baines et al., 2001 | ||
| genetic reagent (D. melanogaster) | UAS-mcd8GFP | Bloomington Drosophila Stock Center | BDSC:32185 | |
| genetic reagent (D. melanogaster) | UAS-mCherry.FRT.eGFP-Kir2.1 | David Anderson; Watanabe et al., 2017 | ||
| genetic reagent (D. melanogaster) | UAS-NiPP1 | Bloomington Drosophila Stock Center | BDSC:23711 | |
| genetic reagent (D. melanogaster) | UAS-PdfRg/CyO; UAS-Cas9/TM6B | Matthias Schlichting; Schlichting et al., 2019 | ||
| genetic reagent (D. melanogaster) | UAS-syt-eGFP, DenMark | Bloomington Drosophila Stock Center | BDSC:33064 | |
| genetic reagent (D. melanogaster) | UAS-TNT-C | Bloomington Drosophila Stock Center | BDSC:28996 | |
| genetic reagent (D. melanogaster) | UAS-TNT-E | Bloomington Drosophila Stock Center | BDSC:28837 | |
| genetic reagent (D. melanogaster) | UAS-TNT-G | Bloomington Drosophila Stock Center | BDSC:28838 | |
| genetic reagent (D. melanogaster) | UAS-TrpA1 | Bloomington Drosophila Stock Center | BDSC:26263 | |
| genetic reagent (D. melanogaster) | VT002215-Gal4 | Janelia Research Center | JRC:VT002215 | |
| genetic reagent (D. melanogaster) | VTDh44-Gal4/TM3, Sb | VT039046 (via Daniel Cavanaugh) | ||
| genetic reagent (D. melanogaster) | w; Aug21-Gal4, UAS-GFP/CyO | Derived from C(1)Dxyfv(X^X)/Y; Aug21-Gal4, UAS-GFP/CyO | ||
| other | Acetone | Sigma | Cat#: 179124 | Vehicle for Methoprene and Prococene I |
| other | Fluvastatin | Sigma | Cat: PHR1620 | Mevalonate synthesis pathway inhibitor |
| other | Hoechst 33342 | Thermo Fisher | Cat#: H-3570 | IF(1:1000) |
| other | Methoprene | Sigma | Cat#: 33375 | JH analog |
| other | Precocene I | Sigma | Cat#: 195855 | CA inhibitor |
| other | Pyriproxyfen | Sigma | Cat#: 34174 | JH analog |
| other | RhoB | Sigma | Cat#: R6626 | (1.44 mg/mL) |
| peptide, recombinant protein | Phalloidin | Thermo Fisher | Cat#: A-12380 | IF(1:400) |
| software, algorithm | MARGO | Werkhoven et al., 2019 | ||
| strain (Entomophthora muscae) | Entomophthora muscae | Elya et al., 2018 | ARSEF #13514 |
Additional files
-
Supplementary file 1
Fly strains tested in summit inactivation screen.
Genotypes are abbreviated for lines deposited at stock centers, for clarity. Stock centers are as follows: BDSC = Bloomington Drosophila Stock Center; KDSC = Kyoto Drosophila Stock Center; JRC = Janelia Research Campus. Functional/morphological annotations for the summit screen are abbreviated as follows: AM = AMMC; Ar = arousal; Ci = circadian; CX = central complex; Gr = gravitaxis; SO = subesophageal ganglion; MB = mushroom body; NM = neuromodulator & neurotransmitter; NP = neuropeptide; PI = pars intercerebralis.
- https://cdn.elifesciences.org/articles/85410/elife-85410-supp1-v2.docx
-
Supplementary file 2
Genotypes of flies in manuscript figures.
Genotypes for deposited lines are abbreviated for clarity (i.e. interrupted alleles are designated [-], most y and w alleles have been omitted). Stock centers are as follows: BDSC = Bloomington Drosophila Stock Center; KDSC = Kyoto Drosophila Stock Center; JRC = Janelia Research Campus.
- https://cdn.elifesciences.org/articles/85410/elife-85410-supp2-v2.docx
-
Supplementary file 3
Metabolomics data for both manual and classifier-assisted experiments.
Sheets with prefix ‘All_’ contain all of the data from the indicated metabolomics experiment. Sheet with prefix ‘Overlap_’ contains data for compounds that were observed in both experiments (compounds whose molecular weight corresponded within five parts per million [ppm]). Column headers ending in (M) indicate results from the manually-staged experiment; (C) from the classifier-staged experiments. Annotations for columns A, B, C, K, and N are as follows (with values of 1 or TRUE indicating the condition is met, 0 or FALSE that the condition is not met): ‘Significant’ = significantly different between NS and S across both experiments; ‘Infected-specific’ = compound only present in NS and S, but not C, samples, per manual chromatogram inspection; ‘Summiting-specific’ = compound only present in S, but not NS or C, samples, per manual chromatogram inspections; ‘Same tentative formula?’ = predicted formulae for given compound match exactly across experiments; ‘Same sign?’ = log2 fold change of compound abundance between S and NS samples is consistently positive or negative across experiments. Abbreviations: S = summiting, NS = exposed but non-summiting, C = unexposed control.
- https://cdn.elifesciences.org/articles/85410/elife-85410-supp3-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85410/elife-85410-mdarchecklist1-v2.docx





