Biallelic variants in MAD2L1BP (p31comet) cause female infertility characterized by oocyte maturation arrest
Figures
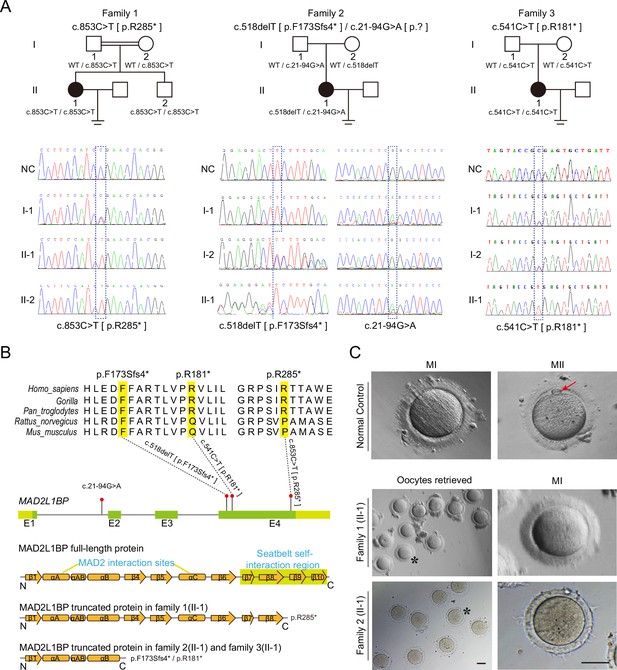
Identification of pathogenic variants in MAD2L1BP from three unrelated families.
(A) Pedigrees of three female patients with oocyte maturation arrest at the MI stage from three unrelated families. The first case was from a consanguineous family, while the other two were not. The patients carried homozygous c.853C>T [p.R285*] and c.541C>T [p.R181*] variants from Family 1 and Family 3, respectively, while the patient from Family 2 had a compound heterozygous mutation c.518delT and c.21–94G>A as indicated. The chromatograms below show the Sanger sequencing results of the PCR-amplified fragments containing the respective variants in each family. (B) Phylogenetic conservation of the identified amino acid mutations in MAD2L1BP. The positions of all mutations are indicated in the schematic genomic structure of MAD2L1BP. The MAD2-interacting site and the seatbelt self-interaction region for MAD2L1BP are labeled in the secondary structure of MAD2L1BP protein at the bottom. All identified mutations residing in Exon 4 of MAD2L1BP generated the premature STOP codon, likely resulting in truncated proteins. (C) The representative morphology of normal and affected individual oocytes. The normal MI and MII oocytes are shown in the top panel. The red arrow points to the first polar body (PB1). Oocytes retrieved from Family 1 (II-1) and Family 2 (II-1) were arrested at the MI stage. The asterisks indicate the oocytes with a magnified view in the right panel. MI: metaphase I; MII: metaphase II. Scale bar, 80 μm.

Multi-sequence alignment of amino acids for MAD2L1BP (p31comet) (A) and its interacting partner MAD2 (B) orthologs from eutherian species with Jalview.
Pathogenic mutations in affected individuals reported in this study are marked in red box.

Semen analysis of the individual II-2 in Family 1 with a homozygous mutation (c.853C>T [p.R285*]) in MAD2L1BP.
(A–C) Photomicrographs of the ejaculated semen smear stained by papanicolaou. Few normally shaped spermatozoa can be observed in the ejaculated semen from the individual II-2 in Family 1. (D) The control sperm morphology by papanicolaou staining from a fertile man. Scale bars, 10 μm. See Supplementary file 1A for semen parameters of the individual II-2 in Family 1.
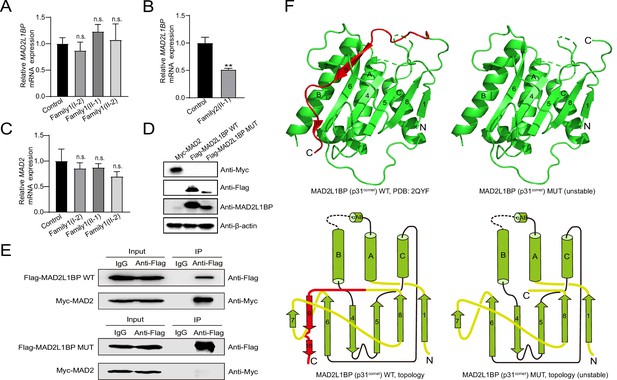
Adverse impacts of MAD2L1BP variants on mRNA expression and protein function.
(A–B) Quantitative PCR (qPCR) assays comparing the mRNA expression levels of MAD2L1BP in peripheral blood samples from Family 1 and Family 2 as indicated. The experiments were performed in technical triplicates. (C) qPCR assay showing the mRNA expression levels of MAD2 in individual blood samples from Family 1. The experiments were performed in technical triplicates. (D) The ectopic protein expression levels of Myc-MAD2, Flag-MAD2L1BP WT(wild-type), and Flag-MAD2L1BP MUT (p.R285*) by immunoblotting following transfection of individual plasmids into 293T cells in culture. Full-length and truncated MAD2L1BP protein can be seen in the panel. (E) Co-immunoprecipitation (Co-IP) assays demonstrating that the truncated MAD2L1BP MUT (p.R285*) lost interaction with MAD2, as compared with MAD2L1BP WT, when co-transfected into 293T cells in vitro. (F) Ribbon and topological diagrams showing the structure of MAD2L1BP WT (PDB ID: 2QYF) and the predicted MAD2L1BP MUT (p.R285*). Notably, MAD2L1BP MUT (p.R285*) lacks a C-terminal seatbelt configuration (highlighted in red) resulting in structural instability.
-
Figure 2—source data 1
Blots for detecting Myc-MAD2, Flag-MAD2L1BP WT, Flag-MAD2L1BP MUT protein expression.
- https://cdn.elifesciences.org/articles/85649/elife-85649-fig2-data1-v2.zip
-
Figure 2—source data 2
Co-immunoprecipitation (Co-IP) blots for detecting interaction between MAD2 and MAD2L1BP WT or MAD2L1BP MUT.
- https://cdn.elifesciences.org/articles/85649/elife-85649-fig2-data2-v2.zip
-
Figure 2—source data 3
The original file of blots for Figure 2D.
- https://cdn.elifesciences.org/articles/85649/elife-85649-fig2-data3-v2.zip
-
Figure 2—source data 4
The original file of blots for Figure 2E.
- https://cdn.elifesciences.org/articles/85649/elife-85649-fig2-data4-v2.zip
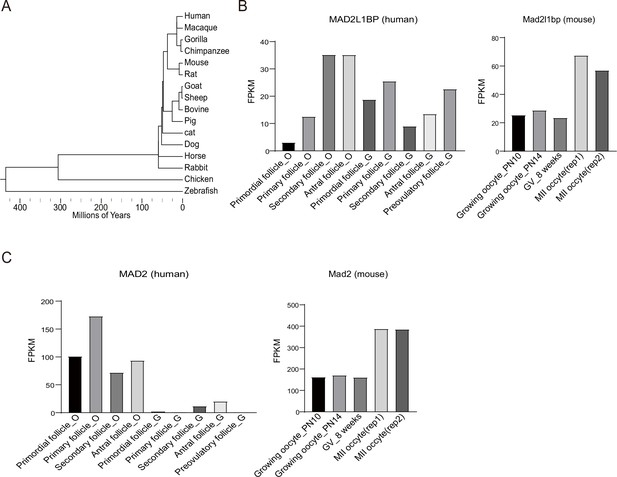
Phylogenetic tree of MAD2L1BP and mRNA expression pattern of MAD2L1BP and MAD2 in human follicles and mouse oocytes.
(A) Evolution of the MAD2L1BP gene orthologs in metazoan species. Sequence conservation analysis was performed with Jalview software, and processed on Evolview website. (B–C) mRNA expression levels of MAD2L1BP (B) and MAD2L1(MAD2) (C) in developing human follicles and mouse oocytes. RNA-seq data was downloaded from GSE71434 and GSE107746. Reads were mapped to the mouse genome (mm10) or human genome (hg38) using STAR. Relative expression quantification of genes and transcripts was performed using RSEM. PN, postnatal; GV, germinal vesicle; MII, metaphase II; rep, replicate. O, oocyte; G, granulosa cell.
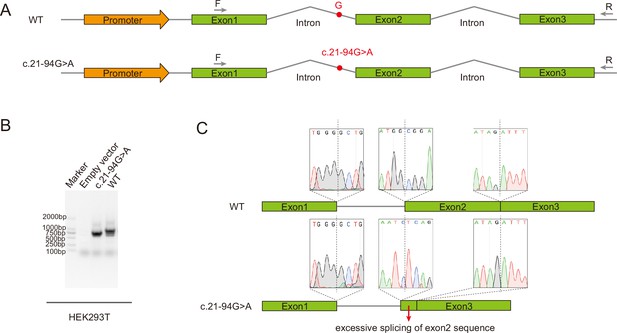
Minigene assay showing the aberrant alternative splicing of MAD2L1BP exons associated with the variant c.21-94G>A from the Family 2.
(A) Schematic diagram of minigene splicing assay. The G→A substitution in intron 1 produced a novel acceptor site for Exon 2, leading to the generation of a premature termination codon (PTC). HEK293T cells were transfected with minigene constructs (WT or c.21-94G>A), RT-PCR was performed with the primers as indicated. (B) Different splicing patterns in WT and c.21-94G>A group were shown by the agarose gel. (C) Representative Sanger sequencing chromatogram validating the excessively splicing of the exon 2 in MAD2L1BP mRNA with c.21-94G>A and thus caused deletion of large fragment of MAD2L1BP protein.
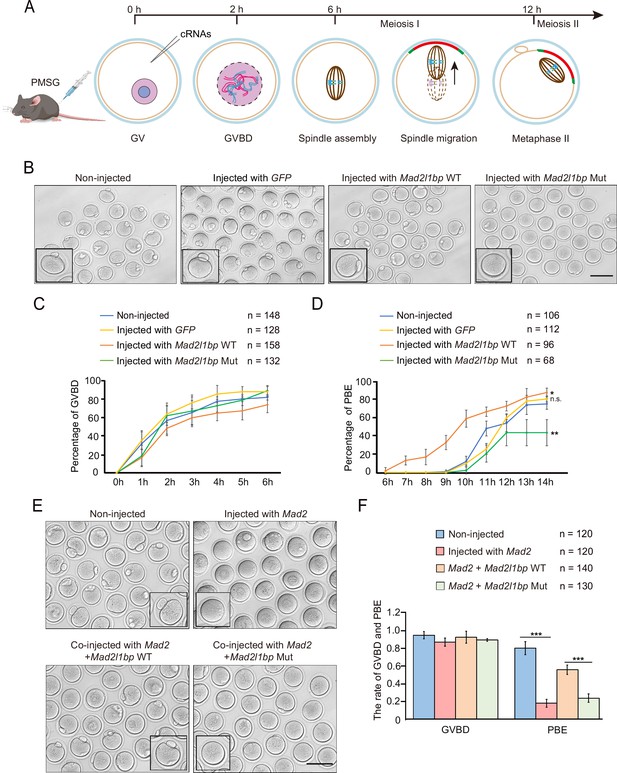
cRNA microinjection of Mad2l1bp accelerated or rescued the meiotic division in mouse germinal vesicle (GV) oocytes.
(A) A schematic diagram depicting the time points of distinctive events through meiotic progression in mouse oocytes. cRNAs were microinjected into GV oocytes after 44 hr of PMSG priming. (B) Representative morphology of oocytes after microinjection with cRNAs encoding mock GFP, mouse Mad2l1bp WT, and Mad2l1bp Mut (equivalent to human p.R285*). Non-injected group was treated similarly to the other three groups without cRNA microinjection. Images were taken 14 hr following release from milrinone inhibitor. Scale bar, 100 μm. (C and D) Kinetic recordings showing the percentages of oocytes with GV breakdown (GVBD) (C), and the oocyte polar body extrusion (PBE) rate (D) through the time-lapse imaging experiment. The total numbers of oocytes microinjected were labeled as indicated. The experiments were performed in biological triplicates. Data are presented as mean ± SEM. (E) Representative morphology of oocytes after cRNA co-microinjection of Mad2 supplemented with WT or Mad2l1bp Mut (equivalent to human p.R285*) into the mouse GV oocytes. Images were taken 14 hr after release from milrinone. Scale bar, 100 μm. (F) Comparison of the percentages of oocytes with GVBD and PBE among the four microinjection groups in (E). The total numbers of oocytes microinjected were labeled as indicated. The experiments were performed in biological triplicates. The statistics were performed with Student’s t-test. Data are presented as mean ± SEM. ‘***‘ indicates p<0.001.
-
Figure 3—source data 1
cRNA microinjection of full-length or truncated Mad2l1bp uncovered their discordant roles in driving the extrusion of polar body 1 (PB1) in mouse oocytes.
- https://cdn.elifesciences.org/articles/85649/elife-85649-fig3-data1-v2.zip
-
Figure 3—source data 2
Co-injection of Mad2 cRNAs with Mad2l1bp cRNAs (WT vs p.R285* mutant) indicated the Mad2l1bp variant lost its function in vivo as compared with its WT counterpart.
- https://cdn.elifesciences.org/articles/85649/elife-85649-fig3-data2-v2.zip
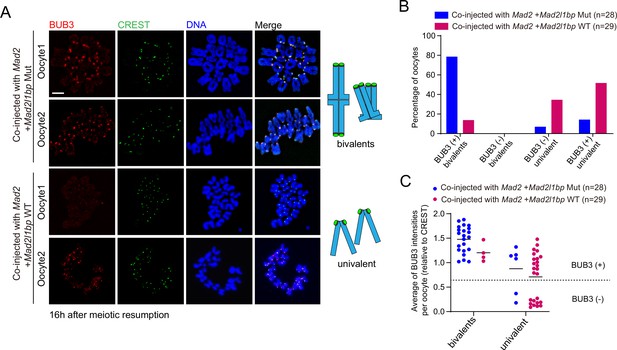
Mad2l1bp variant lost its function in silencing the spindle assembly checkpoint (SAC) in oocytes.
(A) Immunofluorescence co-staining of BUB3 (mitotic checkpoint complex [MCC]) and CREST (kinetochore) using chromosomes spreads from oocytes co-injected with Mad2l1bp WT and oocytes co-injected with Mad2l1bp Mut, at 16 hr after release from milrinone. Schematic diagrams of bivalents at metaphase I (MI) or univalent chromosomes at MII are shown on the right side to aid interpretation. Scale bar, 5 μm. (B) Calculation of the percentages of BUB3 (+) bivalents, BUB3 (-) bivalents, BUB3 (-) univalent, and BUB3 (+) univalent for oocytes at 16 hr after release from milrinone. The numbers of oocytes in each group were indicated. (+), positive; (-), negative. (C) Comparison of the average BUB3 signal intensities among oocytes co-injected with Mad2l1bp WT and Mad2l1bp Mut.
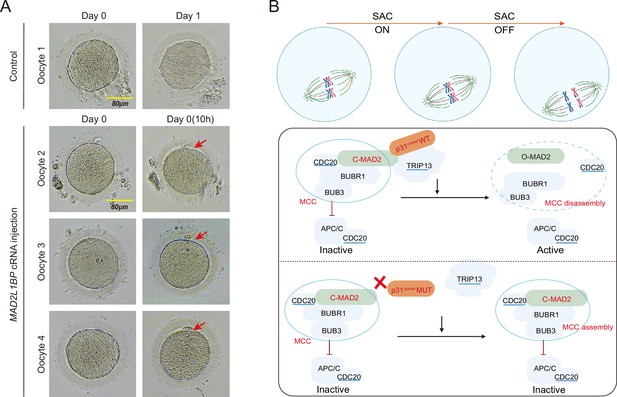
Meiotic rescue of the metaphase I (MI)-arrested frozen human oocytes by MAD2L1BP cRNA microinjection.
(A) The resumption of polar body extrusion through MAD2L1BP cRNA microinjection into the frozen oocytes of the patient in Family 1 (II-1). The extrusion of the first polar body indicates the completion of meiosis I of the oocytes. In the control group, the retrieved MI oocytes remained arrested at MI after sham microinjection (upper panel). Six oocytes from the patient in Family 1 (II-1) were microinjected with full-length MAD2L1BP cRNAs, of which four successfully finished extrusion of polar body 1 (PB1) (lower panel). The red arrows point to PB1. Scale bar, 80 μm. (B) A summarized working model depicting how MAD2L1BP(p31comet) deficiency causes human oocyte meiotic arrest. The mitotic checkpoint complex (MCC) is well known to comprise four core members: BUBR1, BUB3, CDC20, and MAD2. MAD2L1BP(p31comet) is known as the core adaptor that bridges the interaction between MAD2 and TRIP13 (Yang et al., 2007; Alfieri et al., 2018). The MAD2L1BP (p31comet)-mediated interplay between MAD2 and TRIP13, known as the MAD2•MAD2L1BP(p31comet)•TRIP13 axis, disassembles the MCC signaling, which consequently drives the meiotic progression during oocyte meiosis. Top panel: The spindle assembly checkpoint (SAC) is switched ‘ON’ until all the chromosome kinetochores are correctly connected to the spindles. Bottom panel: In the presence of WT MAD2L1BP, SAC is turned ‘OFF’ through the recruitment of MAD2-MAD2L1BP-TRIP13 signaling to silence MCC, leading to the meiotic cell-cycle progression. By contrast, in the presence of mutated MAD2L1BP, MCC is unable to be silenced, resulting in persistent SAC activation and oocyte meiotic arrest.
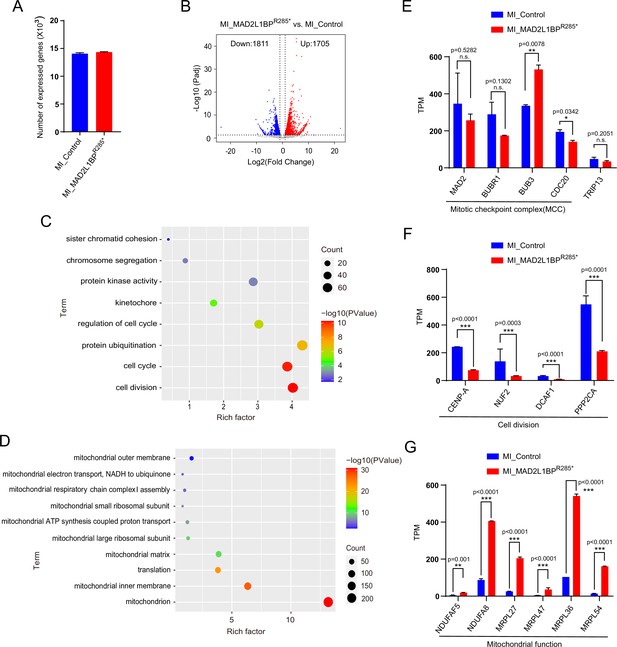
Modified Smart-seq2 analysis revealed that MAD2L1BP variant impaired human oocyte meiotic progression.
(A) Bar graph showing the average numbers of transcripts detected in two MI_Control and two MI_MAD2L1BPR285* oocytes (TPM >1). Data are presented as mean ± SEM, n=2. (B) Volcano plot of Smart-seq2 data obtained from human oocytes in MI_Control and MI_MAD2L1BPR285* groups. ‘Up’ and ‘Down’ represent the up-regulated and down-regulated transcripts in MI_MAD2L1BPR285* oocytes. Cutoff: fold change (FC) ≥2, adjusted p≤0.05. (C–D) Gene Ontology (GO) enrichment analysis of down-regulated (C) and up-regulated (D) genes in arrested MAD2L1BPR285* oocytes compared with healthy oocytes. (E) The expression levels of mitotic checkpoint complex (MCC) components and TRIP13 in both groups. Data are presented as mean ± SEM, n=2. Statistical analyses were performed using R software. Detailed p-value as indicated. (F) The expression levels of selected gene transcripts relevant to cell division. Data are presented as mean ± SEM, n=2. Statistical analyses were performed using R software. Detailed p-value as indicated. (G) The expression levels of selected genes related to mitochondria. Data are presented as mean ± SEM, n=2. Statistical analyses were performed using R software. Detailed p-value as indicated.
Tables
Clinical characteristics of affected individuals and their retrieved oocytes.
| Age (years) | Duration of infertility (years) | IVF/ICSI cycles | Protocol | Total number of oocytes retrieved | GV oocytes | MI oocytes | PB1 oocytes | Fertilized oocytes | Cleaved embryos |
|---|---|---|---|---|---|---|---|---|---|
| Family 1 (II-1) | |||||||||
| 22 | 2 | 1(IVF) | Modified ultra-long | 11 | 0 | 11 | 0 | 0 | 0 |
| 2(IVF) | Mild stimulation | 6 | 1 | 5 | 0 | 0 | 0 | ||
| 3(IVF) | PPOS | 13 | 0 | 13 | 0 | 0 | 0 | ||
| Family 2 (II-1) | |||||||||
| 31 | 6 | 1(IVF) | PPOS | 9 | 0 | 9 | 0 | 0 | 0 |
| Family 3 (II-1) | |||||||||
| 34 | 9 | 1(IVF) | Natural | 1 | 0 | 1 | 0 | 0 | 0 |
| 2(IVF) | Short | 4 | 0 | 4 | 0 | 0 | 0 | ||
-
GV, geminal vesicle; MI, metaphase I; PB1, first polar body; PPOS, progestin-primed ovarian stimulation.
Summary of the variants identified after filtering by whole-exome sequencing (WES) in patients from three families (F1: II-1, F2: II-1, and F3: II-1).
| F1: II-1 | F2: II-1 | F3: II-1 | |
|---|---|---|---|
| Total variants | 134,159 | 392,392 | 66,914 |
| After excluding variants reported in dbSNP, 1000 genomes, EVS, ExAC, and gnomAD (MAF <0.01) | 3619 | 14,023 | 22,832 |
| Exonic nonsynonymous or splicing variants, or coding indels | 273 | 571 | 9660 |
| Homozygous or compound heterozygous (excluding X and Y chromosomes) | 12 | 14 | 11 |
| Homozygous | 6 | 5 | 3 |
| In homozygous region >2 Mb | 1 | – | – |
-
Table 2—source data 1
Homozygous and compound heterozygous variants identified by whole-exome sequencing (WES) that survived filtering in patient F1: II-1, F2: II-1, and F3: II-1.
- https://cdn.elifesciences.org/articles/85649/elife-85649-table2-data1-v2.zip
MAD2L1BP pathogenic variants observed in the three families.
| Families | Genomic position | cDNA change | Protein change | Mutation type | SIFT* | PPH2* | Mutation taster* | NNsplice* | ASSP* | gnomAD† allele | gnomAD† homozygotes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Chr6: 43608202 | c.853C>T | p.R285* | Nonsense | NA | NA | D | NA | NA | 3/248598 | 0/248598 |
| 2 | Chr6: 43607867 | c.518delT | p.F173Sfs4* | Frameshift deletion | NA | NA | D | NA | NA | Not found | Not found |
| Chr6: 43600837 | c.21-94G>A | – | Splicing | NA | NA | NA | 0.13>0.0‡ | 7.1>2.2‡ | 111/31250 | 2/31250 | |
| 3 | Chr6: 43607890 | c.541C>T | p.R181* | Nonsense | NA | NA | D | NA | NA | 2/250850 | 0/250850 |
-
Abbreviations are as follows: D, deleterious; NA, not available.
-
*
Mutation assessment by SIFT, Polyphen-2 (PPH2), Mutation Taster, NNsplice, and ASSP.
-
†
Frequency of corresponding mutations in all population of gnomAD.
-
‡
The variant c.21-94G>A is predicted to introduce an alternative splice acceptor by NNsplice (the increase of the acceptor site score from 0 up to 0.13) and by ASSP (the increase of the acceptor site score from <2.2 up to 7.1).
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | DH5α competent E. coli | Thermo Fisher | Cat # 18265017 | |
| Genetic reagent (Mus musculus) | C57BL/6J | Jackson Laboratories | RRID: IMSR_JAX:000664 | |
| Cell line (Homo sapiens) | HEK293T cells | ATCC | Cat # CRL-3216; RRID: CVCL_0063 | Medium: DMEM+10% FBS |
| Recombinant DNA reagent | pcDNA3.1-human MAD2L1BP isoform1 | This paper | pcDNA3.1-hMAD2L1BP iso1 | Synthesized from Genewiz. Available from Jianqiang Bao’s lab |
| Recombinant DNA reagent | pcDNA3.1-human MAD2L1BP isoform2 | This paper | pcDNA3.1-hMAD2L1BP iso2 | Synthesized from Genewiz. Available from Jianqiang Bao’s lab |
| Recombinant DNA reagent | M46-mouse Mad2l1bp full-length-Flag | This paper | M46-mMad2l1bp-Flag | Available from Jianqiang Bao’s lab |
| Recombinant DNA reagent | p3xFLAG-CMV-24-mouse Mad2l1bp truncated (1-254aa, equivalent to human p. R285*) | This paper | p3xFLAG-mMad2l1bp Mut | Available from Jianqiang Bao’s lab |
| Recombinant DNA reagent | p3xFLAG-CMV-24-mouse Mad2l1bp full-length(1-276aa) | This paper | p3xFLAG-mMad2l1bp WT | Available from Jianqiang Bao’s lab |
| Recombinant DNA reagent | pcDNA3.1-mouse Mad2 full-length-Myc | This paper | pcDNA3.1-mMad2-Myc | Available from Jianqiang Bao’s lab |
| Recombinant DNA reagent | pcDNA-human MAD2L1BP-WT minigene | This paper | pcDNA-hMAD2L1BP-WT minigene | Available from Jianqiang Bao’s lab |
| Recombinant DNA reagent | pcDNA-human MAD2L1BP-c.21-94G>A minigene | This paper | pcDNA-hMAD2L1BP-c.21–94G>A minigene | Available from Jianqiang Bao’s lab |
| Antibody | Anti-MAD2L1BP (Mouse monoclonal) | Santa Cruz | Cat # sc-134381 | WB (1:200) |
| Antibody | Anti-Flag (Mouse monoclonal) | Proteintech | Cat # 66008-3-Ig | WB (1:2000) |
| Antibody | Anti-Myc (Mouse monoclonal) | Proteintech | Cat # 60003-2-Ig | WB (1:5000) |
| Antibody | Anti-Beta Actin (Mouse monoclonal) | Proteintech | Cat # 66009-1-Ig | WB (1:10,000) |
| Antibody | HRP-conjugated goat anti-mouse IgG heavy chain (Goat polyclonal) | ABclonal | Cat # AS064 | WB (1:10,000) |
| Antibody | HRP-conjugated goat anti-mouse IgG light chain (Goat polyclonal) | ABclonal | Cat # AS062 | WB (1:10,000) |
| Antibody | Anti-BUB3 (Rabbit polyclonal) | Santa Cruz | Cat # sc-28258 | IF (1:300) |
| Antibody | Anti-CREST (Human, unknown clonality) | Fitzgerald Industries International | Cat # 90C-CS1058 | IF (1:100) |
| Antibody | Goat Anti-Human lgG H&L (FITC) (Goat polyclonal) | ZENBIO | Cat # 550022 | IF (1:500) |
| Antibody | CoraLite 594 Anti-Rabbit (Goat polyclonal) | Proteintech | Cat # SA00013-4 | IF (1:500) |
| Software, algorithm | R software | R Project for Statistical Computing | RRID:SCR_001905 http://www.r-project.org/ | |
| Software, algorithm | DAVID | DAVID | RRID:SCR_001881 https://david.ncifcrf.gov/ | |
| Software, algorithm | Fiji/ImageJ | Fiji | RRID:SCR_002285 http://fiji.sc | |
| Software, algorithm | Jalview | Jalview | RRID:SCR_006459 https://www.jalview.org/ | |
| Software, algorithm | Prism | GraphPad Prism | RRID:SCR_002798 http://www.graphpad.com/ | |
| Software, algorithm | PyMOL | PyMOL | RRID:SCR_000305 http://www.pymol.org/ | |
| Other | GenBank | NIH | RRID:SCR_002760 https://www.ncbi.nlm.nih.gov/genbank/ | Database |
| Other | gnomAD | Genome Aggregation Database | RRID:SCR_014964 http://gnomad.broadinstitute.org/ | Database |
| Other | OMIM | OMIM | RRID:SCR_006437 http://omim.org | Database |
| Other | ASSP | ASSP | http://wangcomputing.com/assp/ | Web resource |
| Other | EvolView | EvolView | http://evolgenius.info | Web resource |
| Other | Mutation Taster | MutationTaster | RRID:SCR_010777 http://www.mutationtaster.org/ | Web resource |
| Other | NNSplice | NNSplice | http://www.fruitfly.org/seq_tools/splice.html | Web resource |
| Other | PolyPhen-2 | PolyPhen: Polymorphism Phenotyping | RRID:SCR_013189 http://genetics.bwh.harvard.edu/pph2/ | Web resource |
| Other | SIFT | SIFT | RRID:SCR_012813 https://sift.bii.a-star.edu.sg/ | Web resource |
Additional files
-
Supplementary file 1
The supplementary file includes file (A and B).
- https://cdn.elifesciences.org/articles/85649/elife-85649-supp1-v2.zip
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85649/elife-85649-mdarchecklist1-v2.docx





