Staphylococcus aureus FtsZ and PBP4 bind to the conformationally dynamic N-terminal domain of GpsB
Figures
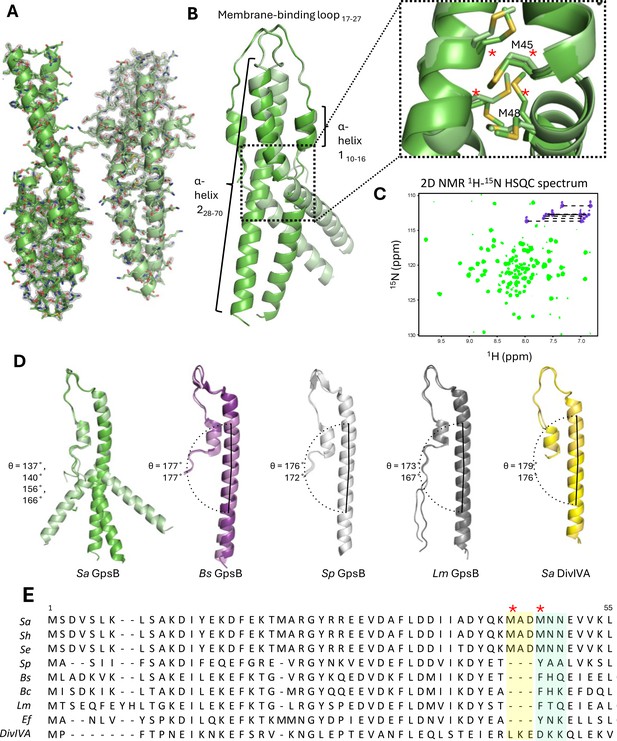
Crystal structure of the N-terminal domain of Sa GpsB.
(A) Two dimers lie antiparallel in the asymmetric unit from the P21 space group. The 2Fo-Fc electron density map, shown in gray, is contoured at 1σ with a resolution of 1.95 Å. (B) Superimposition of GpsB dimers reveals the dimers splay at a hinge region, formed by a cluster of four interlocked Met sidechains. *designates position in multisequence alignment shown in panel E. (C) The 1H-15N HSQC spectrum of the GpsB N-terminal domain, Sa GpsBWT1-70, shows a significantly higher number of signals compared to what is expected of a symmetric dimer based on the sequence of the domain, indicating the presence of conformational heterogeneity. Backbone and Asn/Gln sidechain amide signals are shown in green and purple respectively. (D) Comparison of different GpsB/DivIVA monomers from previously solved structures. Pitch angles were determined by placing a marker atom at the centroid of the beginning, midpoint ‘hinge’, and end of each helix, then measuring the angle. (E) Multisequence alignment of GpsB within select members of the Firmicutes phylum and with S. aureus DivIVA. Staphylococci GpsB contain a three-residue insertion that forms the hinge region, either MAD or MNN, depending on the sequence alignment parameters. The two Met residues (four per dimer) of the hinge region are designated with a red *. Sa - S. aureus, Sh - S. haemolyticus, Se - S. epidermidis, Sp - S. pneumoniae, Bs - B. subtilis, Bc - B. cereus, Lm - L. monocytogenes, Ef - E. faecalis, DivIVA - S. aureus DivIVA.
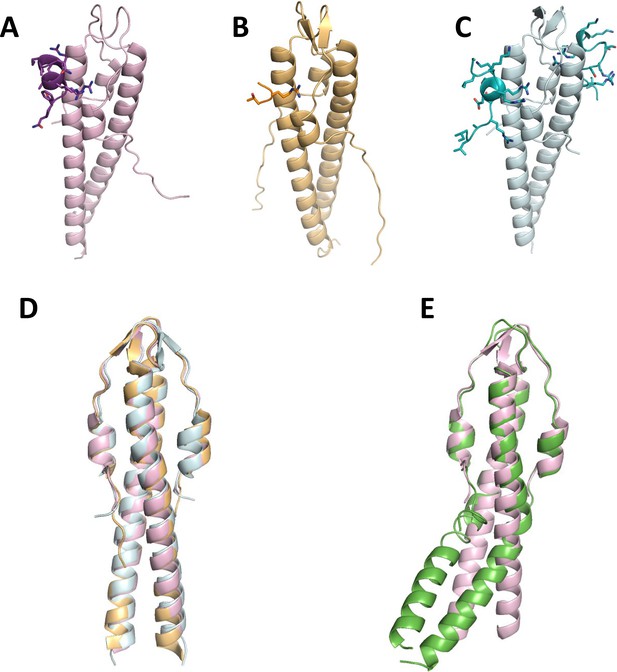
Crystal structures of penicillin-binding protein (PBP) mini-domains in complex with the N-terminal domain of their cognate GpsB, published by Cleverley et al., 2019.
(A) Bs PBP1, (B) Lm PBPA1, (C) Sp PBP2a. (D) Superimposition of previously solved GpsB structures from subpanels A–C. (E) Superimposition of Sa GpsB with Bs GpsB from subpanel A.
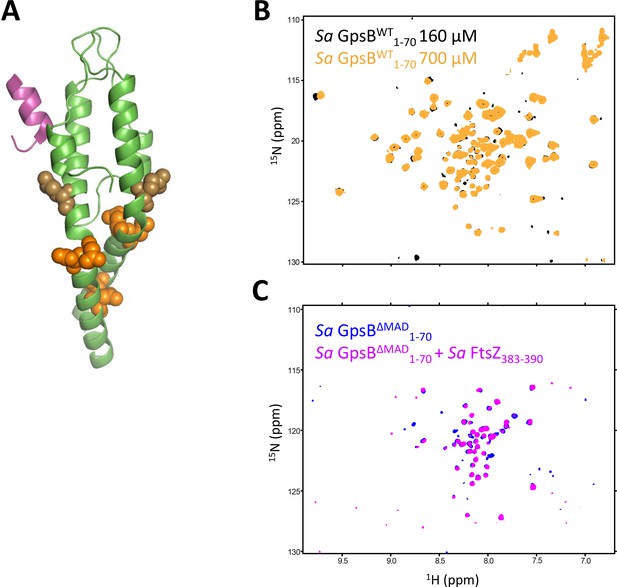
1H-15N HSQC HSQC identifies conformational heterogeneity within the N-terminal domain of Sa GpsB.
(A) Asn (orange) and Gln (brown) residues mapped on the structure of GpsB, modeled with a penicillin-binding protein (PBP)-derived peptide (purple) from Bs PBP1A (PDB ID 6GP7). (B) Overlays of the 1H-15N HSQC of spectrum of Sa GpsBWT1-70 acquired at different concentrations. (C) The 1H-15N HSQC of Sa GpsBΔMAD1-70 in the absence (blue) and presence of 1.5 equivalents of Sa FtsZ383-390 peptide (magenta).

Molecular dynamics simulations and homology models of GpsB mutants reveal potential structural origins of GpsB stability and flexibility.
(A) Molecular dynamics simulation of the wildtype (WT) GpsB N-terminal domain. Significant fluctuations in pitch angle are observed within 100 ns for both dimers from the crystal structure that underwent separate simulations, and for even longer periods of time for dimer 2. (B) Superimposed homology model of Sa GpsB ΔMNN (tan), Sa GpsB ΔMAD (dark gray), and the crystal structure of Lm GpsB (purple; PDB ID 4UG1). Residues following the MNN or MAD deletion are depicted, which are more consistent with the multiple sequence alignment presented in Figure 1E. (C) Several residues form preferential interactions following the MAD and MNN deletions. Shown here is an example of a potentially new electrostatic interaction between Asp47 and Lys51 in ΔMNN mutant. This +/- pair replaces the unfavorable -/- Asp/Glu pair found in Sa GpsB WT. Asp47 is replaced by Asn47 in ΔMAD mutant.
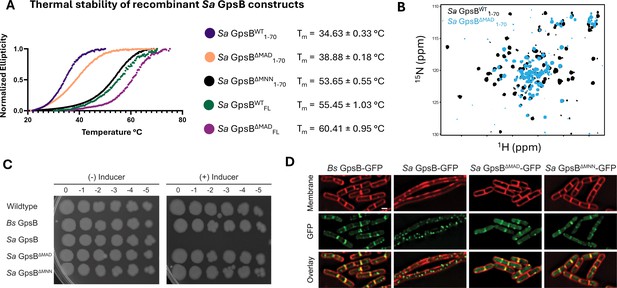
Deletion of a three-residue insertion in Sa GpsB increases thermal stability in solution and abolishes toxicity in B. subtilis.
(A) Circular dichroism (CD) melt profiles of recombinantly expressed Sa GpsB constructs reveal ΔMAD and ΔMNN mutants have increased Tm, compared to wildtype (WT) Sa GpsB. (B) Overlays of the 1H-15N HSQC of spectrum of Sa GpsBWT 1-70 and Sa GpsBΔMAD 1-70 suggest there are significant differences in the conformational properties of these two proteins. (C) Serial dilutions of B. subtilis strains harboring inducible Bs GpsB (GG18), Sa GpsB (GG7), Sa GpsB ∆MAD (LH119), and Sa GpsB ∆MNN (LH115), plated on LB plates without (left) and with (right) 1 mM IPTG demonstrate WT Sa GpsB is lethal, but ΔMAD Sa GpsB and ΔMNN Sa GpsB are not. (D) Fluorescence micrographs showing the protein localization of Bs GpsB-GFP (GG19), Sa GpsB-GFP (GG8), Sa GpsB-GFP ∆MAD (LH126), and Sa GpsB-GFP ∆MNN (LH116). Cell membrane was visualized using SynaptoRed membrane dye (1 µg/mL). Scale bar, 1 µm. In contrast to WT Sa GpsB, strains of B. subtilis that overexpress ΔMAD and ΔMNN Sa GpsB have similar cellular morphology to WT B. subtilis and these proteins localize to the division septum.

Deletion of MNN residues more drastically affects the function of Sa GpsB than deletion of MAD residues.
A) Growth assay: serial dilutions of Sa strains harboring inducible Sa gpsB (GGS2), Sa gpsB∆MAD (LH129), Sa gpsB∆MNN (LH127), and the empty vector (EV) control (PE355), plated on TSA plates supplemented with 10 µg/mL chloramphenicol both without (left) and with (right) 1 mM IPTG. (B) Bacterial two-hybrid analysis. Pairwise interactions of wildtype Sa GpsB (WT) (LH39/LH40) with the ∆MAD (LH164/MA1) and ∆MNN (LH170/LH168) mutants as well as the ∆MAD and ∆MNN self-interactions plated on McConkey agar plates supplemented with 1% maltose (top). Interactions were also tested by β-galactosidase assay (bottom). Assay was done in triplicate and the dashed line shows the average Miller Unit level of the negative control. *p<0.05 and **p<0.01. (C) Micrographs of S. aureus cells containing plasmids with inducible Sa GpsB (PES13), Sa GpsB ∆MAD (LH135), and Sa GpsB ∆MNN (LH127), and the EV control (PES5), imaged 2 hr post induction. Cells were visualized with 1 µg/mL SynaptoRed membrane dye. Scale bar is 1 µm. (D) Quantification of micrographs shown in panel C. n=500 cells; ***p<0.001 and ****p<0.0001. (E) Western blot of strains shown in panel C. Note: The GpsB band in EV lane is of native GpsB. (F) Micrographs showing the localization of Sa GpsB-GFP (PES6), Sa GpsB ∆MAD (LH133), and Sa GpsB ∆MNN (LH132). Cells visualized with 1 µg/mL SynaptoRed membrane dye. Scale bar is 1 µm.
-
Figure 2—figure supplement 1—source data 1
Images of total protein gel and anti-GpsB western blot.
- https://cdn.elifesciences.org/articles/85579/elife-85579-fig2-figsupp1-data1-v2.zip

Deletion of MAD/MNN residues renders Sa GpsB less functional, but only minimally affects the function of heterocomplex with unmutated counterpart.
(A) Growth assay: serial dilutions of Sa ΔgpsB strains harboring inducible Sa gpsB (LM75), Sa gpsB∆MAD (LM76), Sa gpsB∆MNN (LM77), and the empty vector (EV) control (LM74) as well as wildtype (WT) strain harboring inducible Sa gpsB (GGS1) were plated on TSA plates supplemented with 10 µg/mL chloramphenicol both without (left) and with (right) 1 mM IPTG. (B) Strains used in the spot titer assay including WT EV control (PE355) were observed under the microscope and the diameter of the cells were quantified and plotted. n=100 cells; ****p<0.0001 and ***p<0.0007. (C) Western blot of strains used in panels A and B. Coomassie staining of the gel is shown to serve as loading control. (D) Serial dilutions of B. subtilis strains PY79 (WT), inducible Sa gpsB (GG7), or the ones harboring inducible Sa gpsB (DB44), Sa gpsBΔMAD (DB46), Sa gpsBΔMNN (DB45), or Sa gpsBΔLEE (LH78) in inducible Sa gpsB-gfp background plated on LB plates without (left) and with (right) 1 mM IPTG.
-
Figure 2—figure supplement 2—source data 1
Images of total protein gel and anti-GpsB western blot.
- https://cdn.elifesciences.org/articles/85579/elife-85579-fig2-figsupp2-data1-v2.zip
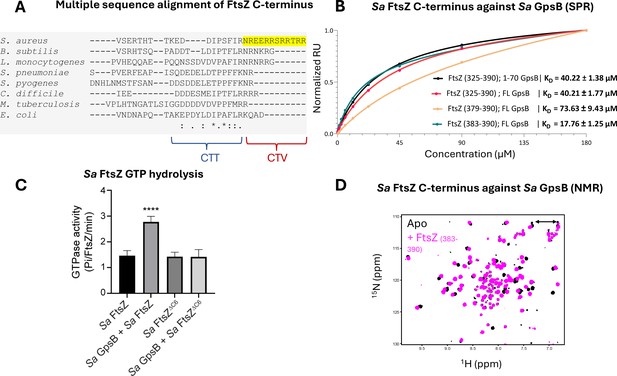
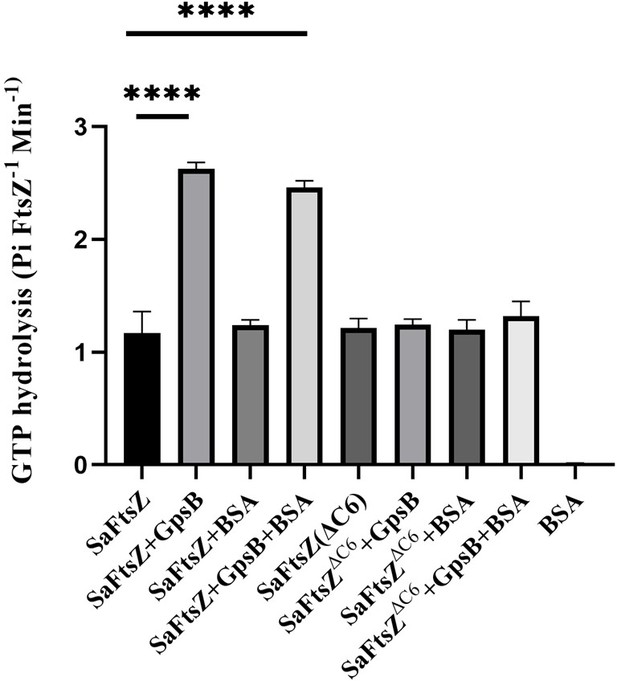
Sa FtsZ contains a repeated GpsB recognition motif at its C-terminus.
(A) A multiple sequence alignment of the FtsZ C-terminus from different representative bacteria reveals that there is a repeated GpsB recognition motif in Sa FtsZ (highlighted region) and that it is unique to this bacterium. (B) Surface plasmon resonance (SPR) titration of peptides corresponding to several segments of Sa FtsZ against Sa GpsB (residues 1–70 or full length). A titration of Sa FtsZ (residues 325–390) against 1–70 GpsB (black), corresponding to the N-terminal domain, shows binding can be isolated to this region. (C) When incubated with Sa GpsB, a Sa FtsZ mutant with a C-terminal truncation (SRRTRR, FtsZΔC6) has significantly lower GTP hydrolysis compared to its full-length counterpart. GTP hydrolysis was measured by monitoring inorganic phosphate (Pi) released (µmoles/min) by either FtsZ or FtsZΔC6 (30 μM) in the absence and presence of GpsB (10 μM). The plot is the average of n=6 independent data sets. p-Value for **** is <0.0001. (D) Overlays of the 1H-15N HSQC of spectrum of Sa GpsB (1–70) in the absence (black) and in the presence of FtsZ (383–390; pink). The boxed region highlights the only sidechain pair of signals that becomes affected by the addition of Ftsz. Based on a model derived from the structure of Bs GpsB in complex with a penicillin-binding protein (PBP)-derived peptide (Figure 1—figure supplement 2), it is tentatively assigned to Q43 of Sa GpsB.
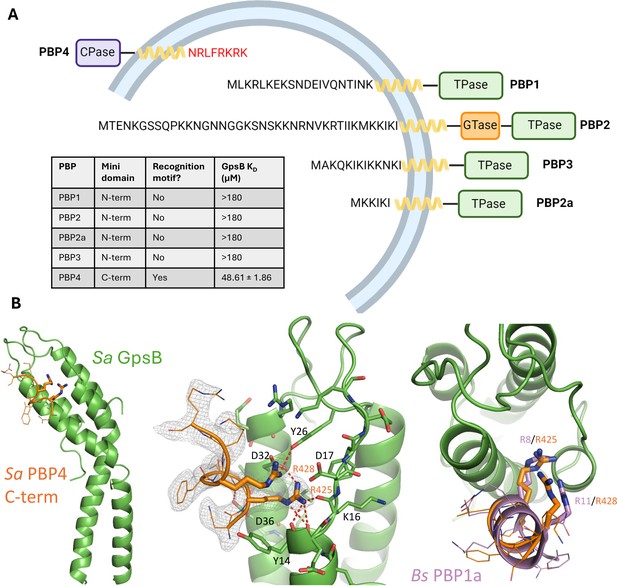
The C-terminal mini-domain of PBP4 directly interacts with GpsB.
(A) Domain representation of the four/five S. aureus (COL)/methicillin-resistant S. aureus (MRSA) (USA300) penicillin-binding proteins (PBPs). Each protein is shown from the N-terminus (left) to the C-terminus (right). All four transpeptidase PBPs - PBP1, PBP2, PBP3, and PBP2a - lack a GpsB recognition motif on their N-terminal, cytosolic mini-domain. In contrast, the C-terminal mini-domain of PBP4, the sole S. aureus class C PBP, contains this motif (NRLFRKRK, red). The dissociation constants were determined with SPR (n=2). (B) Crystal structure of Sa GpsB R24A in complex with PBP4 C-terminal peptide fragment at 2.40 Å resolution. The middle panel includes the electron density map of the Sa PBP4 heptapeptide, 2Fo-Fc=1.0σ. The right panel shows a superimposition of the Bs PBP1 mini-domain from the Bs GpsB+PBP1 complex (PDB ID 6GP7, purple) highlighting similar binding features.

The crystal packing interface of Sa GpsB dimers form interactions that mimic those between GpsB-PBP pairs.
(A) Sa GpsB dimers assemble in a head-to-head, antiparallel arrangement, where the membrane-binding loop interacts with the PBP-binding groove of the other. (B) Enhanced viewpoint of the crystal packing interactions. 2Fo-Fc map, shown in gray, is contoured at 1.0σ. R28, R24, and K18 adopt similar interactions with the negatively charged PBP-binding groove, as the arginine fingers of (C, D) of Bs PBP1a with Bs GpsB (6GP7). PBP, penicillin-binding protein.
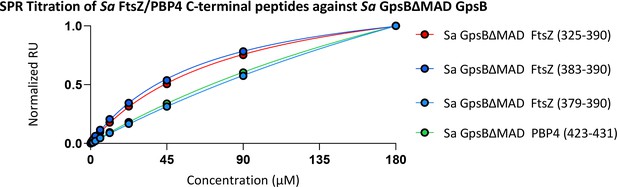
Surface plasmon resonance (SPR) titration of Sa FtsZ C-terminal peptides against Sa GpsBΔMADFL.
KD values are reported in Supplementary file 2.

AlphaFold2-Multimer prediction of GpsB:FtsZ385-390 complex (cyan) superimposed onto the GpsB:PBP4 peptide crystal structure (green:orange).
FtsZ residues are labeled. PBP, penicillin-binding protein.
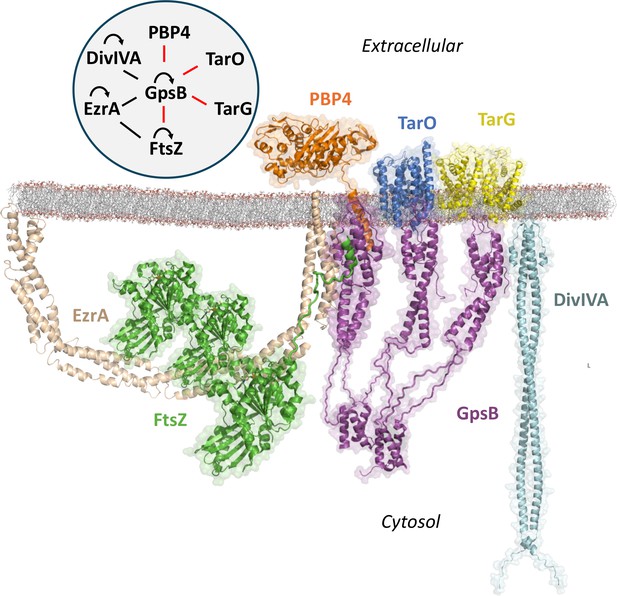
Known interactome of Sa GpsB and putative arrangement at the division septum in graphical and diagram (upper left) format.
In this paper, we demonstrate that the C-terminal mini-domain of PBP4 (orange) and the C-terminus of FtsZ (green) bind to the N-terminal domain of GpsB (purple). The regions within GpsB responsible for interacting with other partners remain to be elucidated. For interaction diagram, the red lines indicate interactions putatively unique to S. aureus, black lines indicate interactions found in both S. aureus and B. subtilis, and curved arrows represent self-interaction.
Additional files
-
Supplementary file 1
Table of crystallographic statistics.
*Values in parentheses indicate those for the highest resolution shell.
- https://cdn.elifesciences.org/articles/85579/elife-85579-supp1-v2.docx
-
Supplementary file 2
Surface plasmon resonance (SPR) dissociation constants (KD) of Sa FtsZ and Sa PBP4 derived peptides for Sa GpsBWTFL and Sa GpsBΔMADFL.
Concentration response sensorgrams are shown in Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/85579/elife-85579-supp2-v2.docx
-
Supplementary file 3
The genotypes of strains used in the cell-based studies.
- https://cdn.elifesciences.org/articles/85579/elife-85579-supp3-v2.docx
-
Supplementary file 4
The oligonucleotide and geneblock sequences used in the cell-based studies.
- https://cdn.elifesciences.org/articles/85579/elife-85579-supp4-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85579/elife-85579-mdarchecklist1-v2.docx